ABSTRACT
Lectin-like bacteriocins (LlpAs) are secreted by proteobacteria and selectively kill strains of their own or related species, and they are composed of two B-lectin domains with divergent sequences. In Pseudomonas spp., initial binding of these antibacterial proteins to cells is mediated by the carboxy-terminal domain through d-rhamnose residues present in the common polysaccharide antigen of their lipopolysaccharide, whereas the amino-terminal domain accounts for strain selectivity of killing. Here, we show that spontaneous LlpA-resistant mutants carry mutations in one of three surface-exposed moieties of the essential β-barrel outer membrane protein insertase BamA, the core component of the BAM complex. Polymorphism of this loop in different Pseudomonas groups is linked to LlpA susceptibility, and targeted cells all share the same signature motif in this loop. Since heterologous expression of such a bamA gene confers LlpA susceptibility upon a resistant strain, BamA represents the primary bacteriocin selectivity determinant in pseudomonads. Contrary to modular bacteriocins that require uptake via the Tol or Ton system, parasitism of BamA as an LlpA receptor advocates a novel bacteriocin killing mechanism initiated by impairment of the BAM machinery.
KEYWORDS: BamA, LlpA, kin discrimination, outer membrane proteins, polymorphism, target receptor
IMPORTANCE
Bacteria secrete a variety of molecules to eliminate microbial rivals. Bacteriocins are a pivotal group of peptides and proteins that assist in this fight, specifically killing related bacteria. In Gram-negative bacteria, these antibacterial proteins often comprise distinct domains for initial binding to a target cell’s surface and subsequent killing via enzymatic or pore-forming activity. Here, we show that lectin-like bacteriocins, a family of bacteriocins that lack the prototypical modular toxin architecture, also stand out by parasitizing BamA, the core component of the outer membrane protein assembly machinery. A particular surface-exposed loop of BamA, critical for its function, serves as a key discriminant for cellular recognition, and polymorphisms in this loop determine whether a strain is susceptible or immune to a particular bacteriocin. These findings suggest a novel mechanism of contact-dependent killing that does not require cellular uptake. The evolutionary advantage of piracy of an essential cellular compound is highlighted by the observation that contact-dependent growth inhibition, a distinct antagonistic system, can equally take advantage of this receptor.
INTRODUCTION
The active elimination of competing microorganisms is primordial in the fight for ecologic space. Bacteriocins are secreted protein antibiotics that act as key mediators of antagonistic interactions between bacteria (1, 2). Studies in the Gram-negative model organisms Escherichia coli and Pseudomonas aeruginosa have identified compounds very diverse in molecular size, ranging from small peptides (e.g., microcins) to modular multidomain proteins (e.g., colicins, S-type pyocins) and large multiprotein complexes resembling bacteriophage tails (tailocins) (3–6). Bacteriocins usually only kill close phylogenetic relatives, typically belonging to the same species or genus (2, 7). These allelopathic molecules are currently receiving renewed interest for combating multidrug-resistant pathogens, mainly in the framework of food and medical applications (8–13). Their high efficacy and selectivity, natural presence in the environment, biodegradability, and nonreplicative behavior (compared to phages, in the case of tailocins) make them attractive drug leads.
Pseudomonads are prominent producers of both ribosomally and nonribosomally synthesized antibacterial molecules. Their genome plasticity has enabled them to acquire a diversified bacteriocin arsenal (5, 14), facilitating colonization of different environments, such as rhizospheres and lungs (15–18). Lectin-like bacteriocins, also called LlpAs, are midsize bacteriotoxins (~28 kDa) that lack an enzymatic or pore-forming toxin-immunity pair characteristic for modular bacteriocins and other polymorphic toxins (4, 5, 19, 20). They adopt an architecture comprising tandem B-lectin domains followed by a short nonconserved carboxy-terminal extension (21, 22). Compared to B-lectins from plants, the affinity of Pseudomonas LlpAs for d-mannose is low, reflecting a much higher affinity of the bacteriocin for d-rhamnose (22), a 6-deoxy-mannose that is omnipresent in the common polysaccharide antigen (CPA) of Pseudomonas lipopolysaccharide (LPS) (23). No association between LlpA selectivity and O serotype of target cells could be detected for lectin-like bacteriocins from Pseudomonas aeruginosa (also designated l-type pyocins) (24). CPA binding capacity is hosted by the carboxy-terminal lectin domain; however, lack of LPS binding is not sufficient for provoking full LlpA resistance toward susceptible strains (22). Via the construction of engineered LlpA chimeras, strain selectivity of killing by LlpAs was attributed to the amino-terminal lectin domain (21).
Here, we identify the essential outer membrane protein insertase BamA as the major selectivity determinant of LlpA susceptibility in Pseudomonas spp. We show that LlpA-resistant mutants arise by mutations in a particular surface-exposed hypervariable loop of BamA, and we link LlpA-selective killing to amino acid sequence polymorphisms in this BamA loop.
RESULTS
LPS binding by LlpA cannot fully account for the bacteriocin’s strain-specific lethality. Given that CPA-deficient mutant strains display a (partial) bacteriocin-inhibited phenotype (22), we anticipated that additional factors must contribute to the toxin’s selective action. To identify such determinants, LlpA-resistant mutants of a bacteriocin-susceptible Pseudomonas strain were isolated, arising as colonies within halos produced by spotting concentrated bacteriocin onto a lawn of late-stationary-phase cells. Pseudomonas fluorescens Pf0-1 was chosen as an indicator. This strain is highly susceptible to recombinant LlpA1 from the biocontrol model strain Pseudomonas protegens Pf-5 (25, 26) and its genome has been completely sequenced (27), facilitating single nucleotide polymorphism (SNP) analysis. A single colony was selected from each of four independent treatments/halos (mutants B1 to B4). In a fifth independent exposure experiment, two slightly different morphotypes could be distinguished (mutants B5a and B5b). After validation of resistance toward LlpA1, the parental origin of the mutants was confirmed via BOX-PCR. The genomes of these six mutants and the Pf0-1 wild type were sequenced and analyzed for SNPs (Table 1; Fig. 1).
TABLE 1 .
SNPs detected in LlpA1-resistant P. fluorescens Pf0-1 mutantsa
| Mutant | Mutation type |
Coding region change |
Amino acid change |
Gene | Protein |
|---|---|---|---|---|---|
| B1 | SNV |
ABA77421.1, 316 C → T |
Gln106* |
pfl01_RS28605
(pfl01_5685) |
Wzt |
| B2 | SNV |
ABA72852.1, 2000 G → T |
Gly667Val |
pfl01_RS05600
(pfl01_1109) |
BamA |
| B3 | Ins |
ABA72221.1, 857_858 InsC |
Gly286Fs |
pfl01_RS02405
(pfl01_0477) |
FuzY |
| B4 | SNV |
ABA72852.1, 2011 A → G |
Thr671Ala |
pfl01_RS05600
(pfl01_1109) |
BamA |
| B5a | SNV |
ABA72852.1, 1996 C → T |
Arg666Cys |
pfl01_RS05600
(pfl01_1109) |
BamA |
| B5b | SNV |
ABA72852.1, 1996 C → T |
Arg666Cys |
pfl01_RS05600
(pfl01_1109) |
BamA |
| Δ |
ABA77417.1, 4 ΔA |
Lys2Fs |
pfl01_RS28585
(pfl01_5681) |
Rmd |
Abbreviations and symbols: Δ, deletion; Fs, frameshift; Ins, insertion; SNV, single nucleotide variation. An asterisk specifies the introduction of a stop codon.
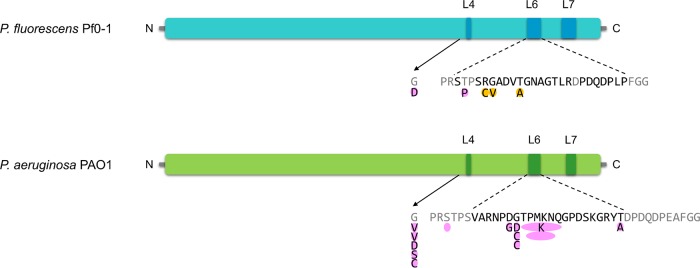
FIG 1 .
Schematic representation of BamAs from P. fluorescens Pf0-1 and P. aeruginosa PAO1, which are susceptible to LlpA1 and PyoL1, respectively, and the detected SNPs in bacteriocin-resistant mutants. Conserved surface-exposed residues in the respective Pseudomonas species are colored gray, and surface-exposed polymorphic residues of L6 in P. fluorescens and P. aeruginosa strains are black. Mutations identified by whole-genome sequencing are highlighted in orange, and others are pink. Amino acid deletions are shown as empty ovals. N and C specify the amino- and carboxy-terminal ends, respectively.
