ABSTRACT
Endosymbiotic bacteria associated with eukaryotic hosts are omnipresent in nature, particularly in insects. Studying the bacterial side of host-symbiont interactions is, however, often limited by the unculturability and genetic intractability of the symbionts. Spiroplasma poulsonii is a maternally transmitted bacterial endosymbiont that is naturally associated with several Drosophila species. S. poulsonii strongly affects its host’s physiology, for example by causing male killing or by protecting it against various parasites. Despite intense work on this model since the 1950s, attempts to cultivate endosymbiotic Spiroplasma in vitro have failed so far. Here, we developed a method to sustain the in vitro culture of S. poulsonii by optimizing a commercially accessible medium. We also provide a complete genome assembly, including the first sequence of a natural plasmid of an endosymbiotic Spiroplasma species. Last, by comparing the transcriptome of the in vitro culture to the transcriptome of bacteria extracted from the host, we identified genes putatively involved in host-symbiont interactions. This work provides new opportunities to study the physiology of endosymbiotic Spiroplasma and paves the way to dissect insect-endosymbiont interactions with two genetically tractable partners.
KEYWORDS: Spiroplasma, endosymbiosis, host-symbiont interaction
IMPORTANCE
The discovery of insect bacterial endosymbionts (maternally transmitted bacteria) has revolutionized the study of insects, suggesting novel strategies for their control. Most endosymbionts are strongly dependent on their host to survive, making them uncultivable in artificial systems and genetically intractable. Spiroplasma poulsonii is an endosymbiont of Drosophila that affects host metabolism, reproduction, and defense against parasites. By providing the first reliable culture medium that allows a long-lasting in vitro culture of Spiroplasma and by elucidating its complete genome, this work lays the foundation for the development of genetic engineering tools to dissect endosymbiosis with two partners amenable to molecular study. Furthermore, the optimization method that we describe can be used on other yet uncultivable symbionts, opening new technical opportunities in the field of host-microbes interactions.
INTRODUCTION
Insects frequently maintain symbiotic relationships with vertically transmitted bacterial partners that live within their body, called endosymbionts. Some endosymbionts provide a direct benefit to the host’s development and fertility by complementing its diet. Others grant their host with a conditional benefit that arises only in given contexts, for example by providing resistance to heat, parasites, or viruses (1). Deciphering the molecular dialogue that underlies host-endosymbiont interactions is thus of major importance to better understand the physiology and evolution of insects. However, functional studies are often focused on the host side, because nearly all endosymbiotic bacteria are uncultivable, and thus genetically intractable. As a consequence, the bacterial determinants that affect the interaction remain largely unknown. To date, only four endosymbionts, Sodalis glossidinus (tsetse flies), Arsenophonus arthropodicus (louse flies), Serratia symbiotica (aphids), and Hamiltonella defensa (aphids), have been cultivated in cell-free media (2–5). Systems of coculture with insect cell lines have also been used successfully for some endosymbionts (6), but such techniques are difficult, and they do not always allow genetic engineering.
The Spiroplasma genus comprises diverse bacteria, including commensal, pathogenic, and mutualistic species, most of them being obligate associates with arthropod or plant partners (7). Spiroplasma cells are long, helical, and devoid of a cell wall. Extensively studied species include pathogens of crustaceans (8), insects (e.g., the bee pathogen Spiroplasma melliferum [9]), and plants. Plant pathogens proliferate in phloem and are vectored by phloem-feeding insects (10–12). Some, notably Spiroplasma citri, can be grown in vitro and are amenable to genetic studies. In addition to strains that are infectious and transmitted horizontally between hosts, many Spiroplasma are facultative inherited endosymbionts of insects (i.e., with transovarial transmission).
Along with Wolbachia, Spiroplasma bacteria are the only known inherited symbionts of Drosophila (13). By far the best-studied species (and strain) is Spiroplasma poulsonii MSRO (MSRO for melanogaster sex ratio organism), which infects Drosophila melanogaster and is the focus of this study. As other facultative endosymbionts, S. poulsonii is transmitted vertically with high efficiency, causes reproductive manipulation (male killing), and confers protection to its Drosophila host against parasitoid wasps (14, 15).
Taking advantage of the genetic tools available in Drosophila, current work has started to investigate the molecular mechanisms underlying Drosophila-Spiroplasma symbiosis (16–19). The study of the bacterial determinants, however, has been hampered by the fact that the endosymbiont was unculturable. To expand the toolbox with which to study this endosymbiosis, we designed a method to optimize the Barbour-Stoenner-Kelly H (BSK-H) medium (21) so it allows a sustainable in vitro culture of Spiroplasma. We also resequenced the S. poulsonii MSRO genome in order to provide a complete draft of the chromosome, as well as the first complete sequence of a natural plasmid in this species. By comparing the transcriptome of the bacterium in vitro and in the host, we identified genes potentially involved in the interaction with the host.
