ABSTRACT
The Salmonella cytolethal distending toxin (S-CDT), first described as the “typhoid toxin” in Salmonella enterica subsp. enterica serotype Typhi, induces DNA damage in eukaryotic cells. Recent studies have shown that more than 40 nontyphoidal Salmonella (NTS) serotypes carry genes that encode S-CDT, yet very little is known about the activity, function, and role of S-CDT in NTS. Here we show that deletion of genes encoding the binding subunit (pltB) and a bacteriophage muramidase predicted to play a role in toxin export (ttsA) does not abolish toxin activity in the S-CDT-positive NTS Salmonella enterica subsp. enterica serotype Javiana. However, S. Javiana strains harboring deletions of both pltB and its homolog artB, had a complete loss of S-CDT activity, suggesting that S. Javiana carries genes encoding two variants of the binding subunit. S-CDT-mediated DNA damage, as determined by phosphorylation of histone 2AX (H2AX), producing phosphorylated H2AX (γH2AX), was restricted to epithelial cells in S and G2/M phases of the cell cycle and did not result in apoptosis or cell death. Compared to mice infected with a ΔcdtB strain, mice infected with wild-type S. Javiana had significantly higher levels of S. Javiana in the liver, but not in the spleen, ileum, or cecum. Overall, we show that production of active S-CDT by NTS serotype S. Javiana requires different genes (cdtB, pltA, and either pltB or artB) for expression of biologically active toxin than those reported for S-CDT production by S. Typhi (cdtB, pltA, pltB, and ttsA). However, as in S. Typhi, NTS S-CDT influences the outcome of infection both in vitro and in vivo.
KEYWORDS: DNA damage, Salmonella, nontyphoidal, typhoid toxin
IMPORTANCE
Nontyphoidal Salmonella (NTS) are a major cause of bacterial food-borne illness worldwide; however, our understanding of virulence mechanisms that determine the outcome and severity of nontyphoidal salmonellosis is incompletely understood. Here we show that S-CDT produced by NTS plays a significant role in the outcome of infection both in vitro and in vivo, highlighting S-CDT as an important virulence factor for nontyphoidal Salmonella serotypes. Our data also contribute novel information about the function of S-CDT, as S-CDT-mediated DNA damage occurs only during certain phases of the cell cycle, and the resulting damage does not induce cell death as assessed using a propidium iodide exclusion assay. Importantly, our data support that, despite having genetically similar S-CDT operons, NTS serotype S. Javiana has different genetic requirements than S. Typhi, for the production and export of active S-CDT.
INTRODUCTION
Infections with nontyphoidal Salmonella (NTS) account for an estimated 93.8 million illnesses and 155,000 deaths per year globally (1), making NTS the third leading cause of bacterial food-borne disease worldwide (2). The Salmonella cytolethal distending toxin (S-CDT) (called the “typhoid toxin”) was first characterized in Salmonella enterica subsp. enterica serotype Typhi, the causative agent of typhoid fever (3, 4). However, recent studies have shown that S-CDT is not unique to S. Typhi, as >40 NTS serotypes are known to carry genes that encode S-CDT (5–7). Furthermore, in vitro characterizations have shown that these S-CDT-positive NTS serotypes produce active toxin (6, 8, 9).
S-CDT is an A2B5 toxin, composed of a pentameric ring of (i) PltB subunits which interact with host cell glycans (4, 10), (ii) an ADP-ribosyltransferase (PltA) with homology to the S1 subunit of the pertussis toxin (3, 4), and (iii) CdtB, which has nuclease activity (11). Infection with S-CDT-positive strains results in the accumulation of eukaryotic cells in the G2/M cell cycle phase (3, 6, 9, 12) and activation of the host cell’s DNA damage response (8). In vivo, administration of purified S-CDT from S. Typhi has been shown to partially recapitulate signs of typhoid fever in a mouse model (4, 10). S-CDT has also been shown to prolong carriage of Salmonella in 129S6/SvEvTac mice (13). Del Bel Belluz et al. showed that genetically engineered S-CDT-positive S. enterica subsp. enterica serotype Typhimurium (a naturally S-CDT-negative serotype [8]) persisted for a longer period of time in vivo than the S-CDT-negative parent strain did, which suggests that S-CDT alters the host-pathogen interaction in vivo, enabling S-CDT-positive Salmonella to persist in the host (13).
S. enterica subsp. enterica serotype Javiana is the fourth most commonly isolated NTS serotype causing infections in the United States (14) and is the most common S-CDT-positive NTS serotype (8). The majority of studies characterizing S-CDT activity have been performed using Salmonella serotype Typhi, which has several important genotypic and phenotypic differences from NTS serotypes (15–17). Furthermore, previous characterizations in vitro (3, 8, 9, 12, 18, 19), which were critical for asserting the activity of NTS S-CDT, were conducted using tumor-derived (cancerous) cell lines, which often have an altered DNA damage response (20). Use of a nontransformed cell line is therefore necessary to accurately reflect how infection with S-CDT-positive NTS impacts the cellular outcome of infection.
To better understand the mechanisms by which S-CDT-mediated intoxication occurs both in vitro and in vivo, we characterized the outcome of infection with S-CDT-positive S. Javiana at the cellular level, using a non-cancer-derived human intestinal epithelial cell line, and at the organismal level, by infecting C56BL/6 mice. We found that infection of human epithelial cells resulted in an altered cell cycle progression characterized by an accumulation of cells in the G2/M phase associated with DNA damage response activation in the absence of cell death. While cdtB and pltA were essential for toxin activity in vitro, pltB, STY1887, and ttsA, which are also carried within the S-CDT islet in S. Javiana, were not. Despite colonizing the ilea and ceca of infected mice at similar levels, infection with wild-type S. Javiana was associated with a higher bacterial load in the liver, suggesting that S-CDT might contribute to bacterial evasion of host innate inflammatory response mechanisms and allow for systemic infection with S. Javiana.
RESULTS
Infection of human epithelial cells with S-CDT-positive serotype S. Javiana induces a DNA damage response that significantly alters cell cycle progression.
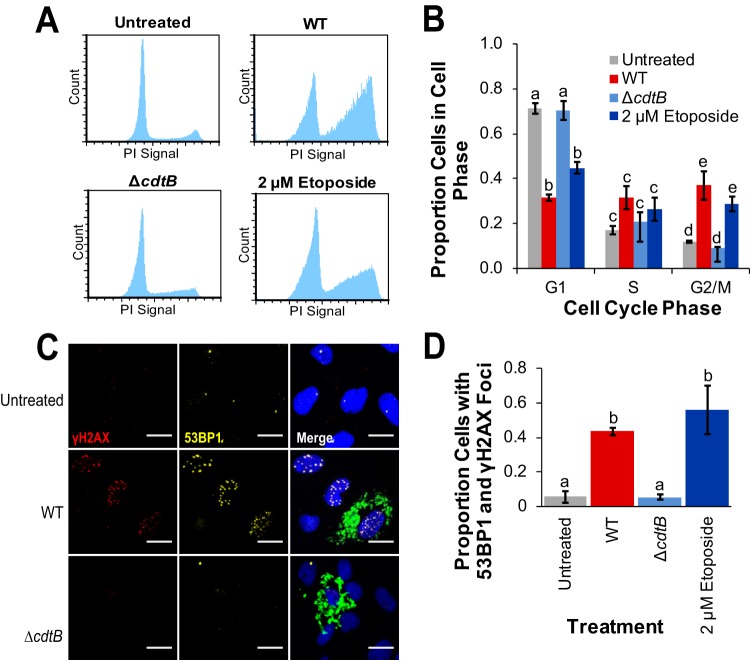
We previously established in a HeLa cell model that infection with S-CDT-positive NTS strains activates a DNA damage response and induces an accumulation of cells in the G2/M phase, while infection with either wild-type S-CDT-negative serotypes or ΔcdtB mutants does not do so (8). To overcome the limitations of using cell lines with altered DNA damage repair pathways, we infected normal (noncancerous) human intestinal epithelial cells (HIEC-6 cells [21]) with wild-type and ΔcdtB strains of S. Javiana to determine whether normal epithelial cells are also susceptible to S-CDT-mediated DNA damage. HIEC-6 cells infected with wild-type S. Javiana displayed a characteristic accumulation of cells in the G2/M phase of the cell cycle (average of 37% of the cells compared to 12% in uninfected controls [Fig. 1A and B]). Similar to uninfected controls, cells infected with the S. Javiana ΔcdtB strain had an average of 9% of cells in the G2/M phase, suggesting that the accumulation of cells in the G2/M phase is dependent on cdtB (Fig. 1A and B). Furthermore, HIEC-6 cells infected with wild-type S. Javiana also had a significantly lower proportion of cells in the G1 phase (Fig. 1B), compared to both uninfected controls (P = 0.0022) and cells infected with the ΔcdtB strain (P = 0.0008). Interestingly, the proportion of cells in S phase did not differ between ΔcdtB or wild-type strain-infected cells and uninfected controls (P > 0.05 for all comparisons). The altered cell cycle progression was consistent with induction of a DNA damage response, as infection with wild-type S. Javiana resulted in 43% of cells having at least four p53-binding protein 1 (53BP1) foci that colocalized with phosphorylated histone 2AX (γH2AX) foci (both DNA damage markers), compared to just 5% in uninfected control cells (P = 0.0113) or in cells infected with the ΔcdtB strain (P = 0.0213; Fig. 1C and D). Because active S-CDT is produced only by intracellular Salmonella (3, 8), we examined whether deletion of cdtB affected invasion efficiency as was reported previously for infection of HeLa cells with wild-type and ΔcdtB S. Javiana (9). Deletion of cdtB did not significantly affect invasion efficiency (P = 0.7113) in the ΔcdtB strain (0.59% versus 0.62% intracellular wild-type and ΔcdtB S. Javiana, respectively, normalized by CFU in the inoculum used for infection).