Escape from LlpA1 killing by mutations in LPS genes.
Whereas the Pf0-1 parent did not contain any mutation compared to the reference genome, the spontaneous mutants all contained one SNP, except for B5b. Three of the mutations observed (in mutants B1, B3, and B5b) were directly linked to LPS biosynthesis. In B1 and B5b, mutations were found in pfl01_5685 (nonsense mutation) and pfl01_5681 (frameshift), which encode orthologues of Wzt and Rmd from P. aeruginosa PAO1, respectively (82.2% amino acid identity to PA5450 and 73.6% amino acid identity to PA5454). These Pf0-1 genes are part of an LPS biosynthetic gene cluster that is syntenic to the CPA biosynthetic cluster of P. aeruginosa PAO1 (rmd-gmd-wbpW-wzm-wzt-wbpX-wbpY-wbpZ) (23). Mutations in several of these genes (wzm, wzt, and wbpZ) confer high tolerance to the P. aeruginosa PAO1-specific LlpA, pyocin L1 (PyoL1), due to lack of CPA assembly and, consequently, absence of the appropriate d-rhamnose-carrying LPS moiety recognized by LlpA’s C-terminal domain (22). This extends the role of CPA in LlpA binding onto LPS, as demonstrated for P. aeruginosa and Pseudomonas syringae (22), to a third phylogenetic group of pseudomonads.
The other mutants are affected in genes that were previously not linked to LlpA function. In B3 cells, a frameshift mutation occurred in pfl01_0477, specifying a putative glycosyl transferase homologous to FuzY (70% amino acid identity) encoded by the fuzVWXYZ operon that accounts for the fuzzy spreader phenotype of P. fluorescens SBW25 and is predicted to modify LPS O antigens (see Fig. S1 in the supplemental material) (28). A fuz-like operon is present in P. fluorescens Pf0-1; however, it has an additional glycosyl transferase gene (pfl01_0474) located between the fuzV and fuzW homologues. The morphotype resulting from a fuzY mutation provokes a highly aggregative phenotype in strain SBW25, which was also evident for the B3 mutant, based on rapid sedimentation of planktonic cells. The exact function of FuzY remains elusive to date. Partly similar clusters can be detected in Pseudomonas syringae and Pseudomonas aeruginosa strains, although an equivalent of pfl01_0477 is consistently lacking. An extra gene involved in the fuzzy spreader phenotype and associated with a partially similar cluster was detected in P. syringae pv. actinidiae ICMP 18884 (Fig. S1) (29). Together, these findings suggest that the carbohydrate binding by LlpAs may involve accessory moieties in addition to d-rhamnose, and they hint at subtle differences in LlpA-LPS interactions that depend on the particular bacteriocin-target strain combination. In general, the affinity of lectins for oligomeric carbohydrates is higher than that for monomers (30). Similarly, secondary LPS carbohydrate binding by LlpA might strengthen the interaction and ensure optimal positioning of LlpA close to other surface constituents.
Genetic organization of the fuzVWXYZ locus in the genome of P. fluorescens SBW25 in comparison with corresponding regions in P. aeruginosa PAO1, P. fluorescens Pf0-1 and P. syringae pv. syringae ICMP 18884. Synteny is visualized by sequence conservation (grey shading) for the corresponding orthologues, if present. Genes (displayed as arrows) of the fuzVWXYZ cluster in P. fluorescens SBW25 are colored blue and an extra gene involved in the fuzzy spreader phenotype in P. syringae pv. syringae ICMP 18884 is shown in yellow. The location of the fuzY mutation (insertion) in P. fluorescens Pf0-1 mutant B3 (red) is indicated with a black triangle. Dotted lines indicate the absence of an equivalent sequence. Download FIG S1, TIF file, 0.2 MB (199.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
LlpA-resistant mutants carry a modified surface loop in BamA.
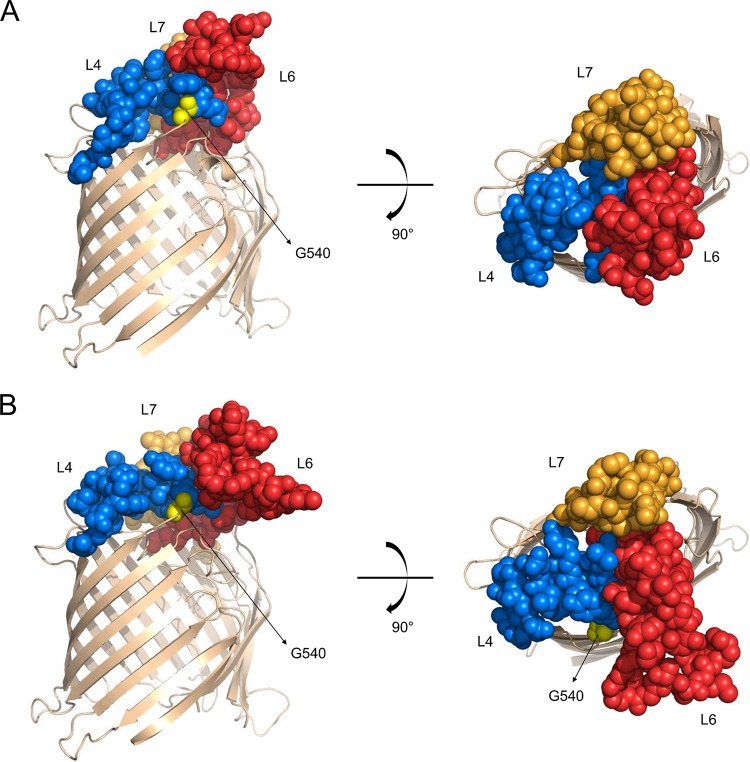
The non-LPS mutants (B2, B4, and B5a/B5b [identical transition in B5a and B5b]) all carried missense mutations in pfl01_1109, which encodes the essential β-barrel membrane protein BamA (Fig. 1; Table 1). In mutant B5b, this was accompanied by the aforementioned rmd mutation, which affected CPA metabolism. BamA, together with the lipoproteins BamB, BamC, BamD, and BamE, constitutes the BAM complex that ensures the integration and folding of proteins in the outer membrane via a mechanism requiring a lateral gate and exit pore formation (31–34). Pfl01_1109 includes four POTRA domains (polypeptide transport associated; Pfam PF07244) and a characteristic conserved VRGF motif in the β-barrel (35). A fifth POTRA domain that is typically present in BamAs from proteobacteria (33, 36) was not detected in the ~70-amino-acid amino-terminal region of mature Pfl01_1109. Distinct BamA crystal structures from Neisseria meningitidis, Haemophilus ducreyi, and Escherichia coli have been solved (37–40) and revealed dome-like structures exposed to the extracellular surface, essentially consisting of three large loops, termed loops 4, 6, and 7 (further abbreviated L4, L6, and L7, respectively), that are stabilized by a salt bridge network (41). The BamA mutations that were retrieved in the LlpA1-resistant Pf0-1 mutants could all be localized to L6 (B2, B4, and B5a/b) (Fig. 1), suggesting L6 as a candidate interaction hot spot in the potential susceptibility determinant BamA. The BamA barrel sequences of an additional set of 10 mutants isolated from halos of independent Pf0-1 cultures were determined, and 2 of these mutants carried BamA mutations as well: T663P and G540D (Fig. 1). The former mutation is located in the predicted surface-exposed L6 loop of BamAPf0−1, whereas G540 is part of L4 and positioned near L6 and possibly assists in its stabilization (based on the structural prediction) (Fig. 2).
FIG 2 .
Cartoon representation of homology models of BamA barrel domains from P. fluorescens Pf0-1 (A) and P. aeruginosa PAO1 (B). Residues composing surface-exposed loops L4, L6, and L7 are shown as spheres colored blue, red, and orange, respectively. G540 residues in the respective BamAs are yellow.