RESULTS
Design and optimization of a culture medium for S. poulsonii.
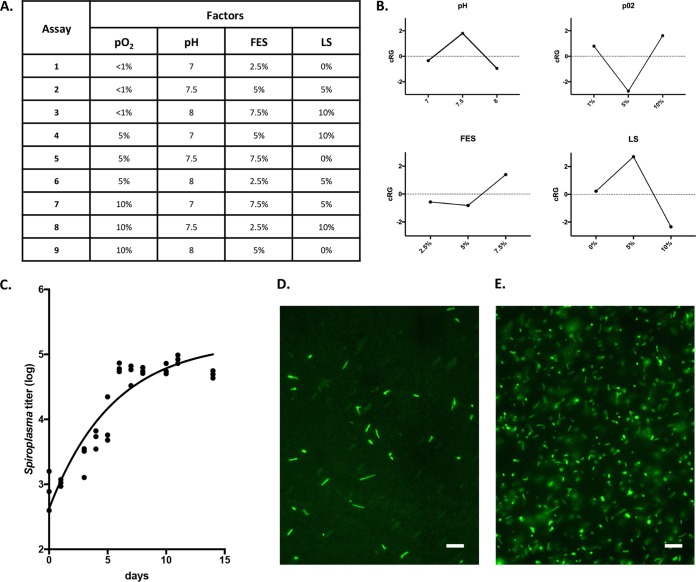
Unlike pathogenic Spiroplasma species, S. poulsonii has a partially degenerated genome (20), leading to poor adaptability to environmental changes. In vitro, it results in the inability of S. poulsonii to grow in culture media designed for pathogenic Spiroplasma, such as SP4 medium (60). We thus developed a new medium using as a starting point the commercial medium Barbour-Stoenner-Kelly H (BSK-H). This standardized complex medium has been designed for the culture of the spirochete Borrelia burgdorferi (21) and is enriched in nutrients that are predicted to be required by S. poulsonii based on its genome. The base medium allows for survival of S. poulsonii for several days but does not sustain its growth. To optimize the medium composition, we elaborated an experimental design to assess the effects of four factors on growth: pH, partial pressure in O2 (pO2), fly extract supplementation (FES), and lipid supplementation (LS). Three levels for each factor were cross-tested against each other following an orthogonal array of assays in BSK-H medium (Fig. 1A). To penalize factor levels that bring in high variability, we computed a growth indicator called contribution to reproducible growth (cRG; mathematical details in Materials and Methods). The analyses of cRG values predicted the best medium to be BSK-H medium supplemented with 7.5% fly extract and 5% lipid mix at pH 7.5 and under 10% pO2 (Fig. 1B). Experimental validation of this predicted best medium, named BSK-H-spiro, yielded sustained growth of S. poulsonii for 6 to 7 days before the bacterial titer reaches a plateau (Fig. 1C). The growth was confirmed by microscopy observations (Fig. 1D and E). S. poulsonii culture in BSK-H-spiro medium could be maintained for more than a year by twofold or threefold dilutions in fresh medium every week. Repeated greater dilutions progressively lead to the collapse of the culture (see Fig. S1 in the supplemental material). Interestingly, the culture can also be frozen at −80°C for more than a year and revived without adding any cryoprotectant. This singularity is likely due to their lack of a cell wall that makes them more deformable, thus more resilient to freezing-induced mechanical stress.
FIG 1 .
(A) Orthogonal matrix of assays for optimization of the BSK-H-spiro medium. Each assay was independently repeated three times. pO2, partial pressure in O2; FES, fly extract supplementation; LS, lipid supplementation. (B) cRG values computed from the assays for each factor. (C) Growth curve of S. poulsonii in BSK-H-spiro medium at 25°C under 10% O2 and 5% CO2. Each point represents one quantitative PCR (qPCR) measurement of Spiroplasma titer in one repetition. The line represents a one-phase exponential fit computed on three independent repetitions. (D and E) Freshly diluted (D) and 2-week-old (E) cultures stained with Syto9. Bars, 10 µm.
S. poulsonii titer in BSK-H-spiro medium diluted weekly with fresh medium. Each bar represents the average titer in three independent replicate experiments. Error bars represent standard deviations. S. poulsonii was not detected by qPCR from passage 5 on when the culture was diluted by 10. Download FIG S1, TIF file, 0.4 MB (469KB, tif) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The BSK-H-spiro medium allowed a doubling time of around 30 h with no difference between a 1-month-old culture and a 1-year-old culture. Infection of naive flies with the culture resulted in a 100% transmission success (24/24 flies) for the 1-month-old culture and 96% infection success (23/24 flies) for the 1-year-old culture, although the amount of bacteria injected (102/fly) is lower than the amount usually injected during hemolymph transfer infections (104/fly). All culture-infected flies transmitted the bacteria to their offspring and displayed a male-killing phenotype, suggesting that prolonged in vitro culture does not significantly alter the host-interacting abilities of S. poulsonii.