FIG 1.
Infection with wild-type S. Javiana activates a DNA damage response and results in an accumulation of cells in the G2/M phase. HIEC-6 cells were infected with S. Javiana for 48 h prior to immunofluorescence staining or flow cytometry analyses. Cells treated with the topoisomerase inhibitor etoposide (at a final concentration of 2 µM) for 24 h served as a positive control. Data are from three independent experiments. (A) Representative histograms showing the cell cycle progression of HIEC-6 cells infected with wild-type (WT) and ΔcdtB S. Javiana strains; an untreated control is included to show a normal cell cycle progression, and cells treated with 2 µM etoposide demonstrate the accumulation of cells in G2/M phase due to DNA damage. (B) Quantification of cell cycle analyses shown in panel A. Histogram bars that do not share letters within a given cell cycle phase group (i.e., G1, S, and G2/M) are significantly different (P < 0.05). (C) Representative images of cells infected with S. Javiana strains (antibody stain shown in green) and positive controls (treated with 2 µM etoposide) and negative controls (untreated). The nuclei of HIEC-6 cells were stained with DAPI (blue). Scale bars represent 20 microns. (D) Quantification of the proportions of cells with at least four 53BP1 foci (shown in yellow) that colocalized with γH2AX foci (shown in red). Treatments that do not share letters have significantly different (P < 0.05) proportions of cells with 53BP1 foci that colocalized with γH2AX foci. P values were corrected for multiple comparisons using the Tukey honestly significant difference (HSD) correction method. Values in panels B and D are means ± standard errors of the means (error bars).
pltA and cdtB, but not pltB, ttsA, or STY1887 are required for expression of active S-CDT in vitro.
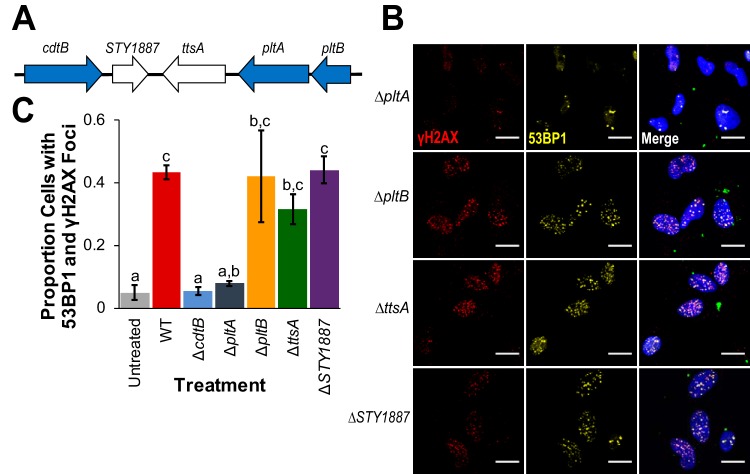
S-CDT is encoded by genes in two adjacent operons (Fig. 2A) (3, 18). In addition to genes encoding S-CDT (pltB, pltA, and cdtB), the islet also carries genes encoding a putative bacteriophage muramidase (ttsA), which is hypothesized to play a role in toxin secretion (18), and STY1887, which has an unknown function in S. Typhi (18). To determine which genes in the S-CDT islet are essential for S-CDT-mediated DNA damage response activation in HIEC-6 cells infected with S. Javiana, we constructed strains with deletions of individual genes in the S-CDT islet.
FIG 2.
pltB, ttsA, and STY1887 are not required for S-CDT-mediated DNA damage in HIEC-6 cells. HIEC-6 cells were infected with S. Javiana strains harboring single-gene deletions in the S-CDT islet. Immunofluorescence staining was performed for DNA damage response foci γH2AX and 53BP1. (A) Organization of the S-CDT islet in S. Javiana. Genes shown in blue represent genes encoding S-CDT subunits; genes shown in white are contained within the islet but do not encode protein products that constitute part of the S-CDT holotoxin. (B) Representative images of cells infected with wild-type S. Javiana and S-CDT single gene deletion strains (both colored green in the merged image); HIEC-6 cell nuclei are shown in blue. (C) Quantification of the proportions of cells with at least four 53BP1 foci that colocalized with γH2AX foci. Treatments that do not share letters have significantly different (P < 0.05) proportions of cells with 53BP1 foci that colocalized with γH2AX foci. Data represent three independent experiments. P values were corrected, to account for multiple testing, using Tukey’s HSD test. Error bars represent standard errors of the means.
Deletion of cdtB and pltA abolished the ability of S. Javiana to induce DNA damage response foci in HIEC-6 cells (Fig. 1C and D and Fig. 2B and C). HIEC-6 cells infected with the ΔcdtB strain or ΔpltA strain had an average of 5% and 8% of cells with at least four 53BP1 foci that colocalized with γH2AX foci, respectively, which did not differ significantly from uninfected controls (ΔcdtB strain versus control, P = 0.9835; ΔpltA strain versus control, P = 0.6469). In contrast, HIEC-6 cells infected with STY1887 or pltB gene-deleted strains had an average of 44% and 42% of HIEC-6 cells with at least four 53BP1 and γH2AX foci, respectively, which did not differ from HIEC-6 cells infected with wild-type S. Javiana (STY1887 versus wild-type, P > 0.9999; ΔpltB strain versus wild-type, P > 0.9999). Deletion of ttsA resulted in slightly lower levels of HIEC-6 cells with DNA damage response foci compared to wild-type S. Javiana (i.e., 32% of cells versus 43%), but this was not significantly different (P = 0.9795). Because ttsA was previously shown to be essential for S-CDT-mediated intoxication for S. Typhi (18), we aligned ttsA from typhoidal strains (i.e., S. Typhi strains CT-18 and Ty2 and S. Paratyphi A strain ATCC 11511) and nontyphoidal strains (e.g., Salmonella serotypes Javiana, Montevideo, and Schwarzengrund) to determine whether sequence diversity could partially explain the discrepant requirement for ttsA between S. Typhi and S. Javiana. Overall, ttsA was highly conserved (98.2% DNA sequence similarity) between typhoidal and nontyphoidal serotypes (see Fig. S1 in the supplemental material), with two nonsynonymous substitutions among 180 amino acids. Interestingly, these two amino acid substitutions were unique to the S. Montevideo ttsA sequence; the ttsA sequences from S. Javiana and the other serotypes (including both typhoidal and nontyphoidal serotypes) were identical, suggesting that sequence diversity in ttsA is unlikely to explain the lack of a ttsA requirement for production of active S-CDT in S. Javiana.
Amino acid alignment of TtsA from typhoidal and nontyphoidal serotypes. Amino acid sequences were predicted from ttsA sequences extracted from whole-genome sequence data. Strains included in analyses follow: S. Typhi strain CT-18, S. Typhi strain Ty2, S. Montevideo strain USDA-ARS-USMARC-1903, S. Paratyphi A strain ATCC 11511, S. Schwarzengrund strain CVM19633, and S. Javiana strain CFSAN0001992. Asterisks represent conserved amino acid residues. Amino acids highlighted in red represent amino acid changes present in TtsA from S. Montevideo compared to all other sequences of TtsA from all other serotypes. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. Javiana carries a pltB homolog, artB, which may substitute for PltB in vitro.
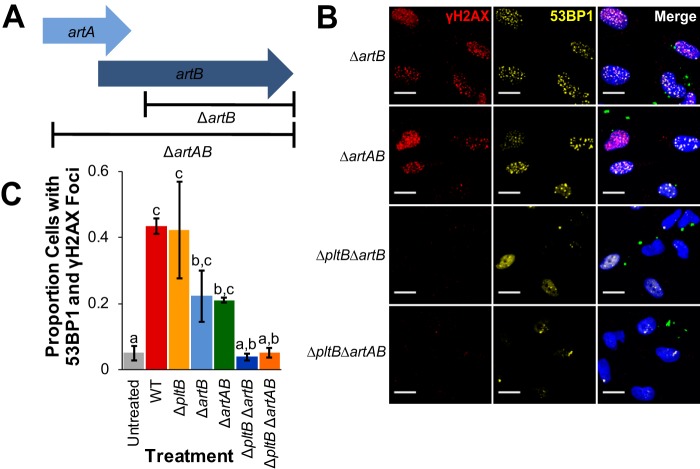
The S-CDT-negative (5, 8) S. enterica subsp. enterica serotype Typhimurium carries genes that encode an ADP-ribosylating toxin (called ArtAB) (22); these genes (artA and artB) are homologous to S-CDT subunits PltA and PltB (4–6). The ArtAB toxin is encoded by artA (encoding an ADP-ribosyltransferase) and artB (encoding the binding subunit) (22). Because deletion of pltB, encoding the binding subunit of S-CDT, did not have a significant effect on S-CDT-induced intoxication of infected HIEC-6 cells, we hypothesized that ArtB, which has homology to PltB (23), could potentially substitute for PltB in vitro. S. Javiana carries a truncated artA gene that overlaps with the full-length artB gene (Fig. 3A). To determine the importance of artA and artB in S-CDT intoxication, we constructed strains with deletions of (i) the entire artAB operon or (ii) just artB (Fig. 3A). HIEC-6 cells infected with ΔartAB or ΔartB strains had an average of 21% and 22% of cells with at least four 53BP1 foci that colocalized with γH2AX foci (Fig. 3B and C), respectively, which did not differ from HIEC-6 cells infected with either the wild-type or ΔpltB strain (P > 0.6 for all pairwise comparisons of wild-type, ΔpltB, ΔartB, and ΔartAB strains). However, infection with strains harboring deletions of both pltB and either artB or artAB abolished S. Javiana’s ability to induce DNA damage response foci in HIEC-6 cells (Fig. 3B and C). Compared to HIEC-6 cells infected with the ΔpltB strain, which had an average of 42% of cells with DNA damage response foci, infection with the ΔpltB ΔartB strain resulted in an average of 4% of cells with DNA damage response foci (P = 0.008; Fig. 3). Similarly, infection with the ΔpltB ΔartAB strain resulted in a significantly lower level of HIEC-6 cells with evidence of S-CDT intoxication (5% of cells with DNA damage response foci, P = 0.0198 compared to the ΔpltB strain).