To further explore this potential targeting of BamA by use of a different indicator strain-bacteriocin combination, a similar experimental setup was applied to P. aeruginosa PAO1, which is susceptible to PyoL1 of P. aeruginosa C1433 (22, 24), a lectin-like bacteriocin sharing only 32% amino acid identity with LlpA1. A set of spontaneous mutants growing inside halos was randomly selected (1 mutant per halo; 30 independent cultures), validated for altered PyoL1 susceptibility, and their β-barrel bamA gene fragments were sequenced. Isolates showing diminished halo sizes did not contain mutations in bamA, whereas fully resistant isolates (13/30) were all affected in residues of L6 or in close proximity to L6 (Fig. 1). In addition to the large set of L6 mutations, G540 was affected in several mutants (Fig. 2), as observed for the corresponding residue in Pf0-1, whereas no other mutations in the covered bamA barrel sequence could be retrieved. Originating from independent cultures, two mutations (G540V and G672C) were retrieved twice. Most likely, the subset of mutants without detectable bamA mutations includes strains affected in LPS metabolism, but this was not further investigated. This convergent genetic data set from independent analysis of two distinct bacteriocin-target combinations strongly suggests that, in addition to CPA, a surface-exposed epitope of BamA, including (part of) L6, functions as a key interaction partner for lectin-like bacteriocins on the surface of susceptible cells.
Heterologous expression of cognate BamA confers susceptibility to LlpA.
Assuming that BamA acts as a LlpA bacteriocin target, one expects a strain to become resistant when deprived from its (cognate) bamA gene. To explore this hypothesis, the bamA insertase from a PAO1 deletion mutant—expressing its bamA gene from a plasmid to retain viability (42)—was transformed with shuttle vector pJB3Tc20 encoding a bamA orthologue originating from strain P. aeruginosa Br670 (43). The latter strain is not killed by PyoL1 but instead is inhibited by PyoL2 (24). When supplemented with this second plasmid, the PAO1 deletion mutant was sensitive to both PyoL1 and PyoL2. Conversely, when cured from its original plasmid, PyoL1 susceptibility was completely abrogated and only PyoL2 susceptibility remained (Fig. 3). These phenotypic alterations point out that BamA indeed acts as a selective killing partner for LlpA, in line with the SNP analysis of resistant mutants and supporting the proposed selectivity hypothesis.
FIG 3 .
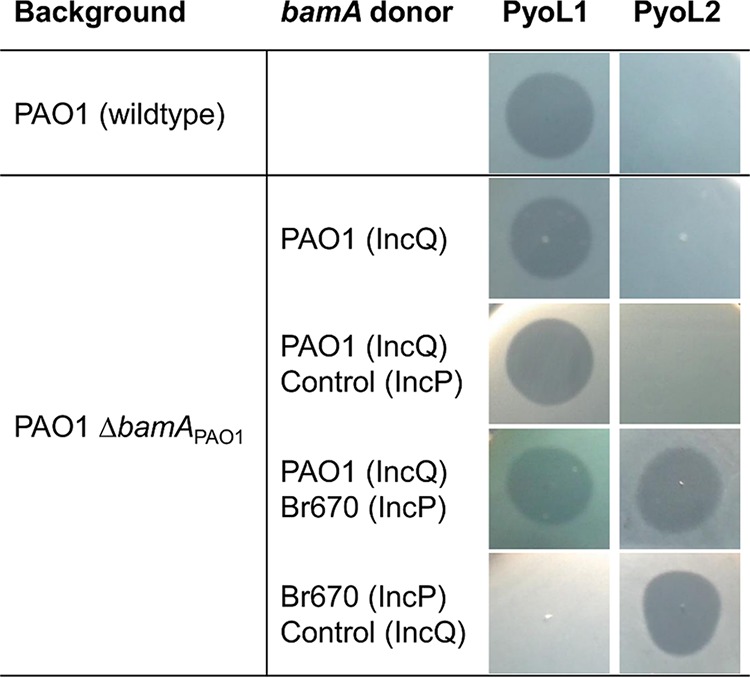
Susceptibilities of P. aeruginosa PAO1 (wild-type and ΔbamAPAO1 background), heterologously expressing bamA genes from P. aeruginosa PAO1 or Br670, for lectin-like bacteriocins PyoL1 and PyoL2. Wild-type P. aeruginosa strains PAO1 and Br670 are killed by PyoL1 and PyoL2, respectively. Controls were plasmids without the insert. The presence of a halo is indicative of growth inhibition due to bacteriocin activity.
To explore this further, bamA genes from Pseudomonas strains targeted by different LlpAs were expressed in select Pseudomonas host strains. In addition to the plasmid expressing bamABr670, corresponding genes from P. aeruginosa PAO1, P. fluorescens Pf0-1, and P. savastanoi pv. glycinea LMG 5066, strains that are killed by PyoL1, LlpA1, and LlpABW11M1, respectively (22, 24–26), were cloned in pJB3Tc20. Pseudomonas acceptor strains were chosen based on their established susceptibility for one of these bacteriocins. Spot assays revealed that PyoL1-susceptible P. aeruginosa strains equipped with a bamA originating from a PyoL2-targeted strain became fully sensitive to PyoL2 (as could be inferred from the plasmid exchange setup in the PAO1 deletion background), and vice versa (Fig. S2). Furthermore, when equipped with the P. fluorescens or P. savastanoi bamA, P. aeruginosa strains became susceptible to LlpA1 and LlpABW11M1, respectively. As expected, when bamA genes were heterologously expressed, a mixed phenotype was obtained.
Susceptibility of Pseudomonas aeruginosa strains, heterologously expressing bamA genes from different donors, to lectin-like bacteriocins PyoL1, PyoL2, LlpA1, and LlpABW11M1. Controls are strains transformed with a wild-type plasmid (no insert). Sensitivity pairs: P. aeruginosa PAO1/PyoL1, P. aeruginosa Br670/PyoL2, P. fluorescens Pf0-1/LlpA1, P. savastanoi pv. glycinea LMG 5066/LlpABW11M1. The presence of a halo is indicative of growth inhibition due to bacteriocin activity. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We also examined whether such bamA-triggered phenotypic conversion in bacteriocin susceptibility may occur in non-P. aeruginosa strains. Pseudomonas lini VA1.2 and Pseudomonas brassicacearum WCS365, both sensitive to LlpABW11M1, could be sensitized to the other LlpAs (Fig. S3). L pyocin action was elicited in Pseudomonas migulae ZA 45.2, a strain killed by LlpA1 and LlpA (26), when it expressed the respective P. aeruginosa bamA genes. For other strains (mainly) originating from the P. fluorescens species group, the bacteriocin phenotype switch was not always conclusive. For example, Pseudomonas fluorescens LMG 1794, which is killed by LlpA1, can only be (partially) sensitized to LlpABW11M1. It should be emphasized that this strain (as well as some other P. fluorescens strains) does not host a (complete) CPA cluster in its genome (44), which may impact LPS-mediated LlpA docking. Another possible reason is that the heterologously expressed bamA is not fully compatible with the BAM machinery of the host strain, given the intimate character of its different accessory partners (31, 45).
Susceptibility of Pseudomonas strains (non P. aeruginosa), heterologously expressing bamA genes from different donors, to lectin-like bacteriocins PyoL1, PyoL2, LlpA1, and LlpABW11M1. Controls are strains transformed with a wild-type plasmid (no insert). Sensitivity pairs: P. aeruginosa PAO1/PyoL1, P. aeruginosa Br670/PyoL2, P. fluorescens Pf0-1/LlpA1, P. savastanoi pv. glycinea LMG 5066/LlpABW11M1. The presence of a halo is indicative of growth inhibition due to bacteriocin activity. Download FIG S3, TIF file, 1.6 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pseudomonas BamA polymorphism is confined to surface-exposed loops.
Given the observations that BamA was key to LlpA selectivity and that BamA mutations resulting in LlpA resistance were mainly retrieved in the surface-exposed L6 loop, sequence polymorphism of BamA in Pseudomonas was further dissected. Representatives from Pseudomonas with abundant genome information (P. aeruginosa, P. fluorescens, P. putida, and P. syringae) were selected for further analysis. Considering that species delineation within pseudomonads is blurry, species groups were considered for P. fluorescens, P. putida, and P. syringae (46, 47). An interesting observation is that a number of pseudomonads encode a second BamA, likely resulting from a recent duplication event (36). P. aeruginosa consistently hosts one bamA, whereas strains with a second bamA are occasionally found in P. fluorescens and P. putida species groups and to a lesser extent in P. syringae. Strains previously used as bamA donors (Br670, LMG 5066, PAO1, and Pf0-1) only encode one bamA, and in our analysis only the ubiquitous gene located between Zn metalloprotease gene rseP and periplasmic chaperone gene hlpA (skp in E. coli) was considered.