S. poulsonii genome sequence update.
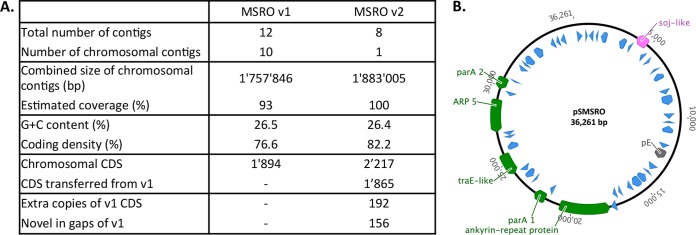
The S. poulsonii MSRO genome was first sequenced and annotated in 2015 (20). However, the presence of repeated sequences complicated the assembly of this draft genome that covered only 93% of the estimated chromosome size. Furthermore, there was doubt about the nature of two extrachromosomal contigs that could have been either plasmids or misassembly products. To complete the genome sequence, we took advantage of recent upgrades in PacBio technology and performed a second sequencing. The new assembly produced eight contigs, including a large contig of 1.8 Mb corresponding to the full circular chromosome of S. poulsonii. A total of 2,217 coding sequences (CDS) were identified, of which 1,865 are identical to those in the first assembly prediction (Fig. 2A). Seven smaller contigs were also produced, of which one (contig 7) could be circularized. We aligned this contig to the reference sequences of plasmids from S. citri and Spiroplasma kunkelii, for which plasmids have been well characterized (Fig. S2). The alignment revealed two conserved synteny blocks between contig 7 and the references. We also detected a coding sequence with 87% homology to the plasmid replication protein sequence pE, proven to code for a plasmid replication protein on S. citri plasmids (22). The presence of those genes on the circular sequence of contig 7 strongly suggests that it is the first full sequence of a plasmid in group IV of Spiroplasma, to which S. poulsonii belongs, and hereafter called pSMSRO (Fig. 2B). The analysis of the remaining extrachromosomal contigs did not allow circularizing any of the contigs or detecting any conserved genes with other Spiroplasma plasmids, although we cannot exclude the possibility that other plasmids were present but not detected in the sequencing data.
FIG 2 .
(A) Comparison between the first draft genome (version 1 [v1]) of S. poulsonii MSRO (20) and this work (version 2 [v2]). CDS, coding sequence. (B) Graphic map of contig 7 after circularization (plasmid pSMSRO). Blue unnamed arrows are hypothetical protein-coding sequences without annotation. Green arrows are annotated genes. The pink arrow indicates a pseudogene, and the gray arrow indicates the coding sequence of the Spiroplasma plasmid replication protein pE.
Multiple alignment of contig 7 with reference plasmid sequences of S. citri and S. kunkelii using the MAUVE progressive algorithm. Conserved synteny blocks (red arrows) contain an ARP operon (green block) and a traE operon (blue block). Download FIG S2, PDF file, 0.7 MB (777.8KB, pdf) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
This new draft confirmed the metabolic landscape already described by Paredes et al. (20), with no obvious difference regarding the presence or absence of metabolic genes. However, the full coverage and extended annotation of the new draft allowed us to compile a comprehensive list of Spiroplasma poulsonii virulence factors (Table 1). Five virulence factors were initially reported in the first genome: two spiralins (Spiralin A and B), a chitinase (ChiD), a cardiolipin synthase (Cls), and a glycerol-3-phosphate oxidase (GlpO). Spiralin A is found in other Spiroplasma species, including S. citri, while Spiralin B is found only in S. poulsonii (20). We found a third gene coding for a spiralin-like protein, Spiralin C, which shares only 15% homology with spiA and spiB but has a conserved spiralin domain. We also identified a group of five genes coding for adhesion-related proteins (ARPs) that were present in the first draft but misannotated. Intriguingly, these genes include one sequence located on pSMSRO, but also four sequences located on the chromosome, while all S. citri ARPs are extrachromosomal (23). A sixth chromosomal ARP pseudogenized by an insertion sequence was identified, as well as two shorter genes partially homologous to ARPs. We also identified a gene containing a Clostridium epsilon toxin (Etx) conserved domain. Etx are major toxins of Clostridium perfringens and cause a variety of symptoms in mammals, including brain damage (24). The results of our genome analysis confirm the presence of five sequences coding for ribosome-inactivating proteins (RIPs), that were initially identified by Hamilton et al. (25). Last, the plasmid bears a coding sequence for an ankyrin repeat protein (Ank). Ank repeats are found in many virulence effector proteins (26). Remarkably, they are widely found in the genome of Wolbachia, another widespread endosymbiont that manipulates insect reproduction, and their large number and diversity suggest that they could play a crucial role in host-symbiont interactions (27).