FIG 3.
The presence of either artB or pltB is essential for S-CDT-mediated intoxication. Normal human intestinal epithelial HIEC-6 cells were infected with S. Javiana strains harboring either single gene deletions in artB or artAB or double deletions in pltB and either artB or artAB. At 48 hpi, cells were stained for the DNA damage response foci γH2AX and 53BP1. (A) Schematic of the gene deletions for ΔartB and ΔartAB strains. (B) Representative immunofluorescence images of HIEC-6 cells infected with S. Javiana deletion strains. S. Javiana cells (green) and HIEC-6 cell nuclei (blue) are shown. (C) Quantification of the proportions of cells with colocalized foci of at least four 53BP1 foci and γH2AX foci. Treatments that do not share letters are significantly different (P < 0.05). Results are from three independent experiments. P values were adjusted using Tukey’s HSD test to correct for multiple-comparison testing. Error bars represent standard errors of the means.
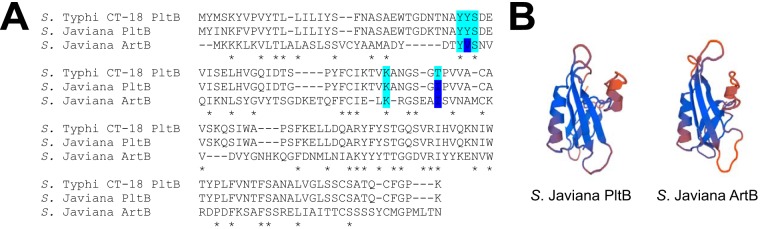
To further explore whether ArtB could substitute for PltB as the binding subunit, we performed in silico comparisons of ArtB and PltB using an amino acid sequence alignment of ArtB with PltB and mapped the five amino acid residues that are predicted to interact with host cell receptors for toxin binding (24). Overall, three out of the five amino acid residues (Tyr33, Ser35, and Lys59), which have been predicted to make contact with host cell Neu5Ac-terminated glycans in S. Typhi (24), are conserved in ArtB from S. Javiana (Fig. 4A). While PltB from both S. Javiana and S. Typhi had Tyr34, S. Javiana ArtB had Val34. Interestingly, the Thr65 residue in S. Typhi PltB was mutated in both PltB and ArtB from S. Javiana; both ArtB and PltB contained isoleucine residues at this position. Finally, the predicted three-dimensional (3D) structures of the PltB and ArtB subunits from S. Javiana were structurally similar (Fig. 4B).
FIG 4.
Key amino acid residues necessary for PltB binding to sugar moieties are also present in ArtB. (A) Alignments of translated amino acid sequences of PltB and ArtB from S. Javiana strain CFSAN0001992 and PltB from S. Typhi strain CT-18. Key residues that are conserved are shown on light blue background. Residues that differ between S. Typhi PltB and S. Javiana ArtB or PltB (24) are shown on dark blue background. Amino acid residues conserved in all three sequences (59) are indicated by an asterisk below the sequence alignment. (B) Predicted 3D structure of PltB and ArtB based on translated amino acid sequences of pltB and artB extracted from S. Javiana strain CFSAN0001992, generated using SWISS-MODEL online software (60).
artB is highly conserved among S-CDT-positive NTS serotypes.
To determine whether artB was conserved among S-CDT-positive serotypes, we aligned the predicted amino acid sequence of ArtB from S. Typhimurium DT104 (where it was first reported [22]) with those from other nontyphoidal and typhoidal S-CDT-positive serotypes. Predicted amino acid sequences of ArtB from S-CDT-positive serotypes clustered into three groups (Fig. S2). ArtB from nontyphoidal Salmonella serotypes Minnesota, Javiana, Rubislaw, Montevideo, and Schwarzengrund were highly similar, with just 2 out of 141 amino acids different (98.6% conserved among these serotypes). Similarly, predicted amino acid sequences for ArtB from S. Typhi CT-18 and S. Paratyphi ATCC 11511 strain had just one amino acid difference (99.3% identical among these serotypes). The predicted ArtB amino acid sequence from S. Typhimurium DT104 was distinct from the other sequences included in the alignment (73.2% identical to ArtB from S. Typhi CT-18).
Alignment of ArtB from typhoidal and nontyphoidal serotypes. (A) Phylogenetic tree of predicted ArtB amino acid sequences constructed using the James-Thorton-Thompson (JTT) model with 1,000 bootstrap repetitions in RAxML and rooted by midpoint. Bar represents 0.05 substitutions per amino acid residue. (B) Alignment of ArtB amino acid sequences. Asterisks represent amino acid residues that are conserved in all sequences in the alignment; amino acids highlighted in red represent amino acid substitutions compared to S. Typhimurium DT104 ArtB. Strains included in analyses follow: S. Typhimurium DT104, S. Javiana strain CFSAN0001992, S. Minnesota strain ATCC 49284, S. Montevideo strain USDA-ARS-USMARC-1921, S. Paratyphi A strain ATCC 11511, S. Rubislaw strain ATCC 10717, S. Schwarzengrund strain CVM19633, and S. Typhi strain CT18. Download FIG S2, EPS file, 1.5 MB (1.6MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S-CDT-mediated activation of a DNA damage response occurs primarily in the S and G2/M phases of the cell cycle in epithelial cells infected with wild-type S. Javiana.
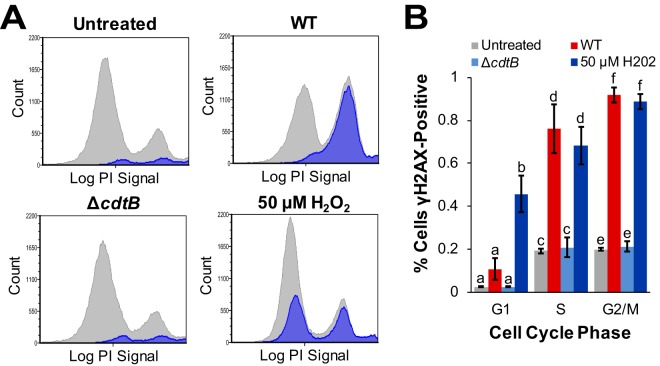
It is well established that the phase of the cell cycle in which DNA damage occurs will determine the DNA repair pathway that will be activated (25). Therefore, we infected HIEC-6 cells with wild-type and ΔcdtB strains of S. Javiana and used γH2AX staining to determine at which phase of the cell cycle S-CDT induced activation of a DNA damage response. Treatment with 50 µM H2O2, included as a positive control, consistently induced γH2AX activation (Fig. 5) throughout all phases of the cell cycle (46% to 89% cells γH2AX positive). HIEC-6 cells infected with wild-type S. Javiana had significantly higher proportions of γH2AX-positive cells only in the S phase (76% γH2AX-positive; P = 0.0175 compared to untreated controls) and G2/M phase (92% γH2AX-positive; P = 0.0044 compared to untreated controls) of the cell cycle. In the G1 phase, only 11% of cells infected with wild-type S. Javiana were γH2AX-positive, which was not significantly different from uninfected HIEC-6 cells (P = 0.1243) (Fig. 5). For a control, we also infected HIEC-6 cells with S. Javiana ΔcdtB strain, which did not result in higher mean proportions of γH2AX-positive cells compared to uninfected control cells; this result was consistent for all of the cell cycle phases (P > 0.999 for all comparisons with untreated cell populations).
FIG 5.
S-CDT-mediated DNA damage primarily occurs in the S and G2/M phases of the cell cycle of HIEC-6 human intestinal epithelial cells. HIEC-6 cells infected with S. Javiana wild-type (WT) and ΔcdtB strains for 48 h were stained to detect γH2AX and DNA content to determine the specific phases of the cell cycle during which S-CDT-mediated DNA damage occurred. (A) Representative histograms of DNA cell cycle analysis of HIEC-6 cells (gray) with γH2AX-positive cells overlaid (blue). (B) Quantification of γH2AX-positive HIEC-6 cells by cell cycle phase. Results are from two independent experiments. Treatments that do not share letters represent significantly different mean proportions (P < 0.05) of cells positive for a given cell cycle phase (e.g., proportions of γH2AX-positive cells in G1 were compared for the four treatment groups). P values were corrected for multiple comparisons using Tukey’s HSD test. Error bars represent standard errors of the means.
Infection with wild-type S. Javiana does not induce apoptosis or cell death.