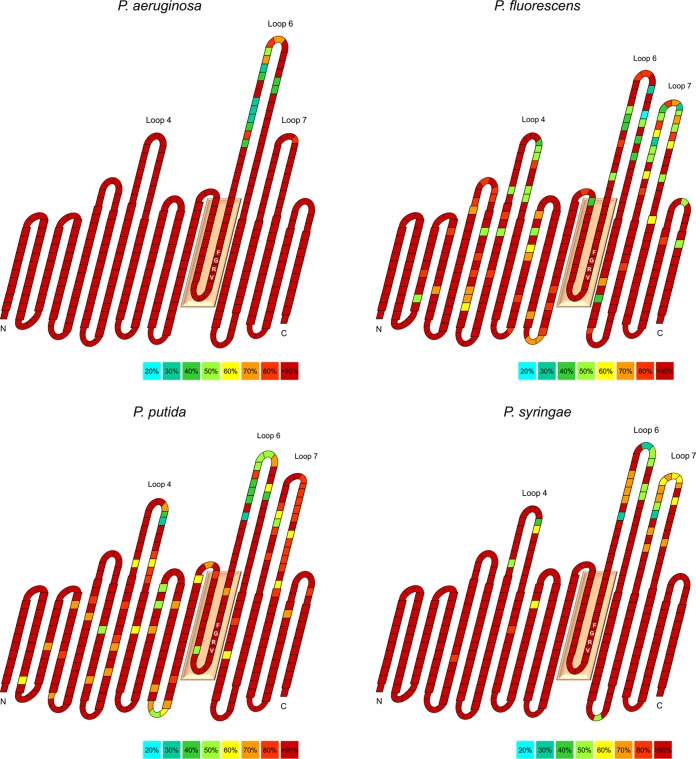
At the species (group) level, BamA polymorphism is essentially confined to loops L4, L6, and L7. Variations may equally occur in the β-barrel sequences but are only present at relevant frequencies in BamAs from the polyphyletic P. fluorescens and P. putida groups (Fig. 4). Polymorphism is restricted to only part of the surface-exposed moieties, although the extent to which this can be observed largely depends on the species (group) of interest. All pseudomonads display polymorphism in L6, with certain loop sequence variants occurring more frequently. L4 and L7 show different levels of polymorphism: the P. aeruginosa and the P. syringae groups display good conservation in L4 (but the latter to a lesser extent); conservation in L7 is only high for P. aeruginosa. In general, the hypervariable part of L4 is rather small and limited to 8 to 9 residues, with (virtually) all Pseudomonas isolates sharing a central EGD triplet. In L7, two conserved cysteines consistently arise, suggesting the formation of a disulfide bridge, as observed in different crystal structures from E. coli BamA (37, 40). There also appears to be no strict coupling of different loops for any of the species groups, i.e., different L4 sequences may be combined with different L6 and L7 sequences and vice versa (not relevant for P. aeruginosa).
FIG 4 .
Transmembrane topology models of the BamA β-barrel domains of P. aeruginosa, P. fluorescens, P. putida, and P. syringae. For each of the species groups, a BamA consensus sequence was derived by using all unique sequences present in GenBank. The crystal structure of E. coli BamA was used as a model (PDB 5EKQ) to delineate β-strands. The weighted percentage of conservation for each amino acid residue, indicated as one block, is indicated according to relative abundance (see the color legend at the bottom right of each panel). The conserved VRGF motif is highlighted with residues inside in the blocks and is part of a residue stretch residing in the lumen of BamA (salmon-colored box). The different lengths for L4, L6, and L7 were derived from the observation that the corresponding sequences may differ greatly, depending on the species group of interest. Residues predicted to be exposed to the periplasm or extracellular medium are shown in smaller blocks. N and C specify the amino- and carboxy-terminal ends of the BamA barrel domains, respectively.
LlpA is directed to a specific loop.
We previously evaluated a randomly chosen panel of P. aeruginosa strains for their susceptibility to pyocins L1 and L2 (24). Analysis of the different BamA β-barrel sequences of this strain set (n = 80) allowed us to retrieve the same L6 subtypes as obtained from reference sequences at similar frequencies (Table 2), whereas polymorphism was barely present in L4 and L7 in the strain set, as expected (see above). Strikingly, PyoL1 essentially only killed strains sharing the same L6 sequence. Likewise, the subset of PyoL2-susceptible strains shared a single L6 sequence, different from the one present in PyoL1-killed strains, the sole noticed exception being P. aeruginosa CPHL 6750: this strain does not carry the prototypical PyoL2-targeted loop, although its L6 has several residues in common with the latter (IARDANGKQIPGPDSQGRYT versus the common PyoL2-targeted VARDG[N/K]GNQIVGPDSKGRYT; the identical residues are shown in boldface italics in the sequences). P. aeruginosa strains with a BamA carrying any other L6 sequence all appeared resistant to lectin-like pyocins (n = 59 for PyoL1 and n = 67 for PyoL2). One exception was P. aeruginosa PA1255, which appeared resistant to PyoL2, despite carrying the corresponding bacteriocin-targeted L6 sequence. One possible explanation for this may be the absence of a suitable LPS binding partner. The crucial role of L6 was also evident for the BamAs from P. aeruginosa PAO1 and Br670 (used in the heterologous expression experiments), which are identical except for the hypervariable part of L6 (43, 48). P. aeruginosa strains with a pyoL1 or pyoL2 gene in their genome all feature L6 sequences different from the (sub)type that they target, as would actually be expected in order to avoid kin killing (Table 2). Such built-in BamA-mediated autoimmunity provides an explanation for why LlpA production does not require a cognate immunity protein (21, 25, 49).
TABLE 2 .
Hypervariable BamA L6 sequences in P. aeruginosa isolates and strains used in the test panel
|
Hypervariable loop L6
amino acid sequencea |
Relative frequency (%) of sequence among: |
Test panel strains susceptible to: |
P. aeruginosa
isolates encoding: |
|||
|---|---|---|---|---|---|---|
| P. aeruginosa isolates | Test panel | PyoL1 | PyoL2 | PyoL1 | PyoL2 | |
| PRSTPSRAYKDGKIIPGPDER GRYTDPDQDPEAFGG |
37.3 | 33.75 | 0/27 | 0/27 | 3/12 | 11/18 |
| PRSTPSVARNPDGTPMKNQG PDSKGRYTDPDQDPEAFGG |
18.2 | 26.25 | 21/21 | 0/21 | 0/12 | 7/18 |
| PRSTPSIARDANGKQIPGPDS QGRYTDPDQDPEAFGG |
2.3 | 1.25 | 0/1 | 1/1 | 0/12 | 0/18 |
| PRSTPSVARDG(N/K)GNQIVG PDSKGRYTDPDQDPEAFGG |
20.8 | 15.0 | 0/12 | 11/12 | 9/12 | 0/18 |
| PRSTPSRGEAEGGRPGTVRDP DQDPEAFGG |
17.2 | 18.75 | 0/15 | 0/15 | 0/12 | 0/18 |
| Other | 4.2 | 5.0 | 0/4 | 0/4 | 0/12 | 0/18 |
Residues shown in boldface italics indicate the hypervariable sequence; nonpolymorphic (conserved) residues that are part of L6 are shown in lightface.
When we assessed the possible correlation of L6 sequence types and LlpABW11M1 susceptibility in the P. syringae group, a similar observation as for P. aeruginosa was made, although this was evaluated for a smaller strain panel (n = 17). L6 sequences from the P. syringae group roughly segregated into four major types, two sets of partly related “long” and “short” loops, along with a significant number of more diverse L6 types that occurred at lower frequencies (Table S1). Although bias appears to be present in our available P. syringae panel, all LlpABW11M1-susceptible strains (n = 13) carry one of the short L6 cores (consensus sequence NGSKPGTLA), whereas the four LlpABW11M1-resistant strains feature the longer L6 sequences. It should be emphasized that a number of additional polymorphic residues arise within the other surface-exposed loops of this LlpABW11M1-susceptible strain set, and at this point it cannot be excluded that these may have more subtle quantitative effects toward bacteriocin inhibition, for example, by stabilizing LlpA docking.