TABLE 1 .
Virulence factors of S. poulsonii
| Family and virulence factor | GenBank locus tag | Contig | Coordinates | Signal peptide |
TM domaina |
Predicted location |
Reference commentb |
|---|---|---|---|---|---|---|---|
| Spiralins | |||||||
| SpiA | SMSRO_SF013140 | 1 | 1005468–1004767 | Yes | No | Membrane | A |
| SpiB | SMSRO_SF009660 | 1 | 753920–754717 | Yes | No | Membrane | A |
| SpiC | SMSRO_SF015890 | 1 | 1203572–1204045 | No | No | Unknown | C |
| Adhesion-related proteinsc | |||||||
| SpARP1 | SMSRO_SF002520 | 1 | 205842–206939 | Yes | 1 | Membrane | C |
| SpARP2 | SMSRO_SF011850 | 1 | 908030–909277 | Yes | Unsure | Membrane | C |
| SpARP3 | SMSRO_SF022680 | 1 | 1731722–1730625 | Yes | 1 | Membrane | C |
| SpARP4 | SMSRO_SF024450 | 1 | 1870575–1871513 | Yes | 1 | Membrane | C |
| SpARP5 | SMSRO_SFP00390 | 7 | 12713–12147 | Yes | 1 | Membrane | D |
| Metabolic genes | |||||||
| Cls | SMSRO_SF001010 | 1 | 81414–82952 | No | 3 | Membrane | A |
| ChiD1 | SMSRO_SF008450 | 1 | 671704–672774 | Yes | No | Secreted | A |
| ChiD2 | SMSRO_SF013110d | 1 | 1002344–1002357 | C | |||
| GlpO | SMSRO_SF018440 | 1 | 1400479–1401657 | No | No | Cytosol | A |
| Toxins | |||||||
| RIP1 | SMSRO_SF016530 | 1 | 1253115–1254512 | Yes | No | Secreted | B |
| RIP2 | SMSRO_SF018820 | 1 | 1438476–1439966 | Yes | No | Secreted | B |
| RIP3 | SMSRO_SF023880 | 1 | 1820456–1821802 | Yes | No | Secreted | B |
| RIP4 | SMSRO_SF020720 | 1 | 1584448–1585794 | Yes | No | Secreted | B |
| RIP5 | SMSRO_SF003660 | 1 | 293319–294665 | Yes | No | Secreted | B |
| ETX-like | SMSRO_SF021610e | 1 | C | ||||
| Ankyrin repeat | SMSRO_SFP00290 | 7 | 6975–9461 | Yes | No | Secreted | D |
The presence or absence of a transmembrane domain and if present, the number of transmembrane domains.
Reference comments give additional information about genes as follows: A, the gene was detected and annotated by Paredes et al. (20); B, the gene was detected by Paredes et al. (20) and annotated and discussed by Hamilton et al. (25); C, the gene was detected by Paredes et al. (20) but was not annotated and/or not discussed; D, the gene was detected and annotated in this work for the first time.
SpARP1, S. poulsonii ARP1.
This gene was pseudogenized.
The gene structure of this gene was unclear.
Transcriptome analysis of S. poulsonii in culture versus in host.
To detect genes involved in S. poulsonii interaction with its host, we compared the transcriptome of S. poulsonii collected from Drosophila hemolymph to the transcriptome of S. poulsonii cultured in vitro for 2 months. This reference transcriptome produced by pooling transcripts detected in both conditions contains 1,491 transcripts. Of the reads, 97.18% mapped to the chromosome and 1.74% mapped to contig 7, while no significant signal was detected for any other extrachromosomal contig. This supports the hypothesis that contig 7 is a plasmid of S. poulsonii, while other extrachromosomal contigs are misassembly products. The most expressed gene under all conditions is spiB, followed by housekeeping genes related to cell division, transcription, and translation.