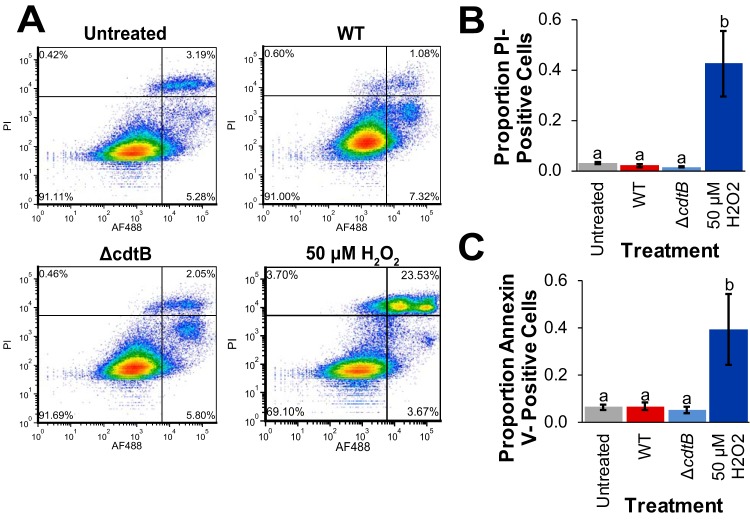
CDTs produced by several Gram-negative bacterial species have previously been shown to induce apoptosis in a variety of cell types (26–31). To determine whether infection with wild-type S. Javiana resulted in cell death (through apoptotic and other cell death pathways), we stained HIEC-6 cells infected with wild-type and ΔcdtB S. Javiana strains with annexin V to detect apoptotic cells and with propidium iodide (PI) to detect dead cells, regardless of the pathway leading to cell death (Fig. 6A). After 48 h, HIEC-6 cells infected with wild-type S. Javiana had an average of 7% cells positive for annexin V staining, which was not significantly different from the value for untreated control cells (10% annexin V-positive cells; Fig. 6B) but was slightly higher than the value for cells infected with the S. Javiana ΔcdtB strain (5% cells positive for annexin V). There was no significant difference in the mean proportion of annexin V-positive cells among untreated cells or cells infected with the wild-type or ΔcdtB strain (P > 0.6 for all comparisons). Because other, nonapoptotic cell death pathways exist (32), we also quantified the number of PI-positive cells (used as a live/dead stain). Cells treated with 50 µM H2O2, included as a positive control (Fig. 6A to C), had an average of 43% PI-positive cells (P = 0.0001 compared to untreated controls). The average proportion of PI-positive cells infected with the wild-type strain or the ΔcdtB strain (both had an average proportion of 2% PI-positive cells) did not differ significantly from uninfected controls (3%; P > 0.3 for all comparisons).
FIG 6.
Infection of HIEC-6 human intestinal epithelial cells with S. Javiana does not induce apoptosis or cell death regardless of S-CDT status. HIEC-6 cells were infected with wild-type or ΔcdtB S. Javiana strains for 48 h or treated with 50 µM H2O2 for 8 h (for a positive control). Cells were stained with annexin V conjugated to Alexa Fluor 488 (AF488); PI was used as a live/dead stain. (A) Representative images of density plots of annexin V and PI staining of HIEC-6 cells infected with S. Javiana, treated with H2O2, or untreated. (B and C) Quantification of the proportions of PI-positive cells (B) and annexin V-positive cells (C) shown in panel A. Treatments that do not share letters have significantly different (P < 0.05) proportions of cells positive for either annexin V or PI staining. Results in panels B and C are from four independent experiments. Error bars represent standard errors of the means.
Infection with wild-type S. Javiana results in higher bacterial counts in the liver, but not in the spleen, ileum, or cecum.
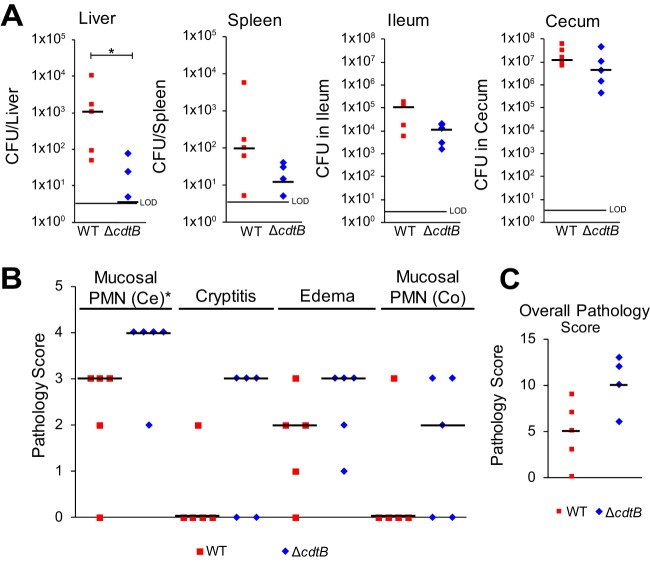
Because S-CDT is predicted to play an important role in the pathogenesis of typhoid fever (4), we infected C56BL/6 mice by pipetting 1 × 109 CFU of wild-type and ΔcdtB S. Javiana strains into the oral cavity and then assessed the severity of infection 48 h postinfection. While mice infected with wild-type S. Javiana had significantly higher bacterial counts in the liver (P = 0.01597; median, 1,050 CFU/liver) than mice infected with ΔcdtB S. Javiana (median, 5 CFU/liver), the levels of Salmonella recovered from the spleen, ileum, and cecum did not differ significantly in mice infected with wild-type and ΔcdtB strains (P > 0.05 for pairwise comparisons of CFU recovered from the ileum and cecum; Fig. 7A).
FIG 7.
Infection of C56BL/6 mice with WT S. Javiana results in a higher bacterial load in the liver, but not in the spleen, ileum, or cecum, compared to infection with a ΔcdtB strain. Eight-week-old female C56BL/6 mice were orally inoculated with 1 × 109 CFU S. Javiana wild-type or ΔcdtB strain (five mice in each group). At 48 hpi, the mice were euthanized, and tissues were harvested for bacteriologic and histological examination. (A) S. Javiana cells were enumerated from the liver, spleen, ileum, and cecum. The average bacterial load was calculated for each tissue. The limit of detection (LOD) for the average bacterial load for all tissues was 5 CFU. Differences in S. Javiana recovered from each tissue were compared using the nonparametric Kruskal-Wallis test. (B and C) Sections of the distal ileum, cecum, and proximal colon were fixed in buffered formaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. The levels of polymorphonuclear leukocytes (PMN) in the cecum (Ce) and proximal colon (Co) were scored on a scale of 0 to 4 (0 representing no PMNs and 4 representing large numbers of PMNs); cryptitis was assessed using the same scale. The degree of edema was scored from 0 (no edema) to 3 (severe edema). (C) The pathology score for typhlocolitis represents the sum of the scores (maximum score is 15) in panel B. Statistical differences in the counts of mice in each category of pathology score were assessed using the nonparametric Kruskal-Wallis test. Each symbol represents the value for an individual mouse. Black bars represent the median bacterial load (A) or median pathology scores (B and C) for mice infected with each S. Javiana strain. Values that are significantly different (P < 0.05) are indicated by a bar and asterisk. Only statistical associations with P < 0.05 are shown.
Histological examination of the distal ilea, ceca, and proximal colons obtained from mice infected with the ΔcdtB strain revealed significantly higher cecal inflammation scores (P = 0.04863) compared to the scores for mice infected with wild-type S. Javiana (Fig. 7B). Mice infected with the ΔcdtB strain also had higher pathology scores for colonic cryptitis and mucosal polymorphonuclear leukocyte (PMN) infiltrate, and cecal submucosal edema, but these scores were not statistically significant (P > 0.05 for all comparisons between mice infected with wild-type and ΔcdtB strains). Overall, the typhlocolitis severity scores of mice infected with the ΔcdtB strain were higher but not significantly different (P = 0.07941) from the scores of mice infected with the wild-type S. Javiana (Fig. 7C).
DISCUSSION
Although infections with S-CDT-positive NTS serotypes greatly outnumber infections with S. Typhi in the United States (14), molecular characterizations have primarily focused on S-CDT (typhoid toxin) produced by S. Typhi. In line with other genetic and phenotypic differences between nontyphoidal and typhoidal serotypes, and despite the fact that the serotypes carry genes encoding genetically similar variants of S-CDT, our data suggest that there are several differences in genes required for expression of biologically active S-CDT produced by S. Javiana, compared to what has been reported previously for the typhoid toxin produced by S. Typhi (3, 8, 18). We also show that S-CDT is an important virulence factor in S. Javiana, both in vitro and in vivo, as infection with wild-type S. Javiana induced a DNA damage response that resulted in an altered cell cycle progression of human intestinal epithelial cells in vitro and higher levels of S. Javiana translocation to the livers of infected mice.
Inactivation of genes in the S-CDT islet for S. Javiana results in different phenotypes compared to S-CDT produced by S. Typhi.
We confirmed that cdtB and pltA are essential for S-CDT-mediated intoxication following infection with wild-type S. Javiana, which has been previously established for the S-CDT produced by S. Typhi (3). Interestingly, deletion of pltB and ttsA in S. Javiana did not abolish toxin activity (Fig. 2), despite the fact that these genes have previously been reported to be essential for toxin activity in S. Typhi (3, 18).