Relative frequencies of occurrence of hypervariable BamA L6 sequences in P. syringae group isolates and strains used in the test panel and LlpABW11M1 susceptibility of strains in the respective set (highly conserved residues that are part of L6 are shown in gray). Download TABLE S1, DOCX file, 0.01 MB (12.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Besides P. syringae group strains, LlpABW11M1 was previously found to also target Pseudomonas strains assigned to other species groups, including P. fluorescens and P. putida (25, 26, 49). Although BamA polymorphism in the latter species groups is much higher and dominant loop signatures are more difficult to discern, strains (highly) susceptible to LlpABW11M1 share an L6 signature sequence that well resembles the targeted short L6 type of P. syringae, despite highly divergent loops 4 and 7: for example, KGTNPGTIE in Pseudomonas capeferrum WCS358 (50), KGTNPGTLR in P. fluorescens BW11P2 (51), SGNKPGTLR in P. mendocina LMG 1223 (52), SGRNPGTQP in P. viridiflava LMG 2352 (53) (the boldface italicized residues are the common residues) (Fig. S4). Conversely, none of the P. fluorescens or P. putida strains that are LlpABW11M1 resistant contain a sequence reminiscent of this more relaxed consensus motif (XGXX[P]GTXX; n = 6 versus n = 20 for the other L6 types). Overall, this P. syringae L6 subtype with the short sequence is more rare in strains of P. fluorescens (~13% of isolates) and P. putida (~27% of isolates), which may explain why fewer strains in these species groups are susceptible for LlpABW11M1 (26). As expected, isolates encoding LlpABW11M1 or a highly homologous sequence (>97.5% amino acid identify) carry a bamA with an L6 type different than the one they target (Table S2).
Multiple sequence alignment of L6 sequences of BamAs from Pseudomonas spp. that are susceptible to LlpABW11M1. Differential shading reflects the degree of sequence conservation. The hypervariable signature motif of LlpABW11M1-susceptible isolates is boxed in blue. Download FIG S4, TIF file, 0.5 MB (521.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Within the P. fluorescens group—the primary species group targeted by LlpA1 (25, 26)—polymorphism appears even more complex. The L6 sequence retrieved in Pf0-1 was reminiscent of the longer subtypes retrieved in P. syringae and appeared in ~82% of the strains belonging to the P. fluorescens species group (PRSTPSRGXXXTGNXGTXXD[S/P]D[N/Q]DPLPFGG; boldface italics indicate [hyper]variable residues in common) (Fig. 4; Table S1). Considering the high number of polymorphic residues in this loop set, no meaningful trend in possible LlpA1 receptiveness can be determined. As suggested above, LPS as a coreceptor may also be more critical for targeting strains in this species group.
List of isolates encoding LlpABW11M1 or a highly homologous sequence (>97.5% amino acid identity; 0 to 2 different amino acid residues in amino-terminal lectin domains) and their corresponding BamA L6 amino acid sequence (nonpolymorphic [conserved] residues that are part of L6 in the Pseudomonas putida species group are shown in gray). Download TABLE S2, DOCX file, 0.01 MB (12.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Our results demonstrated that LlpA parasitizes BamA and that one of its surface-exposed loops (L6) is key to bacteriocin target recognition. This moiety is the prime discriminant for target selectivity of L pyocins in P. aeruginosa. A strong correlation between L6 sequence subtype and susceptibility to a particular LlpA is also evident among other pseudomonads, such as those killed by LlpA of Pseudomonas mosselii BW11M1. Depending on the BamA-LlpA combination, an accessory role for the other exposed loops (L4/L7) cannot be excluded, given that L4 and L7 also display considerable sequence polymorphisms within different Pseudomonas species groups outside the more homogenous P. aeruginosa clade. In contrast to the key determinant L6, we expect that these BamA loops provoke only minor effects on relative susceptibility. Reasons for this are the observation that (i) BamA mutations in spontaneous LlpA-resistant mutants can only be linked to L6, (ii) several L1/L2 pyocin-targeted strains carry identical L4 and L7 sequences and thus only display surface-exposed polymorphism in L6, and (iii) LlpABW11M1-targeted strains belonging to different species only share BamA sequence homology in the surface-exposed region L6, and not in L4 or L7. Given that (i) the amino-terminal lectin domain of lectin-like bacteriocins is responsible for strain selectivity (21) and (ii) highly homologous bacteriocins PyoL1 and PyoL2 display different target spectra and (the majority of) their differential residues form a localized patch in the amino-terminal lectin domain (24), our current hypothesis is that the amino-terminal domain mediates binding to BamA via the L6 loop, and this is facilitated and/or stabilized by interaction of the carboxy-terminal lectin domain of LlpA with specific carbohydrate moieties in LPS. The observation that some pseudomonads host two bamA genes is intriguing as well (36). Expression of both these genes possibly makes isolates susceptible to lectin-like bacteriocins with different target panel specificities. This may explain why P. migulae ZA 45.2 (Fig. S3) and some other strains belonging to the P. fluorescens species group (26) are killed by both LlpABW11M1 and LlpA1.
The receptors of several modular bacteriocins, such as colicins (E. coli) and S-type pyocins (Pseudomonas aeruginosa), have been previously identified and typically function in the uptake of small molecules, such as vitamin B12 or iron-scavenging siderophores (4, 5). After receptor contact, bacteriocins are translocated across the outer membrane, either taking advantage of the receptor channel itself (energized by the Ton system) or via the trimeric OmpF porin (by usage of the Tol system). The dynamics of the latter process have been studied in more detail for colicin E9 of E. coli and involve major structural rearrangements of the bacteriocin to ensure threading through narrow pores of OmpF (54). More recently, the uptake process of pyocin S2 via the TonB-dependent iron transporter FpvAI was elucidated (55). Docking of this bacteriocin is achieved via structural mimicry where pyocin S2 attains a similar structure as the natural siderophore ligand of this receptor. Subsequently, the same conformational changes that would account for uptake of legitimate siderophores are induced, leading to unfolding and displacement of a transporter-occluding plug and hence transfer of the bacteriocin to the periplasm.
Contact-dependent growth inhibition (CDI) cassettes represent a different type of polymorphic toxin widespread among Gram-negative bacteria (20, 56–58). It requires a two-partner secretion system: CdiB is an outer membrane protein that transports the large effector protein CdiA, which is equipped with a toxin module at its carboxy terminus (CdiA-CT). Similar to modular bacteriocins, self-inhibition by the toxin is temporarily impeded by an immunity protein (CdiI). To date, several toxin activities of CdiA-CTs have been described, and a significant portion of these effectors display activities similar to colicins and pyocins (nuclease, pore forming) (20, 56, 59). For example, structural similarities were detected between a CDI toxin from Enterobacter cloacae and the rRNase domain from colicin E3 (60). A central region of CdiA accounts for target recognition and facilitates direct cell-to-cell contact (61). Several CDI receptors have been identified to date: the CdiA from E. coli EC536 interacts with OmpC and OmpF, whereas CdiA from E. coli STEC_O31 binds to the nucleoside transporter Tsx (61). Interestingly, BamA was previously identified as a CDI receptor as well (for CdiA from E. coli EC93) (62), and a specific contribution could be assigned to a combination of L6 and L7 (63). Although a link between loop polymorphism and receptor binding of CDI effectors can be observed, the structural details of the interaction between each of these partners remain elusive at this point.
A similar killing route as that followed by polymorphic toxins is unlikely to be followed by LlpAs. Lectin-like bacteriocins lack an obvious killing domain that needs to be translocated across the outer and/or inner membrane, and in addition, LlpA appears to be a very rigid molecule (21, 22): its two β-prisms are stabilized by conserved tryptophan triads, which are further rigidified by a number of intramolecular interactions. Extensive unfolding during translocation, as postulated for colicins and possibly some CdiAs, seems less plausible for LlpAs, and a “killing-upon-contact” mechanism can be envisaged that somehow interferes with the function of BamA, one of the few essential outer membrane proteins. Within this perspective, the lack of an obvious killer domain in lectin-like bacteriocins could point to a stress-activated killing system. In enterobacteria, the lipoprotein RcsF was previously shown to act as an envelope stress sensor, immediately interacting with BamA (64, 65). It remains unclear, however, whether a similar system is equally active in Pseudomonas.
It will be of particular interest to further explore the role of the carboxy-terminal β-hairpin extension of LlpAs. This stretch is pivotal for obtaining a functional bacteriocin, since a minor shortening (3-amino-acid deletion) results in loss of functionality (21). One possible mechanism of action that is compatible with the overall rigidity imposed by the dual β-prism architecture of LlpA may be that this part of LlpAs interferes with the gating dynamics of BamA, hampering insertion of unfolded outer membrane proteins (37, 66–69). In turn, this may lead to accumulation of unfolded polypeptides in the periplasm and a downstream response. This hypothesis is supported by the observation that L6, the loop selected by LlpAs, is rather rigid and positioned at the same side of BamA’s lateral pore (34). Recently, it was also demonstrated that integration of membrane proteins is highly dependent on the interaction of a buried part of L6 and one of BamA’s β-sheets in the barrel (70); a bacteriocin interaction may therefore also (structurally) interfere with the fine-tuning of this process. Future work will therefore be orientated toward unraveling the three-dimensional interaction of LlpA and BamA, in order to understand the nature of their protein-protein interaction. The direct interference with an essential protein(s) in the outer membrane, such as the BAM complex, is currently being explored as a novel antimicrobial strategy, for example, by designing peptides that actively interfere with the BAM machinery (71, 72) or peptidomimetics that selectively target β-barrel outer membrane proteins (73).