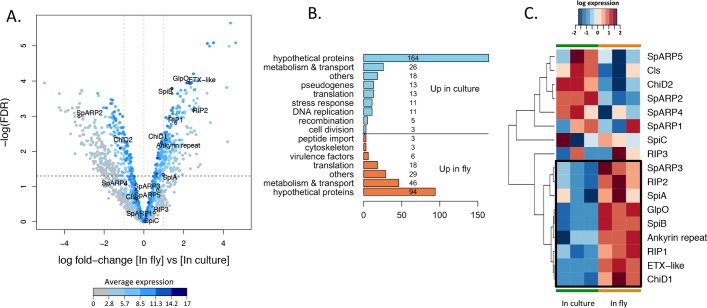
Pairwise comparison between the two experimental conditions identified 465 genes differentially expressed, 201 of the genes being more transcribed in the host, while 264 were significantly more expressed in culture (Fig. 3A). A total of 258 (55%) of the differentially expressed genes were annotated only as “hypothetical proteins” and were not further accounted for in the analysis. Genes that were identified by homology but whose function was very general or unclear were grouped in the category “others.” The remaining sequences have been manually clustered according to their predicted function (Fig. 3B). A majority of identified genes that were found differentially expressed were associated with metabolic pathways and metabolite transport, probably as a consequence of differences between the composition of the medium and the fly hemolymph. Aside from this, a large cluster of genes was related to ribosome assembly and translation, including ribosome structural proteins, tRNA ligases, and translation regulators. Some members of this cluster were found upregulated in culture, while others were upregulated in the fly, suggesting that the switch of environment triggers a qualitative change in the translational activity of the bacterium. The in vitro culture data set also showed enrichment in transcripts related to DNA replication and cell division, consistent with a doubling time of around 30 h in vitro versus 170 h in the adult fly (28). Interestingly, transcripts involved in DNA recombination were also enriched in S. poulsonii grown in culture, including ruvA, ruvB, recR, and recU, as well as genes belonging to the comEC family. The comE operon contributes to the natural competence in Bacillus subtilis, ensuring the binding and uptake of transforming DNA (29), which suggests that S. poulsonii might be naturally competent.
FIG 3 .
(A) Volcano plot of differential gene expression of S. poulsonii in host versus in culture. Each point represents the average value of one transcript in three replicate experiments. The expression difference is considered significant for a log2 fold change of ≥1 (outer light gray broken vertical lines) and for a P value of ≤0.05 [−log(FDR) of ≥1.3, dark broken horizontal line]. Points are colored according to their average expression in all data sets. Names and outlined points represent virulence factors. FDR, false-discovery rate. (B) Manual clustering of the transcripts differentially expressed by S. poulsonii in the fly versus in the culture. The numbers of sequences in the different categories are indicated on the bars or to the right of the bars. (C) Heatmap of S. poulsonii virulence gene expression. Each column represents the value for one replicate experiment in culture or in the fly. The colors represent the log10 level of expression in the corresponding experiment. The cluster of genes that are induced when S. poulsonii is in the host (versus in vitro) is shown enclosed in a black box. SpARP5, S. poulsonii ARP5.
Last, several differentially expressed sequences were identified as pseudogenes resulting either from a frameshift mutation or from the insertion of a mobile element in the coding sequence that causes the protein to be truncated. The active transcriptional regulation of these genes suggests a recent pseudogenization, possibly as a consequence of S. poulsonii switching from a free-living lifestyle to an endosymbiotic lifestyle.
Virulence factors could be classified in two clusters depending on their expression profile (Fig. 3C). spiA, spiB, RIP1, RIP2, ank, etx, glpO, and chiD1 have lower expression levels in culture than in the host, pointing to their role in host-symbiont interaction. Other virulence genes do not display a significant change in their expression level and have low average levels of expression. Such genes include spiC, RIP3, SpARP1, SpARP4, SpARP5, chiD2, and cls. SpARP2 is the only virulence gene that is expressed at higher levels in culture than in the fly, and RIP4 and RIP5 as well as SpARP3 are not detected at all in the transcriptome, implying that they might be pseudogenes, resulting from a duplication of the coding sequence without the regulatory upstream sequence.
Finally, a gene encoding a ferritin, a protein involved in iron sequestration, was expressed at a higher level in S. poulsonii extracted from Drosophila than in S. poulsonii grown in culture, suggesting that iron availability could be a proliferation-limiting factor along with lipids and glucose availability (17, 20).
DISCUSSION
We developed the first reliable method to culture endosymbiotic S. poulsonii from Drosophila in a cell-free medium. This is an important step forward because cultivation is a prerequisite for addressing functional questions on the regulation of this symbiosis via genetic manipulation of the bacterial partner. While pathogenic Spiroplasma bacteria, such as S. citri, can be easily cultivated in standard growth media, no suitable medium was available to grow any endosymbiotic Spiroplasma outside their hosts. This was not for a lack of trying: much work was done in the 1980s to attempt to set up a culture medium for S. poulsonii, until one paper reported in 1986 the successful cultivation of a Drosophila Spiroplasma in a cell-free medium (30). According to the authors, the critical factor was to supply Spiroplasma with growing insect cells in the course of primary isolation, followed by a succession of passages that allowed for Spiroplasma to adapt to an insect cell-free medium. Unfortunately, this work could not be repeated despite several attempts from various laboratories. Spiroplasma thus remained uncultivable in practice.