While deletion of pltB in S. Typhi is sufficient to abolish S-CDT-induced accumulation of cells in the G2/M phase (3), the S. Javiana ΔpltB strains, used in the present study, retained S-CDT activity in vitro, suggesting that PltB is not required for S-CDT expression by S. Javiana, consistent with a previous study by Mezal et al. (9). However, S. Javiana also contains a gene that encodes a pltB homolog (artB). Because deletion of both pltB and artB was required to abolish S. Javiana’s ability to activate a DNA damage response in HIEC-6 cells to a background level, it is likely that for S. Javiana, both ArtB and PltB may play a role in S-CDT-mediated DNA damage response induction in vitro. The conservation of key amino acid residues in ArtB, which have been shown to bind to host glycoproteins for PltB (4, 24), suggests that ArtB may possibly substitute for PltB as the binding subunit or may exist in the form of a heteropentameric binding subunit composed of both ArtB and PltB. Consistent with this hypothesis, recent data have confirmed that homopentamers of ArtB complexing with PltA and CdtB form an active S-CDT in S. Typhi (10). Furthermore, Gao et al. also showed that while PltB preferentially binds to Neu5Ac-terminated glycans (4, 10), ArtB binds both Neu5Ac, found on human cells, and Neu5Gc-terminated glycans, found on most other nonhuman mammalian cells; hence, inclusion of ArtB in the S-CdtB holotoxin could expand the range of host cells susceptible to S-CDT-mediated DNA damage response (10). Future investigations examining the presence of ArtB and PltB in the S-CDT holotoxin (including whether the holotoxin can represent a PltB-ArtB heteropentamer) will be important to understand the potential for ArtB to expand the range of hosts and cell types that can be targeted by S-CDT. Interestingly, S. Typhi strains CT-18 and Ty2 both carry the artB gene, and alignments of predicted amino acid sequences of ArtB for nontyphoidal and typhoidal serotypes show that ArtB is highly conserved (see Fig. S2 in the supplemental material) (6). Herdman et al. further confirmed that purified ArtB cloned from S. Typhi strain Ty2 binds to and induces vacuolization in a variety of host cell types, including Vero, CHO, and U937 cell lines (33).
Deletion of accessory genes STY1887 and ttsA did not alter S-CDT activity in vitro in our study. In S. Typhi, ttsA was characterized as an N-acetyl-β-d-muramidase and was shown to be essential for exporting the toxin out of the host cell, but deletion of STY1887 did not have any effect (18). Although a different cell type was used in that study (Henle cells versus HIEC-6 cells in our study), it is unlikely that this alone accounted for the observed differences, as both epithelial cell lines are of human origin. In their study, Hodak and Galan proposed that S-CDT release requires activity of the N-acetyl-β-d-muramidase (TtsA) and an unidentified holin, although attempts to show in vitro that purified N-acetyl-β-d-muramidase (TtsA) has muramidase activity were unsuccessful (18). Together, these results suggest that the S-CDT produced by nontyphoidal serotypes, such as S. Javiana, utilizes different mechanisms for S-CDT production, binding, and trafficking, compared to the S-CDT produced by S. Typhi. Alternatively, there could be other genes that compensate for the loss of ttsA function, as was observed for pltB and artB.
Infection with wild-type S. Javiana does not induce apoptosis but does induce a DNA damage response that primarily occurs in the S and G2/M phases.
While the mechanism of S-CDT action and outcome of infection following exposure to S-CDT is poorly understood, our data suggest that DNA damage sustained following exposure to S-CDT occurs primarily in the S and G2/M phases of the cell cycle. Importantly, the cell cycle phase in which DNA damage is sustained determines which DNA repair pathway is activated (34). For example, in the G1 phase, homologous recombination is inhibited, and the error-prone nonhomologous end joining (NHEJ) DNA repair pathway is used to repair DNA double-strand breaks. In contrast, double-strand breaks sustained in the S or G2/M phase of the cell cycle are primarily repaired by homologous recombination (34, 35). Our data suggest that DNA damage resulting from infection with S-CDT-producing S. Javiana occurs during the S and G2/M phases of the cell cycle. This is also reflected by the characteristic accumulation of cells in the G2/M phase, which is the hallmark of CDT-induced DNA damage (36–38). Using a slightly different approach, Fedor et al. showed that Escherichia coli CDT induces DNA single-strand breaks that are converted to double-strand breaks in S phase (39). The authors also showed that homologous recombination repair proteins are associated with the double-strand breaks, suggesting that the DNA damage induced by CDT is likely repaired by homologous recombination (39). Although E. coli CDT is structurally different from S-CDT, conservation of CdtB, the active subunit, suggests that both toxins induce DNA damage via a similar mechanism (7, 37). Therefore, we hypothesize that the DNA damage induced by S-CDT from S. Javiana may be repaired in a manner similar to that of the DNA damage sustained following exposure to E. coli CDT, using the high-fidelity homologous recombination pathway rather than the error-prone NHEJ pathway. This would suggest a lower risk of introducing mutations after infection with S-CDT-positive NTS serotypes than would be expected if S-CDT-induced DNA damage were repaired by a more error-prone pathway, such as NHEJ.
S-CDT contributes to S. Javiana systemic infection in C56BL/6 mice.
In contrast to what has been reported previously for S-CDT and other CDTs (19, 28, 40), we found normal human intestinal epithelial cells exposed to S-CDT did not undergo apoptotic cell death, suggesting that following exposure to S-CDT, epithelial cells either initiate a DNA damage response or enter senescence. While apoptosis is an anti-inflammatory cell death pathway (41), senescence is associated with the release of proinflammatory cytokines, known as the senescence-associated secretory phenotype (SASP), which serves as an innate signaling mechanism to recruit inflammatory cells to the site of senescent cells (42, 43). Consistent with our observations, Blazkova et al. showed that human epithelial cells exposed to Haemophilus ducreyi CDT enter a senescent state (44). Future work characterizing whether or not cells become senescent following S-CDT-mediated DNA damage will be necessary to understanding the potential contribution of SASP-associated inflammation in infections with S-CDT-positive NTS.
In mice, both the administration of purified S-CDT and infection with S. Typhimurium expressing S. Typhi S-CDT have been associated with suppression of the immune system (4, 13), which could enable S-CDT-producing NTS to evade host defense mechanisms (45). In our study, we found significantly higher levels of S. Javiana in the livers of mice infected with wild-type S. Javiana than in mice infected with the ΔcdtB strain; although the same trend was also observed for the spleen, differences in the levels of wild-type and ΔcdtB S. Javiana isolated from the spleen were not significant (P = 0.1127). Our data suggest that wild-type S. Javiana could use S-CDT to partially suppress the immune response, thereby allowing the Salmonella cells to spread to extraintestinal sites. In agreement with this, Song et al. (4) showed that mice injected with wild-type typhoid toxin (S-CDT) displayed a decrease in circulating monocytes, neutrophils, and lymphocytes five days after injection; mice injected with a catalytically inactive toxin had levels of white blood cells that were indistinguishable from those of untreated control mice (4). This suggests that S-CDT partially suppresses the innate immune response in vivo, which could enable S-CDT-positive Salmonella to evade immune cell killing and spread to extraintestinal sites. Furthermore, H. ducreyi CDT has been shown to induce apoptosis in human monocytic THP-1 cells (26). Although Salmonella resides primarily within Kupffer cells in the liver (46), it is possible that intoxication of these cells (i.e., exposure to DNA-damaging S-CDT) allows Salmonella to escape, and multiply outside Kupffer cells, in nearby hepatocytes. Importantly, mice infected with the ΔcdtB strain had higher cecal pathology scores than mice infected with the wild-type strain, despite similar S. Javiana colonization levels in the cecum. This suggests that S-CDT might alter the host innate inflammatory and immune responses to allow bacterial dissemination to extraintestinal sites. Although our study did not address the contribution of the adaptive immune response, Del Bel Belluz et al., using a chronic-infection mouse model, showed that S. Typhi S-CDT contributes to establishment of a persistent asymptomatic carrier status (13). We selected the streptomycin-treated mouse model to examine the effect of S-CDT on both gastroenteritis (47) and invasive disease. Streptomycin treatment has been shown to effectively alter the microbiome of mice (48), which enhances Salmonella’s ability to colonize the mouse intestine. Future studies using other mouse models, such as BALB/c mice, will be beneficial for confirming the results obtained in this study, as BALB/c mice are naturally susceptible to Salmonella infection (47). Furthermore, histological examination of liver and spleen samples will further expand our understanding of the mechanism by which S-CDT affects these tissues. While further studies will be helpful in understanding the role of S-CDT during an infection, our results suggest that S-CDT may play a key role in the acute phase of host infection with S. Javiana and contribute to systemic bacterial spread.
Despite an increasingly recognized role for CDTs in acute (4) and chronic (49, 50) disease, the role that S-CDT plays both in nontyphoidal and typhoidal salmonellosis is largely unknown. While our data suggest that a DNA damage response is induced when homologous recombination DNA repair pathways are active, future investigations determining the fate of the S-CDT-intoxicated cells in vivo remains an important gap in our current understanding of S. Javiana pathogenesis. Furthermore, understanding the regulation and in vivo targets of S-CDT will provide critical information concerning S-CDT contribution to the severity and outcome of salmonellosis. Whether S-CDT exposure predisposes individuals for chronic carriage or other conditions associated with the DNA-damaging effects of S-CDT, including genomic instability and certain cancers, is an area of active future investigation.
MATERIALS AND METHODS
Bacterial strains, epithelial cell line, and culture conditions.