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S3. E. coli and P. aeruginosa strains were grown in lysogeny broth (MP Biomedicals) at 37°C. Other Pseudomonas strains were cultured in Trypticase soy broth (TSB; BD Biosciences) at 30°C. All cultures underwent shaking at 200 rpm. Media were solidified with 1.5% agar (Invitrogen). Media were supplemented with filter-sterilized ampicillin (100 to 800 µg/ml; Sigma-Aldrich), isopropyl β-d-1-thiogalactopyranoside (IPTG; 25 µg/ml; ForMedium), kanamycin (50 µg/ml; Sigma-Aldrich), tetracycline (15 to 150 µg/ml; Sigma-Aldrich), or gentamicin (15 to 45 µg/ml; TCI Europe) when required. Bacterial stocks were stored in the appropriate medium at −80°C in 25% (vol/vol) glycerol (VWR).
Strains and plasmids used in this study. Download TABLE S3, DOCX file, 0.02 MB (17.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plasmids were propagated in E. coli TOP10F′ (Invitrogen). E. coli BL21(DE3) (Novagen) was used as a host for generating recombinant bacteriocins. Genomic DNA from Pseudomonas strains was collected with the Puregene Yeast/Bact kit B (Qiagen). Plasmid DNA was isolated using the QIAprep spin miniprep kit (Qiagen).
Expression and purification of lectin-like bacteriocins.
Plasmid construction and production of His-tagged LlpA bacteriocins were described previously (21, 24, 26). Briefly, overnight cell cultures of BL21(DE3) containing the plasmid of interest were subcultured in 500-ml Erlenmeyer flasks, with shaking. After the optical density (600 nm) reached 0.6, cultures were cooled to 16°C, supplemented with IPTG, and induced for an additional 16 h (overnight). The following day, cells were harvested by centrifugation (20 min, 5,000 × g), and pellets were frozen overnight at −20°C. Next, cells were resuspended in lysis buffer containing 300 mM NaCl (VWR International), 5 mM imidazole (Sigma-Aldrich), 50 mM Na2HPO4 (VWR International) at pH of 8.0. Subsequently, cells were lysed by sonication with a Branson digital sonifier (amplitude of 20%, 10 cycles of 30 s on/30 s off), and supplemented with 10 U/ml benzonase nuclease (0.5 h at 37°C; Sigma-Aldrich). Cell debris was removed by centrifugation (10,000 × g, 30 min), and the presence of recombinant protein in soluble fractions was confirmed by SDS-PAGE. The cell-free lysate was loaded on a 5-ml His-trap column (GE Healthcare), equilibrated with lysis buffer, using an Äkta purifier (GE Healthcare). Recombinant proteins were eluted over a 5 to 500 mM imidazole gradient, and bacteriocin fractions were selected by SDS-PAGE. Samples were dialyzed overnight (20 mM Tris-HCl, 200 mM NaCl; pH 7.5) and further polished by gel filtration on a Superdex 200 prep grade column (HiLoad 16/600; GE Healthcare). Recombinant bacteriocins were concentrated with Vivaspin filters (Sartorius Stedim). Protein concentrations were measured at 280-nm absorbance (Genesys 10S UV-Vis spectrophotometer; Thermo Scientific).
Antibacterial spot assay and isolation of mutants with altered LlpA sensitivity.
Strains of interest were grown overnight in LB or TSB (16 h, ~108 to 109 CFU/ml) supplemented with antibiotics and IPTG, if needed. Twenty-microliter aliquots of cell suspensions were added to 5 ml molten soft agar (0.5%) and poured on sterile plates containing the same medium to obtain cell lawns. Subsequently, 10-µl volumes of filter-sterilized purified recombinant bacteriocin (1 mg/ml) were spotted on top of the solidified lawn (dialysis buffer was used as a negative control). After drying, plates were incubated overnight. The following day, cell lawns were scored for the presence of halos (zones of growth inhibition), which are indicative of bacteriocin activity (74).
To obtain bacteriocin-resistant mutants, P. fluorescens Pf0-1 (15 independent cultures) and P. aeruginosa PAO1 (30 independent cultures) were grown for 48 h to obtain late-stationary-phase cultures. Cell lawns were made in a similar way as described above and supplemented with 50-µl spots of LlpA1 (on P. fluorescens Pf0-1) or PyoL1 (on P. aeruginosa PAO1) (5 mg/ml). One colony emerging per halo after 24 h of incubation time was selected, streaked to single colony, and verified for an altered (absent or highly tolerant) bacteriocin susceptibility phenotype. To exclude the presence of nonparental contaminants, genomic DNA of the isolates was collected and their boxA amplicon profile was compared with that of the wild type (BOX-PCR) (75).
Whole-genome sequencing and SNP detection.
High-quality genomic DNA was isolated from overnight cultures of P. fluorescens Pf0-1 wild type and selected Pf0-1 mutants. Concentration and purity of the DNA were determined using Nanodrop analysis (Thermo Fisher Scientific), gel electrophoresis, and Qubit analysis (Thermo Fisher Scientific). Libraries were prepared at GeneCore (EMBL, Heidelberg) using the NEBNext kit with an average insert size of 200 bp. The DNA libraries were multiplexed and subjected to 100-cycle paired-end massive parallel sequencing with the Illumina HiSeq2000 system (GeneCore, EMBL, Heidelberg). CLC Genomics Workbench version 6.5.1 (Qiagen) was used for analysis of the sequences. Following quality assessment of the raw data, reads were trimmed using quality scores of the individual bases. The quality limit was set to 0.01, and the maximum allowed number of ambiguous bases was set to 2. Reads shorter than 15 bases were discarded from the set. After trimming, the average length of the remaining reads was 83.31 bp. The trimmed reads were mapped (mismatch cost of 2, insertion cost of 3, deletion cost of 3, length fraction of 0.8, similarity fraction of 0.8) to the Pseudomonas fluorescens Pf0-1 reference genome (NC_007492) using the CLC Assembly Cell 4.0 algorithm, yielding an average coverage of approximately 154×. Subsequently, mutations in all samples were identified using the CLC quality-based variant detection tool with default parameters. No mutations could be retrieved in P. fluorescens Pf0-1 in comparison to the reference genome.
Recombinant DNA methods.
Preparation of competent E. coli cells, heat shock transformation, and DNA gel electrophoresis were done using standard methods (76). The bamA genes from P. aeruginosa PAO1 (AAG07036), P. aeruginosa Br670 (KSQ40041), P. fluorescens Pf0-1 (ABA72852), and P. savastanoi pv. glycinea LMG 5066 (KPX45010) were amplified by PCR with Q5 DNA polymerase (New England Biolabs) with a C1000 thermal cycler (Bio-Rad), using genomic DNA as a template. Amplicons were purified with the QIAquick PCR purification kit (Qiagen), digested, ligated in shuttle vector pJB3Tc20, and transformed to E. coli TOP10F′. Restriction (New England Biolabs) and ligation (T4 DNA ligase; Invitrogen) enzymes were used according to the supplier specifications. Plasmids with inserts were verified by sequencing (GATC Biotech, Constance, Germany) and subsequently introduced into Pseudomonas cells via electroporation. Electrocompetent cells were prepared by sucrose (Merck) treatment (77). After introduction of pCMPG6279 (pJB3Tc20 plasmid carrying bamABr670) in P. aeruginosa PS186, the original plasmid carrying pMMB67EH with bamAPAO1 was cured by introducing pMMB67EH carrying a Gmr cassette. Loss of bamAPAO1 was verified by PCR.
The bamA fragments encoding β-barrels in P. aeruginosa and P. syringae strains and the 16S rRNA fragments of Pseudomonas sp. strains VA 2.1, WCS365, and ZA 45.2 were amplified with Q5 DNA polymerase, purified, and sequenced. Primers used in this study are summarized in Table S4.
Primers used in this study. Download TABLE S4, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome searches and multiple sequence analysis.
BamAs and lectin-like bacteriocins in Pseudomonas genomes were identified using the National Center for Biotechnology (NCBI) nonredundant database for BLAST searches (sample date 1 July 2017). Sequences of BamAs from P. fluorescens Pf0-1 and P. aeruginosa PAO1, and also previously characterized LlpAs were used as search queries. Domains were predicted with Pfam (78). Multiple sequence alignments were created with MUSCLE (79), and gene synteny was explored with MAUVE 2.3.1 (80), implemented in Geneious v.7.1.9.