An important difference between previous attempts and the present work is that crucial requirements of the symbiont were addressed based on very recent discoveries about S. poulsonii physiology. The need of S. poulsonii for host lipids to synthesize its membrane, for example, was demonstrated only a few years ago (17, 20), and lipid supplementation turned out to be crucial to promote growth in vitro. Another difference was supplementing the medium with fly extract, which was also not undertaken in previous works. Since the unsupplemented medium already contains glucose, essential amino acids, lipids, and vitamins needed by the bacteria, we assume that the growth improvement observed with fly extract supplementation might come either from a fly hormone or neurotransmitter or from a nonorganic growth factor available in the fly. A possible candidate would be iron, as suggested by the overexpression of a ferritin-like coding gene by S. poulsonii in vitro, or another metallic ion, not present in sufficient quantities in the BSK-H base medium (31). Importantly, starting with a high density of bacteria seems to be a key point for the successful establishment of the culture. Beginning with an amount of infected hemolymph that is too small does not allow the culture to thrive, and repeated strong dilutions lead to a collapse of the culture. This suggests the existence of a density threshold below which S. poulsonii growth is inhibited. The exact mechanism leading to this inhibition remains elusive however, as the genome analysis did not highlight any quorum-sensing system.
This work also allowed an initial comparison between the in vivo transcriptome and the in vitro transcriptome of S. poulsonii, identifying genes that are overexpressed when the bacterium is in contact with its host. Membrane proteins are particularly interesting, as they are associated with host infection in pathogenic Spiroplasma. S. citri spiralin for example acts as a lectin and binds to insect host’s glycoproteins to invade cells (32, 33). Its expression is downregulated when the bacterium is in its plant hosts compared to in vitro culture, while its expression is not altered in the insect and in culture (34). In S. poulsonii, spiA, the closest homologue to the S. citri spiralin gene, is only slightly upregulated in the insects, while spiB, which is found only in S. poulsonii, is strongly upregulated. This points to a function of SpiB that is specific to endosymbiosis, possibly related to the bacterial entry in the oocyte during vertical transmission. ARPs are also major lipoproteins involved in S. citri transmission (35, 36), turning out to be essential for insect cell invasion but nonessential for transmission from insects to plants (37). In S. poulsonii, few predicted ARPs have a complete and functional sequence, and their expression is not differentially regulated in vitro compared to in the host. S. citri, as a strictly horizontally transmitted pathogen, could require diverse ARPs to infect new hosts efficiently. S. poulsonii on the other hand is mostly vertically transmitted, although horizontal transmission to new host is possible notably via ectoparasite vectors (38, 39). ARPs in this species could thus be less diverse because of its more limited host range. The chromosomal location of most S. poulsonii ARPs (rather than extrachromosomal as in S. citri [23]) also reflects a lower ability of these genes to be horizontally transferred. This could reflect the fixation of this gene family during the coevolution of vertically transmitted Spiroplasma with its host.
Several genes coding for toxins are overexpressed in the host compared to in vitro, including ribosome-inactivating proteins (RIPs) and two yet uncharacterized toxins (Ank and Etx). RIPs are involved in the protection of Drosophila against nematodes and parasitoid wasps by selectively inactivating the 28S rRNA of the parasites (25, 40). The upregulation of RIP1 and RIP2 when S. poulsonii is in the host compared to in vitro suggests that RIPs could have a function in host-symbiont interactions, regardless of parasite infections, possibly in male killing. Etx might be involved in the neuronal symptoms, notably tremors and dopaminergic neuron degeneration observed in Spiroplasma-infected flies when the flies are old (28). We also cannot exclude the possibility that it functions as a defensive agent against parasites, which would explain the transcription increase when S. poulsonii is in the insect compared to in vitro, although the toxicity of Etx has not yet been investigated in nonmammal models. Further functional studies will be necessary to assess the exact function of these toxins in the S. poulsonii-Drosophila interaction.
It is noteworthy that 2 out of 18 identified virulence genes (SpARP5 and ank) are located on a plasmid, which indicates that extrachromosomal DNA may play an important role in S. poulsonii-Drosophila interactions. Variability in plasmid presence and/or copy number in S. poulsonii strains could be accountable for the variability in the host phenotypes caused by the endosymbiont, including variable male-killing penetrance.