Salmonella strains (Table 1) were preserved at −80°C in 15% (vol/vol) glycerol. Salmonella were grown on brain heart infusion (BHI) (Becton Dickinson, Sparks, MD) agar plates, which were incubated at 37°C. For in vitro infections, single colonies of Salmonella isolates grown on BHI agar plates were inoculated into 5-ml aliquots of LB (pH 8; 0.3 M NaCl), followed by incubation under static conditions at 37°C for 12 to 14 h. These cultures were subsequently subcultured 1:100 into fresh aliquots of LB (pH 8; 0.3 M NaCl), followed by incubation at 37°C under static conditions, until mid-log phase (optical density at 600 nm [OD600] of 0.4 to 0.5).
TABLE 1.
S. Javiana strains used in this study
| Strain | Relevant genotype |
|---|---|
| FSL S5-0395 | Wild-type |
| FSL M8-0532 | S5-0395 ΔpltA |
| FSL M8-0533 | S5-0395 ΔpltB |
| FSL M8-0540 | S5-0395 ΔcdtB |
| FSL M8-0577 | S5-0395 ΔttsA |
| FSL M8-0578 | S5-0395 ΔSTY1887 |
| FSL M8-0582 | S5-0395 ΔartB |
| FSL M8-0583 | S5-0395 ΔartAB |
| FSL M8-0585 | S5-0395 ΔpltB ΔartB::Kanr |
| FSL M8-0586 | S5-0395 ΔpltB ΔartAB::Kanr |
| FSL M8-0590 | S5-0395 ΔcdtB ΔphoN::Kanr |
| FSL M8-0591 | S5-0395 ΔphoN::Kanr |
Human intestinal epithelial cells (HIEC-6 cells; ATCC), derived from human fetal ileum (51), were grown in Opti-MEM medium supplemented with recombinant epidermal growth factor (10 ng/ml; Gibco-Invitrogen, Carlsbad, CA) and 10% (vol/vol) fetal bovine serum (FBS) (Gibco-Invitrogen) and were incubated at 37°C with 5% CO2. HIEC-6 cell supernatants were routinely tested for Mycoplasma infection using the VenorGEM Mycoplasma detection kit (Sigma-Aldrich, St. Louis, MO). For Salmonella enterica serotype Javiana infections, HIEC-6 cells were grown in 6-well or 24-well plates (Corning, Corning, NY). Cells were seeded into 6-well (2 × 105 cells per well) or 24-well (1 × 105 cells) plates 48 h (+4) h before infection.
Strain construction.
Bacterial strains are shown in Table 1. The λ Red recombinase system was used to create all in-frame deletions (52). Briefly, S. Javiana FSL S5-0395 (wild type) containing the pKD46 plasmid (described in reference 52) was grown with shaking at 30°C in LB broth containing 100 μg/ml ampicillin and 0.01% l-arabinose (Sigma) to an OD600 of 0.5 to 0.7. Cells were transformed with kanamycin resistance cassettes containing flanking regions homologous to chromosomal sites of the gene targeted for deletion, using electroporation followed by a 2-h recovery in SOC medium (New England BioLabs [NEB], Ipswich, MA) at 37°C. PCR amplification was performed with the high-fidelity polymerase Q5 (NEB), used to amplify kanamycin cassettes from pKD46 with the primers listed in Table S1 in the supplemental material. Subsequently, cells were plated on LB supplemented with 50 μg/ml kanamycin and were incubated at 37°C for 18 to 24 h. Successful chromosomal integration of the kanamycin resistance cassette was confirmed by PCR. For removal of the kanamycin resistance cassette, cells were grown to mid-log phase (OD600, ~0.5 to 0.7) and were electroporated with pCP20, followed by incubation at 30°C for 2 h; colonies were selected by plating on LB agar, supplemented with 100 μg/ml ampicillin, for 20 to 24 h. For strains requiring multiple deletions, crosses with phage P22 were conducted (52). In-frame deletions were confirmed by Sanger sequencing. To generate the streptomycin-resistant strains for the mouse infection experiments, mid-log-phase wild-type S. Javiana was spread plated onto LB agar plates supplemented with 100 μg/ml streptomycin and incubated at 37°C. A single streptomycin-resistant colony was selected and was used to create the ΔphoN::Kanr strain, so that both strains used for mouse infections (i.e., strains M8-0590 and M8-0591 [Table 1]) were generated from the same parental strain. The ΔcdtB ΔphoN::Kanr strain was generated using transduction with P22 lysates from a ΔcdtB strain. Whole-genome sequencing was performed for both FSL M8-0590 and FSL M8-0591 strains and confirmed the integrity of the mutations generated, and the lack of single nucleotide polymorphisms (SNP); FSL M8-0591 and FSL M8-0590 showed a single SNP difference, located in a noncoding region.
Primers used in this study. Download TABLE S1, DOCX file, 0.02 MB (16.9KB, docx) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Salmonella infection of HIEC-6 cells.
Treatments (i.e., strain used, negative- or positive-control treatments) were randomly assigned to HIEC-6 cells seeded in 6-well or 24-well plates. HIEC-6 cells were infected with approximately 2 × 106 CFU or 4 × 106 CFU Salmonella for HIEC-6 cells seeded in 24-well and 6-well plates, respectively. After incubation of the infected cells for 1 h at 37°C (with 5% CO2), HIEC-6 cells were washed three times with phosphate-buffered saline (PBS), followed by incubation with medium supplemented with 100 μg/ml gentamicin (Gibco) for 1 h at 37°C to kill extracellular bacteria. Subsequently, HIEC-6 cells were washed an additional three times with PBS and were then maintained in medium containing 10 μg/ml gentamicin (Gibco) to prevent recurrent infection and bacterial outgrowth during incubation.
Flow cytometry for DNA content.
Cell cycle analyses were performed on populations of HIEC-6 cells that were infected with Salmonella strains as described above. Briefly, HIEC-6 cells were washed once with PBS and harvested using 0.25% trypsin-EDTA (Gibco). Cells were fixed in ice-cold 70% ethanol and stored at −20°C. Ethanol-fixed cells were permeabilized with PBS containing 0.1% Tween 20 (PBS-T) and bovine serum albumin (BSA) (1 g/100 ml) (Sigma-Aldrich) at room temperature for 10 min. Cells were subsequently stained (10 min at room temperature) with a solution containing propidium iodide (PI) (Thermo Fisher Scientific, Waltham, MA) at a final concentration of 50 μg/ml and RNase A (Sigma-Aldrich) at a final concentration of 100 μg/ml. Stained cells were held at 4°C for no longer than 4 h, prior to DNA content analysis using the BD FACSARIA sorter. Cells were gated to exclude doublets and multiplets (Fig. S3), as described previously (53).
Gating strategies used in this study. Download FIG S3, EPS file, 1.6 MB (1.7MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Immunofluorescence staining.
Immunofluorescence staining for γH2AX and 53BP1 foci was performed at 48 (+2) h postinfection (hpi) as described previously (8). Briefly, HIEC-6 cells grown on 12-mm coverslips (Thermo Fisher Scientific) were washed with PBS and were then fixed with 4% formaldehyde in PBS at room temperature for 10 to 15 min. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min. Permeabilized cells were blocked with 10% (vol/vol) normal (healthy) donkey serum (Sigma) in PBS containing 0.1% Tween 20 (PBS-T) (J. T. Baker, Phillipsburg, NJ) for 1 h at room temperature. Incubation with primary and secondary antibodies was performed for 1 h at room temperature using the following dilution factors in PBS-T: polyclonal goat anti-Salmonella antibody (KPL antibodies; KPL, Gaithersburg, MA; 1:500), mouse anti-γH2AX (EMD Millipore, Billerica, MA; 1:500), rabbit anti-53BP1 (Novus Biologicals, Littleton, CO; 1:500). Incubation with secondary antibodies diluted 1:200, except where noted, in PBS-T was performed for 1 h at room temperature: donkey anti-goat conjugated to Alexa Fluor 488, donkey anti-rabbit conjugated to Alexa Fluor 555 (diluted 1:500), and donkey anti-mouse conjugated to Alexa Fluor 647 (all Thermo Fisher Scientific). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) at a final concentration of 1 μg/ml for 5 min at room temperature. Slides were mounted onto microscope slides with glycergel (Dako, Carpinteria, CA) and imaged using a Zeiss 710 confocal microscope. Images were processed with FIJI software, and cells were counted using the cell counter plug-in (54). At least 50 nuclei were analyzed per slide to identify cells that had at least four 53BP1-positive foci that colocalized with γH2AX foci. The observer was blind to treatments while collecting and analyzing images.
Apoptosis staining.
HIEC-6 cells were infected with Salmonella as described above. Hydrogen peroxide was added as a positive control (final concentration of approximately 50 μM) to uninfected control cells approximately 8 h prior to staining. Detached cells (potentially dead cells) were included in analyses by collecting supernatants of treated HIEC-6 cells via centrifugation at 3,000 rpm for 5 min. The adherent cells were trypsinized with 0.25% trypsin-EDTA (Gibco), washed with PBS (4°C), and subsequently combined with cells collected from supernatants via centrifugation at 3,000 rpm for 5 min. Cells were stained for 15 min at room temperature with annexin V conjugated to Alexa Fluor 488 (Invitrogen) and PI. Cells were analyzed within 2 h of staining using the BD FACSARIA sorter. Refer to Fig. S3 for a description of the gating strategy used.
γH2AX flow cytometry.