Structural modeling of β-barrel domains of Pseudomonas BamAs.
The I-TASSER server (81) was used for structural modeling of the β-barrel domains of BamAs from P. aeruginosa PAO1 and P. fluorescens Pf0-1. Models were visualized with PyMOL.
Accession number(s).
The genome sequencing data set generated and analyzed in this study is available in the SRA repository of NCBI under accession number SRP118490 with BioProject number PRJNA408273.
ACKNOWLEDGMENTS
We thank M. Toyofuku and N. Nomura (University of Tsukuba) for kindly providing P. aeruginosa PS186.
M.G.K.G. is the recipient of a fellowship from the Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen (12M4618N). T.S. was a fellow of IWT Vlaanderen (IWT.121525). The genomic analysis was supported by the KU Leuven Research Council (J.M.; PF/10/010). S.K.B. is supported by the Intramural Research Program of the NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). This research was supported by grants 1523116N, 1525618N, and 1529718N (M.G.K.G., Krediet aan Navorser, FWO Vlaanderen).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Ghequire MGK, Swings T, Michiels J, Buchanan SK, De Mot R. 2018. Hitting with a BAM: selective killing by lectin-like bacteriocins. mBio 9:e02138-17. https://doi.org/10.1128/mBio.02138-17.
Contributor Information
Stephen P. Diggle, Georgia Institute of Technology School of Biological Sciences.
Marvin Whiteley, University of Texas at Austin.
REFERENCES
- 1.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 3.Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 4.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 6.Ghequire MG, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Riley MA, Goldstone CM, Wertz JE, Gordon D. 2003. A phylogenetic approach to assessing the targets of microbial warfare. J Evol Biol 16:690–697. doi: 10.1046/j.1420-9101.2003.00575.x. [DOI] [PubMed] [Google Scholar]
- 8.Schulz S, Stephan A, Hahn S, Bortesi L, Jarczowski F, Bettmann U, Paschke AK, Tusé D, Stahl CH, Giritch A, Gleba Y. 2015. Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc Natl Acad Sci U S A 112:E5454–E5460. doi: 10.1073/pnas.1513311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCaughey LC, Ritchie ND, Douce GR, Evans TJ, Walker D. 2016. Efficacy of species-specific protein antibiotics in a murine model of acute Pseudomonas aeruginosa lung infection. Sci Rep 6:30201. doi: 10.1038/srep30201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. 2015. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6:e02368-14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CL, Smith K, Wall DM, Walker D. 2015. Activity of species-specific antibiotics against Crohn’s disease-associated adherent-invasive Escherichia coli. Inflamm Bowel Dis 21:2372–2382. doi: 10.1097/MIB.0000000000000488. [DOI] [PubMed] [Google Scholar]
- 12.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghequire MG, De Mot R. 2018. Turning over a new leaf: bacteriocins going green. Trends Microbiol 26:1–2. doi: 10.1016/j.tim.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS III, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghoul M, West SA, Johansen HK, Molin S, Harrison OB, Maiden MC, Jelsbak L, Bruce JB, Griffin AS. 2015. Bacteriocin-mediated competition in cystic fibrosis lung infections. Proc Biol Sci 282. doi: 10.1098/rspb.2015.0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce JB, West SA, Griffin AS. 2017. Bacteriocins and the assembly of natural Pseudomonas fluorescens populations. J Evol Biol 30:352–360. doi: 10.1111/jeb.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorosky RJ, Yu JM, Pierson LS III, Pierson EA. 2017. Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilms and on roots. Appl Environ Microbiol 83:e00706-17. doi: 10.1128/AEM.00706-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godino A, Príncipe A, Fischer S. 2016. A ptsP deficiency in PGPR Pseudomonas fluorescens SF39a affects bacteriocin production and bacterial fitness in the wheat rhizosphere. Res Microbiol 167:178–189. doi: 10.1016/j.resmic.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Jamet A, Nassif X. 2015. New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghequire MG, Garcia-Pino A, Lebbe EK, Spaepen S, Loris R, De Mot R. 2013. Structural determinants for activity and specificity of the bacterial toxin LlpA. PLoS Pathog 9:e1003199. doi: 10.1371/journal.ppat.1003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCaughey LC, Grinter R, Josts I, Roszak AW, Waløen KI, Cogdell RJ, Milner J, Evans T, Kelly S, Tucker NP, Byron O, Smith B, Walker D. 2014. Lectin-like bacteriocins from Pseudomonas spp. utilise d-rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathog 10:e1003898. doi: 10.1371/journal.ppat.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. doi: 10.3389/fmicb.2011.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghequire MG, Dingemans J, Pirnay JP, De Vos D, Cornelis P, De Mot R. 2014. O serotype-independent susceptibility of Pseudomonas aeruginosa to lectin-like pyocins. Microbiologyopen 3:875–884. doi: 10.1002/mbo3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parret AH, Temmerman K, De Mot R. 2005. Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 71:5197–5207. doi: 10.1128/AEM.71.9.5197-5207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghequire MG, Li W, Proost P, Loris R, De Mot R. 2012. Plant lectin-like antibacterial proteins from phytopathogens Pseudomonas syringae and Xanthomonas citri. Environ Microbiol Rep 4:373–380. doi: 10.1111/j.1758-2229.2012.00331.x. [DOI] [PubMed] [Google Scholar]
- 27.Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang XX, Moon CD, Gehrig SM, Godfrey SA, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferguson GC, Bertels F, Rainey PB. 2013. Adaptive divergence in experimental populations of Pseudomonas fluorescens. V. Insight into the niche specialist fuzzy spreader compels revision of the model Pseudomonas radiation. Genetics 195:1319–1335. doi: 10.1534/genetics.113.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesarich CH, Rees-George J, Gardner PP, Ghomi FA, Gerth ML, Andersen MT, Rikkerink EH, Fineran PC, Templeton MD. 2017. Transposon insertion libraries for the characterization of mutants from the kiwifruit pathogen Pseudomonas syringae pv. actinidiae. PLoS One 12:e0172790. doi: 10.1371/journal.pone.0172790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis H, Sharon N. 1998. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem Rev 98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 31.Noinaj N, Gumbart JC, Buchanan SK. 2017. The β-barrel assembly machinery in motion. Nat Rev Microbiol 15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyton DL, Belousoff MJ, Lithgow T. 2015. The β-barrel assembly machinery complex. Methods Mol Biol 1329:1–16. doi: 10.1007/978-1-4939-2871-2_1. [DOI] [PubMed] [Google Scholar]
- 33.Webb CT, Heinz E, Lithgow T. 2012. Evolution of the β-barrel assembly machinery. Trends Microbiol 20:612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Noinaj N, Rollauer SE, Buchanan SK. 2015. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr Opin Struct Biol 31:35–42. doi: 10.1016/j.sbi.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard-Rivera M, Misra R. 2012. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol 194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz E, Lithgow T. 2014. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Front Microbiol 5:370. doi: 10.3389/fmicb.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. 2016. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 38.Albrecht R, Schütz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K. 2014. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr 70:1779–1789. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 39.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakelar J, Buchanan SK, Noinaj N. 2016. The structure of the β-barrel assembly machinery complex. Science 351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y. 2014. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 28:2677–2685. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 42.Tashiro Y, Nomura N, Nakao R, Senpuku H, Kariyama R, Kumon H, Kosono S, Watanabe H, Nakajima T, Uchiyama H. 2008. Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J Bacteriol 190:3969–3978. doi: 10.1128/JB.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Belkum A, Soriaga LB, LaFave MC, Akella S, Veyrieras JB, Barbu EM, Shortridge D, Blanc B, Hannum G, Zambardi G, Miller K, Enright MC, Mugnier N, Brami D, Schicklin S, Felderman M, Schwartz AS, Richardson TH, Peterson TC, Hubby B, Cady KC. 2015. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. mBio 6:e01796-15. doi: 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flury P, Aellen N, Ruffner B, Péchy-Tarr M, Fataar S, Metla Z, Dominguez-Ferreras A, Bloemberg G, Frey J, Goesmann A, Raaijmakers JM, Duffy B, Höfte M, Blom J, Smits TH, Keel C, Maurhofer M. 2016. Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J 10:2527–2542. doi: 10.1038/ismej.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. 2016. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 46.Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. 2015. Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 48.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 49.Parret AH, Schoofs G, Proost P, De Mot R. 2003. Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J Bacteriol 185:897–908. doi: 10.1128/JB.185.3.897-908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berendsen RL, van Verk MC, Stringlis IA, Zamioudis C, Tommassen J, Pieterse CM, Bakker PA. 2015. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics 16:539. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen DD, Melnik AV, Koyama N, Lu X, Schorn M, Fang J, Aguinaldo K, Lincecum TL Jr., Ghequire MG, Carrion VJ, Cheng TL, Duggan BM, Malone JG, Mauchline TH, Sanchez LM, Kilpatrick AM, Raaijmakers JM, De Mot R, Moore BS, Medema MH, Dorrestein PC. 2016. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat Microbiol 2:16197. doi: 10.1038/nmicrobiol.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonezuka K, Shimodaira J, Tabata M, Ohji S, Hosoyama A, Kasai D, Yamazoe A, Fujita N, Ezaki T, Fukuda M. 2017. Phylogenetic analysis reveals the taxonomically diverse distribution of the Pseudomonas putida group. J Gen Appl Microbiol 63:1–10. doi: 10.2323/jgam.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Thakur S, Weir BS, Guttman DS. 2016. Phytopathogen genome announcement: draft genome sequences of 62 Pseudomonas syringae type and pathotype strains. Mol Plant Microbe Interact 29:243–246. doi: 10.1094/MPMI-01-16-0013-TA. [DOI] [PubMed] [Google Scholar]