In conclusion, the method described in this work is the first protocol to allow cultivation of an endosymbiotic Spiroplasma in a cell-free medium in almost 30 years. The technical approach that was used to design the BSK-H-spiro medium can be adapted to optimize a medium for other uncultivable bacteria, for which a favorable physicochemical environment can be partially predicted. The expression of comE indicates that S. poulsonii might be naturally competent and the transcriptional regulation findings (up- or downregulation) observed with recombination-related genes suggest that knockout mutants by homologous recombination might be possible despite recA pseudogenization (20), as in pathogenic Spiroplasma species (41, 42). Eventually, several bacterial genes were predicted to have a key function in the interaction between S. poulsonii and its host, including virulence factors. These genes are thus priority candidates for further investigation upon the development of genetic tools to modify S. poulsonii in order to unravel their precise function. Coupled with the powerful genetic tools available on the Drosophila side, the development of genetic tools to modify S. poulsonii will be a major achievement in the field of symbiosis, as it will provide the first insect model where both the host and the endosymbiont are readily transformable.
MATERIALS AND METHODS
Spiroplasma stock.
We used a wild-type Oregon-R (ORR) fly stock that has been cured of Wolbachia by antibiotic treatment and infected by the Spiroplasma poulsonii MSRO strain Uganda (28, 43). The stock has been maintained in the lab for several years between these treatments and the experiments.
Cell-free culture medium design.
The medium basis was the Barbour-Stoenner-Kelly H medium (BSK-H) without l-glutamine from BioSell (Feucht bei Nürnberg, Germany). BSK-H medium from Sigma has also been used successfully. The design of an orthogonal array of growth assays was based on the choice of four factors (pH, partial pressure in oxygen [pO2], fly extract supplementation [FES], and lipid supplementation [LS]) that were a priori expected to affect Spiroplasma growth significantly. The use of an orthogonal array allows the extraction of relevant information from a reduced number of factor level combinations rather than from all possible combinations. For each factor, three levels were arbitrarily chosen around an expected optimal value (e.g., pH 7.5 as the expected optimal value, pH 7, 7.5, and 8 as the tested levels). Cultures were started from hemolymph extracted from the thorax of 1-week-old infected flies by aspiration with a Nanoject II nanoinjector (Drummond Scientific). A preculture was launched 1 week prior to the experiment by adding 8 µl of hemolymph to 3.2 ml of BSK-H medium plus 5% fly extract (1,000 to 5,000 bacteria/µl) without agitation. Aliquots of 100 µl of preculture were then frozen at −80°C before use. For each assay, aliquots were centrifuged for 40 min at 2,000 relative centrifugal force (rcf) at 18°C, and pellets were resuspended in 200 µl of medium. Aliquots (10-µl aliquots) were taken 1 day and 7 days later for growth assessment by quantitative PCR. A linear regression on the log-transformed measures between day 1 and day 7 was computed for each level of each tested factor, and the slope of the regression was used as a growth rate measurement. Three independent replicates were made for each medium testing. Since some combinations of factors yielded high variability in growth, we analyzed the data with a statistical approach inspired from the Taguchi method (44, 45). To penalize factor levels that entail a high variability in growth, a “reproducible growth” (RG) parameter was computed, where S̄ is the average slope with the considered level of the considered factor and σ is the standard deviation. The contribution of one level to the reproducible growth (cRG) was calculated as cRG = RGconsidered level − RGall levels. For each factor, the level with the highest cRG was selected as the optimal value. An experimental validation was then performed in a medium bringing together the best levels for each of the four factors, hereafter designed as BSK-H-spiro, following the same protocol as for the optimization assays. Three independent replicates were made for the validation assay. Protocols for preparing the fly extract, lipid mix, and BSK-H-spiro medium are detailed in Text S1 in the supplemental material. The media of freshly started cultures were completely renewed every three or four passages until at least passage 12, by centrifugation for 20 min at 12,000 rcf at 18°C and replacement of the used medium (supernatant) by an equivalent amount of fresh medium. These replacements are a necessary adaptation step that becomes unnecessary for older cultures. Long-lasting cultures were maintained by a weekly threefold dilution in fresh culture medium. All experiments and cultures were performed at 25°C, which is the temperature at which Drosophila infected stocks are routinely maintained, and yet close to the 26°C predicted optimal temperature for S. poulsonii (46).
Recipes for fly extract, lipid mix, and BSK-H-spiro medium. Download TEXT S1, PDF file, 0.05 MB (52.4KB, pdf) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Culture density measurement.
For DNA extraction for quantitative PCR, bacteria were lysed by osmotic shock by adding 400 µl of distilled water to 10 µl of culture and heated at 95°C for 15 min. This simple method ensures an efficient yield from a small amount of initial bacterial material. DNA was then used for quantitative PCR as described before (47) with primers amplifying a 300-bp fragment of the 16S rRNA gene (primer forward, 5′-TACATGCAAGTCGAACGGGG-3′; primer reverse, 5′-CTACTGCTGCCTCCCGTAG-3′). Microscopic observation was performed as previously described (28).