At 48 (+2) h postinfection, Salmonella-infected HIEC-6 cells and controls were washed once with PBS and subsequently harvested with 0.25% trypsin-EDTA. The positive-control cells were incubated with H2O2 at a final concentration of 200 μM for 1 h at 37°C. Cells were fixed with 4% formaldehyde (Thermo Fisher Scientific) at room temperature for 5 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Cells were then blocked with 3% BSA (Sigma-Aldrich) in PBS for 30 min. Antibody staining was performed at room temperature for 1 h with mouse anti-γH2AX antibody (Millipore) and for 30 min with donkey anti-mouse conjugated to Alexa Fluor 488 (Thermo Fisher Scientific); both antibodies were used at a 1:200 dilution. Labeled cells were subsequently stained for cell cycle analysis by incubation with PI for 10 min at room temperature (using 300 µl of the PI staining solution described above). Cells were analyzed using the BD FACSAria sorter within 2 h of staining. The gating strategy used is shown in Fig. S3.
Mouse infection experiments.
Ten female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All experimental procedures were approved by the Cornell University Institutional Use and Care of Animals Committee. Mice were administered 20 mg streptomycin sulfate by pipetting 50 μl of a 400-mg/ml stock solution into the oral cavity 24 h prior to infection with Salmonella. Strains FSL M8-0590 and FSL M8-0591 (Table 1) were inoculated into 25 ml of LB broth and grown with aeration (200 rpm) at 37°C for 18 h. These bacterial cultures were pelleted, resuspended in PBS, and concentrated to achieve 1 × 1010 CFU in 500 μl of PBS. Mice were orally inoculated by pipetting 50 µl of this inoculum (containing 1 × 109 CFU Salmonella; five mice in each group) into the oral cavity of each mouse. Mice were euthanized 48 h postinfection, and the liver, spleen, ileum, and cecum were aseptically removed at necropsy. The liver and spleen were homogenized and resuspended in 1 ml of PBS. The distal ileum and cecum were transferred to a 50-ml Falcon tube containing 5 ml of PBS followed by vortex agitation for 5 min (i.e., wash 1). A second wash was performed by transferring the organs to a new 50-ml Falcon tube containing 5 ml of PBS, and the tubes were vortexed for an additional 5 min (i.e., wash 2). The organs were removed, and the two corresponding PBS wash solutions were combined. Homogenates were serially diluted and spread plated on LB supplemented with 50 µg/ml kanamycin. The plates were incubated for 20 to 24 h at 37°C. Homogenates were stored at 4°C and replated if the bacterial counts were below detectable levels.
Histological analysis.
The distal ileum (approximately 0.5 cm), cecum, proximal colon (approximately 1.5 cm), and associated mesenteric lymph nodes were harvested, fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 5-µm thickness, and stained with hematoxylin and eosin for histological assessment; liver and spleen samples were not collected for histological analysis. Sections of the cecum and proximal colon were scored by a board-certified veterinary pathologist, in a blind fashion, for the presence and distribution of polymorphonuclear leukocytes within (i) the lamina propria of the intestinal mucosa and (ii) inside intestinal crypts (cryptitis) using a modified version of a previously described (48) scale of 0 to 4 (0, none; 1, rare; 2, few scattered; 3, many groups; 4, large numbers). Additionally, the degree of edema within the submucosa of intestinal sections was scored on a scale of 0 to 3 (0, none; 1, mild; 2, moderate; 3, severe). Typhlocolitis severity was calculated as the sum of the scores for the categorical parameters of the cecum and proximal colon (maximum of 15).
Data analysis.
All raw data are available by request. For codes used in the statistical analyses, refer to Data Set S1. Statistical analyses were performed using R version 3.4.2. with packages lme4 1.1-14 (55), lmerTest (56) version 2.0-33, and lsmeans version 2.27-2 (57).
Codes used in the statistical analyses. Download Data Set S1, PDF file, 0.6 MB (671.7KB, pdf) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For comparing the proportions of (i) cells in a given cell cycle phase (Fig. 1B), (ii) cells with at least four 53BP1foci colocalized with γH2AX foci (Fig. 1D, 2C, and 3C), (iii) cells positive for γH2AX staining by cell cycle phase (Fig. 5B), and (iv) cells positive for annexin V (Fig. 6B) or PI (Fig. 6C) staining, proportions were first logit transformed, and linear mixed effect models were fit (see Data Set S1 for complete model details and output). Pairwise comparisons to determine differences in cellular outcomes dependent on the strain used for infection (e.g., wild-type versus ΔcdtB strain) were performed using the lsmeans package. P values were corrected for multiple comparisons using the false-discovery rate correction in the stats package (58).
For comparing bacterial loads recovered from the spleen and liver and the fecal contents of the ileum and cecum, the nonparametric Kruskal-Wallis test was used. Data that were below the detectable levels were plotted on graphs as being at the limit of detection (LOD) (e.g., 5 CFU in the liver and spleen) and were included as levels at the LOD in statistical analyses. Due to the low variability in the organ weights (see Data Set S1), we compared the overall bacterial load, rather than correcting by organ weight.
Statistical differences in pathology scores were assessed using the Kruskal-Wallis test to determine whether (i) cecal mucosal PMN, (ii) cecal cryptitis, (iii) cecal edema, and (iv) colonic mucosal PMN scores were significantly different between mice infected with wild-type and ΔcdtB strains. Comparison of differences in the mean total pathology scores between mice in the groups inoculated with wild-type and ΔcdtB strains was done using the Kruskal-Wallis test.
For comparing the rates of invasion/adherence between wild-type and ΔcdtB strains, a linear mixed-effect model was fit using logit transformed CFU/milliliter counts, and lsmeans was used to determine whether the mean CFU recovered were statistically different between strains.
ACKNOWLEDGMENTS
R.A.M. was supported by USDA NIFA award 2016-67011-24714. Partial funding was also provided by the U.S. Department of Agriculture Hatch grant NYC-143436.
Footnotes
Citation Miller RA, Betteken MI, Guo X, Altier C, Duhamel GE, Wiedmann M. 2018. The typhoid toxin produced by the nontyphoidal Salmonella enterica serotype Javiana is required for induction of a DNA damage response in vitro and systemic spread in vivo. mBio 9:e00467-18. https://doi.org/10.1128/mBio.00467-18.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spanò S, Ugalde JE, Galán JE. 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Song J, Gao X, Galán JE. 2013. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova E, Furtado MR, Wiedmann M. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425. doi: 10.1186/1471-2164-12-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Rivera LD, Bowen BM, den Bakker HC, Duhamel GE, Wiedmann M. 2015. Characterization of the cytolethal distending toxin (typhoid toxin) in non-typhoidal Salmonella serovars. Gut Pathog 7:19. doi: 10.1186/s13099-015-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller R, Wiedmann M. 2016. Dynamic duo—the Salmonella cytolethal distending toxin combines ADP-ribosyltransferase and nuclease activities in a novel form of the cytolethal distending toxin. Toxins 8:121. doi: 10.3390/toxins8050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller RA, Wiedmann M. 2016. The cytolethal distending toxin produced by nontyphoidal Salmonella serotypes Javiana, Montevideo, Oranienburg, and Mississippi induces DNA damage in a manner similar to that of serotype Typhi. mBio 7:e02109-16. doi: 10.1128/mBio.02109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezal EH, Bae D, Khan AA. 2014. Detection and functionality of the CdtB, PltA, and PltB from Salmonella enterica serovar Javiana. Pathog Dis 72:95–103. doi: 10.1111/2049-632X.12191. [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Deng L, Stack G, Yu H, Chen X, Naito-Matsui Y, Varki A, Galán JE. 2017. Evolution of host adaptation in the Salmonella typhoid toxin. Nat Microbiol 2:1592–1599. doi: 10.1038/s41564-017-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nešić D, Hsu Y, Stebbins CE. 2004. Assembly and function of a bacterial genotoxin. Nature 429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 12.Haghjoo E, Galán JE. 2004. Salmonella Typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, Wrande M, Candela M, Turroni S, Nastasi C, Consolandi C, Peano C, Tebaldi T, Viero G, Gorgoulis VG, Krejsgaard T, Rhen M, Frisan T. 2016. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog 12:e1005528. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boore AL, Hoekstra RM, Iwamoto M, Fields PI, Bishop RD, Swerdlow DL. 2015. Salmonella enterica infections in the United States and assessment of coefficients of variation: a novel approach to identify epidemiologic characteristics of individual serotypes, 1996-2011. PLoS One 10:e0145416. doi: 10.1371/journal.pone.0145416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 16.Atif SM, Winter SE, Winter MG, McSorley SJ, Bäumler AJ. 2014. Salmonella enterica serovar Typhi impairs CD4 T cell responses by reducing antigen availability. Infect Immun 82:2247–2254. doi: 10.1128/IAI.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter SE, Winter MG, Atluri V, Poon V, Romão EL, Tsolis RM, Bäumler AJ. 2015. The flagellar regulator TviA reduces pyroptosis by Salmonella enterica serovar Typhi. Infect Immun 83:1546–1555. doi: 10.1128/IAI.02803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodak H, Galán JE. 2013. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep 14:95–102. doi: 10.1038/embor.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams K, Gokulan K, Shelman D, Akiyama T, Khan A, Khare S. 2015. Cytotoxic mechanism of cytolethal distending toxin in nontyphoidal salmonella serovar (Salmonella Javiana) during macrophage infection. DNA Cell Biol 34:113–124. doi: 10.1089/dna.2014.2602. [DOI] [PubMed] [Google Scholar]