- 54.Housden NG, Hopper JT, Lukoyanova N, Rodriguez-Larrea D, Wojdyla JA, Klein A, Kaminska R, Bayley H, Saibil HR, Robinson CV, Kleanthous C. 2013. Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science 340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White P, Joshi A, Rassam P, Housden NG, Kaminska R, Goult JD, Redfield C, McCaughey LC, Walker D, Mohammed S, Kleanthous C. 2017. Exploitation of an iron transporter for bacterial protein antibiotic import. Proc Natl Acad Sci U S A 114:12051–12056. doi: 10.1073/pnas.1713741114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. 2015. Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J Mol Biol 427:3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danka ES, Garcia EC, Cotter PA. 2017. Are CDI systems multicolored, facultative, helping greenbeards? Trends Microbiol 25:391–401. doi: 10.1016/j.tim.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guérin J, Bigot S, Schneider R, Buchanan SK, Jacob-Dubuisson F. 2017. Two-partner secretion: combining efficiency and simplicity in the secretion of large proteins for bacteria-host and bacteria-bacteria interactions. Front Cell Infect Microbiol 7:148. doi: 10.3389/fcimb.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. 2012. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol 84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. 2014. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure 22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruhe ZC, Nguyen JY, Xiong J, Koskiniemi S, Beck CM, Perkins BR, Low DA, Hayes CS. 2017. CdiA effectors use modular receptor-binding domains to recognize target bacteria. mBio 8:e00290-17. doi: 10.1128/mBio.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. 2008. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruhe ZC, Wallace AB, Low DA, Hayes CS. 2013. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio 4:e00480-13. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov AK, Leverrier P, Van der Henst C, Vertommen D, Typas A, Collet JF. 2014. Detecting envelope stress by monitoring β-barrel assembly. Cell 159:1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 65.Laloux G, Collet JF. 2017. Major Tom to ground control: how lipoproteins communicate extra-cytoplasmic stress to the decision center of the cell. J Bacteriol 199:e00216-17. doi: 10.1128/JB.00216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warner LR, Gatzeva-Topalova PZ, Doerner PA, Pardi A, Sousa MC. 2017. Flexibility in the periplasmic domain of BamA is important for function. Structure 25:94–106. doi: 10.1016/j.str.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. 2016. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sinnige T, Weingarth M, Daniëls M, Boelens R, Bonvin AM, Houben K, Baldus M. 2015. Conformational plasticity of the POTRA 5 domain in the outer membrane protein assembly factor BamA. Structure 23:1317–1324. doi: 10.1016/j.str.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Browning DF, Bavro VN, Mason JL, Sevastsyanovich YR, Rossiter AE, Jeeves M, Wells TJ, Knowles TJ, Cunningham AF, Donald JW, Palmer T, Overduin M, Henderson IR. 2015. Cross-species chimeras reveal BamA POTRA and beta-barrel domains must be fine-tuned for efficient OMP insertion. Mol Microbiol 97:646–659. doi: 10.1111/mmi.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wzorek JS, Lee J, Tomasek D, Hagan CL, Kahne DE. 2017. Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. Proc Natl Acad Sci U S A 114:2598–2603. doi: 10.1073/pnas.1616576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagan CL, Wzorek JS, Kahne D. 2015. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A 112:2011–2016. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mori N, Ishii Y, Tateda K, Kimura S, Kouyama Y, Inoko H, Mitsunaga S, Yamaguchi K, Yoshihara E. 2012. A peptide based on homologous sequences of the β-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa. J Antimicrob Chemother 67:2173–2181. doi: 10.1093/jac/dks174. [DOI] [PubMed] [Google Scholar]
- 73.Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, Robinson JA. 2016. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J Biol Chem 291:1921–1932. doi: 10.1074/jbc.M115.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hockett KL, Baltrus DA. 2017. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp 119:e55064. doi: 10.3791/55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koeuth T, Versalovic J, Lupski JR. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res 5:408–418. doi: 10.1101/gr.5.4.408. [DOI] [PubMed] [Google Scholar]
- 76.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 77.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 78.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic organization of the fuzVWXYZ locus in the genome of P. fluorescens SBW25 in comparison with corresponding regions in P. aeruginosa PAO1, P. fluorescens Pf0-1 and P. syringae pv. syringae ICMP 18884. Synteny is visualized by sequence conservation (grey shading) for the corresponding orthologues, if present. Genes (displayed as arrows) of the fuzVWXYZ cluster in P. fluorescens SBW25 are colored blue and an extra gene involved in the fuzzy spreader phenotype in P. syringae pv. syringae ICMP 18884 is shown in yellow. The location of the fuzY mutation (insertion) in P. fluorescens Pf0-1 mutant B3 (red) is indicated with a black triangle. Dotted lines indicate the absence of an equivalent sequence. Download FIG S1, TIF file, 0.2 MB (199.5KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Susceptibility of Pseudomonas aeruginosa strains, heterologously expressing bamA genes from different donors, to lectin-like bacteriocins PyoL1, PyoL2, LlpA1, and LlpABW11M1. Controls are strains transformed with a wild-type plasmid (no insert). Sensitivity pairs: P. aeruginosa PAO1/PyoL1, P. aeruginosa Br670/PyoL2, P. fluorescens Pf0-1/LlpA1, P. savastanoi pv. glycinea LMG 5066/LlpABW11M1. The presence of a halo is indicative of growth inhibition due to bacteriocin activity. Download FIG S2, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Susceptibility of Pseudomonas strains (non P. aeruginosa), heterologously expressing bamA genes from different donors, to lectin-like bacteriocins PyoL1, PyoL2, LlpA1, and LlpABW11M1. Controls are strains transformed with a wild-type plasmid (no insert). Sensitivity pairs: P. aeruginosa PAO1/PyoL1, P. aeruginosa Br670/PyoL2, P. fluorescens Pf0-1/LlpA1, P. savastanoi pv. glycinea LMG 5066/LlpABW11M1. The presence of a halo is indicative of growth inhibition due to bacteriocin activity. Download FIG S3, TIF file, 1.6 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Relative frequencies of occurrence of hypervariable BamA L6 sequences in P. syringae group isolates and strains used in the test panel and LlpABW11M1 susceptibility of strains in the respective set (highly conserved residues that are part of L6 are shown in gray). Download TABLE S1, DOCX file, 0.01 MB (12.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Multiple sequence alignment of L6 sequences of BamAs from Pseudomonas spp. that are susceptible to LlpABW11M1. Differential shading reflects the degree of sequence conservation. The hypervariable signature motif of LlpABW11M1-susceptible isolates is boxed in blue. Download FIG S4, TIF file, 0.5 MB (521.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of isolates encoding LlpABW11M1 or a highly homologous sequence (>97.5% amino acid identity; 0 to 2 different amino acid residues in amino-terminal lectin domains) and their corresponding BamA L6 amino acid sequence (nonpolymorphic [conserved] residues that are part of L6 in the Pseudomonas putida species group are shown in gray). Download TABLE S2, DOCX file, 0.01 MB (12.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Strains and plasmids used in this study. Download TABLE S3, DOCX file, 0.02 MB (17.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers used in this study. Download TABLE S4, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.