- 20.Dietlein F, Thelen L, Reinhardt HC. 2014. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet 30:326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Pageot LP, Perreault N, Basora N, Francoeur C, Magny P, Beaulieu JF. 2000. Human cell models to study small intestinal functions: recapitulation of the crypt-villus axis. Microsc Res Tech 49:394–406. doi:. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh M, Tanaka K, Nishimori K, Makino S-I, Kanno T, Ishihara R, Hatama S, Kitano R, Kishima M, Sameshima T, Akiba M, Nakazawa M, Yokomizo Y, Uchida I. 2005. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology 151:3089–3096. doi: 10.1099/mic.0.27933-0. [DOI] [PubMed] [Google Scholar]
- 23.Tamamura Y, Tanaka K, Uchida I. 2017. Characterization of pertussis-like toxin from Salmonella spp. that catalyzes ADP-ribosylation of G proteins. Sci Rep 7:2653. doi: 10.1038/s41598-017-02517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galán JE, Varki A. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You Z, Bailis JM. 2010. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol 20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wising C, Azem J, Zetterberg M, Svensson LA, Ahlman K, Lagergård T. 2005. Induction of apoptosis/necrosis in various human cell lineages by Haemophilus ducreyi cytolethal distending toxin. Toxicon 45:767–776. doi: 10.1016/j.toxicon.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Shenker BJ, Hoffmaster RH, Zekavat A, Yamaguchi N, Lally ET, Demuth DR. 2001. Induction of apoptosis in human T cells by Actinobacillus actinomycetemcomitans cytolethal distending toxin is a consequence of G2 arrest of the cell cycle. J Immunol 167:435–441. doi: 10.4049/jimmunol.167.1.435. [DOI] [PubMed] [Google Scholar]
- 28.Liyanage NP, Manthey KC, Dassanayake RP, Kuszynski CA, Oakley GG, Duhamel GE. 2010. Helicobacter hepaticus cytolethal distending toxin causes cell death in intestinal epithelial cells via mitochondrial apoptotic pathway. Helicobacter 15:98–107. doi: 10.1111/j.1523-5378.2010.00749.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato S, Nakashima K, Nagasawa T, Abiko Y, Furuichi Y. 2013. Involvement of Toll-like receptor 2 in apoptosis of Aggregatibacter actinomycetemcomitans-infected THP-1 cells. J Microbiol Immunol Infect 46:164–170. doi: 10.1016/j.jmii.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Cortes-Bratti X, Karlsson C, Lagergård T, Thelestam M, Frisan T. 2001. The Haemophilus ducreyi cytolethal distending toxin induces cell cycle arrest and apoptosis via the DNA damage checkpoint pathways. J Biol Chem 276:5296–5302. doi: 10.1074/jbc.M008527200. [DOI] [PubMed] [Google Scholar]
- 31.Alaoui-El-Azher M, Mans JJ, Baker HV, Chen C, Progulske-Fox A, Lamont RJ, Handfield M. 2010. Role of the ATM-checkpoint kinase 2 pathway in CDT-mediated apoptosis of gingival epithelial cells. PLoS One 5:e11714. doi: 10.1371/journal.pone.0011714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tait SW, Ichim G, Green DR. 2014. Die another way–non-apoptotic mechanisms of cell death. J Cell Sci 127:2135–2144. doi: 10.1242/jcs.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herdman BP, Paton JC, Wang H, Beddoe T, Paton AW. 2017. Vacuolation activity and intracellular trafficking of ArtB, the binding subunit of an AB5 toxin produced by Salmonella enterica serovar Typhi. Infect Immun 85:e00214-17. doi: 10.1128/IAI.00214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 35.Hustedt N, Durocher D. 2016. The control of DNA repair by the cell cycle. Nat Cell Biol 19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 36.Gargi A, Reno M, Blanke SR. 2012. Bacterial toxin modulation of the eukaryotic cell cycle: are all cytolethal distending toxins created equally? Front Cell Infect Microbiol 2:124. doi: 10.3389/fcimb.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. 2011. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 157:1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elwell C, Chao K, Patel K, Dreyfus L. 2001. Escherichia coli CdtB mediates cytolethal distending toxin cell cycle arrest. Infect Immun 69:3418–3422. doi: 10.1128/IAI.69.5.3418-3422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fedor Y, Vignard J, Nicolau-Travers ML, Boutet-Robinet E, Watrin C, Salles B, Mirey G. 2013. From single-strand breaks to double-strand breaks during S-phase: a new mode of action of the Escherichia coli cytolethal distending toxin. Cell Microbiol 15:1–15. doi: 10.1111/cmi.12028. [DOI] [PubMed] [Google Scholar]
- 40.Rabin SD, Flitton JG, Demuth DR. 2009. Aggregatibacter actinomycetemcomitans cytolethal distending toxin induces apoptosis in nonproliferating macrophages by a phosphatase-independent mechanism. Infect Immun 77:3161–3169. doi: 10.1128/IAI.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallach D, Kovalenko A. 2014. Keeping inflammation at bay. eLife 3:e02583. doi: 10.7554/eLife.02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, Kogan SC, Lowe SW. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blazkova H, Krejcikova K, Moudry P, Frisan T, Hodny Z, Bartek J. 2010. Bacterial intoxication evokes cellular senescence with persistent DNA damage and cytokine signalling. J Cell Mol Med 14:357–367. doi: 10.1111/j.1582-4934.2009.00862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galán JE. 2016. Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc Natl Acad Sci U S A 113:6338–6344. doi: 10.1073/pnas.1606335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nnalue NA, Shnyra A, Hultenby K, Lindberg AA. 1992. Salmonella choleraesuis and Salmonella typhimurium associated with liver cells after intravenous inoculation of rats are localized mainly in Kupffer cells and multiply intracellularly. Infect Immun 60:2758–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higginson EE, Simon R, Tennant SM. 2016. Animal models for salmonellosis: applications in vaccine research. Clin Vaccine Immunol 23:746–756. doi: 10.1128/CVI.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. 2009. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect Immun 77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guidi R, Levi L, Rouf SF, Puiac S, Rhen M, Frisan T. 2013. Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cell Microbiol 15:2034–2050. doi: 10.1111/cmi.12172. [DOI] [PubMed] [Google Scholar]
- 50.Ge Z, Rogers AB, Feng Y, Lee A, Xu S, Taylor NS, Fox JG. 2007. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell Microbiol 9:2070–2080. doi: 10.1111/j.1462-5822.2007.00939.x. [DOI] [PubMed] [Google Scholar]
- 51.Perreault N, Beaulieu JF. 1996. Use of the dissociating enzyme thermolysin to generate viable human normal intestinal epithelial cell cultures. Exp Cell Res 224:354–364. doi: 10.1006/excr.1996.0145. [DOI] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler-Stevenson MA, Gabrielson E. 2001. Doublet discrimination in DNA cell-cycle analysis. Cytometry 46:296–306. doi: 10.1002/cyto.1171. [DOI] [PubMed] [Google Scholar]
- 54.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bates D, Maechler M, Bolker B, Walker S, Christensen R, Singmann H, Dai B, Grothendieck G, Green P. 2017. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1-14. [Google Scholar]
- 56.Kuznetsova A, Brockhoff PB, Christensen RHB. 2015. Package “lmerTest”. R package version 2. [Google Scholar]
- 57.Lenth RV. 2016. Least-squares means: the R package lsmeans. J Stat Softw 69:1–33. [Google Scholar]
- 58.R Core Team 2017. R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed 28 9 2017.
- 59.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid alignment of TtsA from typhoidal and nontyphoidal serotypes. Amino acid sequences were predicted from ttsA sequences extracted from whole-genome sequence data. Strains included in analyses follow: S. Typhi strain CT-18, S. Typhi strain Ty2, S. Montevideo strain USDA-ARS-USMARC-1903, S. Paratyphi A strain ATCC 11511, S. Schwarzengrund strain CVM19633, and S. Javiana strain CFSAN0001992. Asterisks represent conserved amino acid residues. Amino acids highlighted in red represent amino acid changes present in TtsA from S. Montevideo compared to all other sequences of TtsA from all other serotypes. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of ArtB from typhoidal and nontyphoidal serotypes. (A) Phylogenetic tree of predicted ArtB amino acid sequences constructed using the James-Thorton-Thompson (JTT) model with 1,000 bootstrap repetitions in RAxML and rooted by midpoint. Bar represents 0.05 substitutions per amino acid residue. (B) Alignment of ArtB amino acid sequences. Asterisks represent amino acid residues that are conserved in all sequences in the alignment; amino acids highlighted in red represent amino acid substitutions compared to S. Typhimurium DT104 ArtB. Strains included in analyses follow: S. Typhimurium DT104, S. Javiana strain CFSAN0001992, S. Minnesota strain ATCC 49284, S. Montevideo strain USDA-ARS-USMARC-1921, S. Paratyphi A strain ATCC 11511, S. Rubislaw strain ATCC 10717, S. Schwarzengrund strain CVM19633, and S. Typhi strain CT18. Download FIG S2, EPS file, 1.5 MB (1.6MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S1, DOCX file, 0.02 MB (16.9KB, docx) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gating strategies used in this study. Download FIG S3, EPS file, 1.6 MB (1.7MB, eps) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Codes used in the statistical analyses. Download Data Set S1, PDF file, 0.6 MB (671.7KB, pdf) .
Copyright © 2018 Miller et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.