Fly infections.
One-week-old female Oregon flies were infected from the S. poulsonii culture by an injection of 23 nl of a dense culture (1 week after the latest dilution with fresh medium) with a Nanoject II nanoinjector (Drummond Scientific). Flies were allowed to recover from the injection in a tube with fresh medium in the absence of males for 1 day. Each female fly was then coupled with a male, and each couple was isolated in a tube in order to monitor the infection status of the progeny of single flies. Couples were flipped onto fresh medium every 2 or 3 days. The eggs laid for the first week following mating were discarded. The progeny was screened for S. poulsonii infection 1 week after hatching by Syto9 staining as previously described (28).
Genome sequencing and analysis.
S. poulsonii DNA was extracted from fly hemolymph as previously described (20). Processing of the samples was performed in the University of Lausanne Genomic Technologies Facility. The DNA was sheared in a Covaris g-TUBE (Covaris, Woburn, MA, USA) to obtain 20-kb fragments. After the DNA was sheared, the size distribution of the DNA fragments was checked on a Fragment Analyzer (Advanced Analytical Technologies, Ames, IA, USA). Five micrograms of the sheared DNA was used to prepare an SMRTbell library with the PacBio SMRTbell template prep kit 1 (Pacific Biosciences, Menlo Park, CA, USA) according to the manufacturer’s recommendations. The resulting library was size selected on a BluePippin system (Sage Science, Inc., Beverly, MA, USA) for molecules larger than 20 kb. The recovered library was sequenced on one SMRT cell with P6/C4 chemistry and MagBeads on a PacBio RSII system (Pacific Biosciences, Menlo Park, CA, USA) in a 240-min movie. Assembly was performed with HGAP (hierarchical genome assembly process) version 2 from the PacBio smrtpipe (v2.3.0). Circularization of main contig 1 was performed using Amos (v3.1.0; Amos Consortium; http://amos.sourceforge.net). Plasmid contig 7 was refined using the PacBio read data from Paredes et al. (20) with Quiver version 1. Genome annotation was performed with Prokka v 1.11 (48) using parameters --addgenes --genus Spiroplasma --species poulsonii --gcode 4 --rawproduct –rfam --rnammer). The Nucmer tool (Mummer suite v3.23 [49]) was used to align coding sequences (CDS) from the annotated version (accession no. JTLV01000000) to the new annotation. Some gene product annotations were refined using NCBI PSI-blast tool. Multiple alignments of Spiroplasma plasmids have been performed using MAUVE version 2.4.0 (50).
RNA sequencing and analysis.
RNA was extracted from (i) the hemolymph of 30 1-week-old infected flies and (ii) 20 ml of 4-month-old in vitro culture pelleted by 30 min of centrifugation at 16,000 × g by the TRIzol method following the manufacturer’s instructions. Three independent replicates were prepared for each condition. Libraries were prepared using the Illumina Truseq RNA kit and sequenced on an Illumina HiSeq 2000 system at the University of Lausanne Genomic Technologies Facility. Purity-filtered reads were adapters and quality trimmed with Cutadapt v.1.8 (51). Reads matching to rRNA sequences were removed with fastq_screen v. 0.9.3 (Babraham Bioinformatics; http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/). Remaining reads were further filtered for low complexity with reaper v. 15-065 (52). Reads were aligned against the Spiroplasma poulsonii MSRO (v2) genome using STAR v. 2.5.2b (53). The number of read counts per gene locus was summarized with htseq-count v. 0.6.1 (54) using Spiroplasma poulsonii MSRO (v2) gene annotation. The quality of the transcriptome sequencing (RNA-seq) data alignment was assessed using RSeQC v. 2.3.7 (55). Statistical analysis was performed for genes in R version 3.4.1 (56). Genes with low counts were filtered out according to the rule of one count per million (cpm) in at least one sample. rRNA and tRNA gene counts were discarded. Library sizes were scaled using TMM normalization with EdgeR package version 3.18.1 (57) and log cpm transformed with limma voom function, limma package version 3.32.5 (58). Differential expression was computed with limma (59) by fitting the samples into a linear model and performing “in fly” versus “in culture” comparison. Moderated t test was used, and adjusted P values were computed by the Benjamini-Hochberg method, controlling for the false-discovery rate.
Accession number(s).
The genome with DDBJ/EMBL/GenBank accession no. JTLV00000000 was updated. The genome version described in this paper has accession no. JTLV02000000. The RNA-seq differential expression analysis can be found in Data Set S1.
Detailed differential expression analysis of S. poulsonii transcriptome in the host versus in vitro. Download DATA SET S1, TXT file, 0.4 MB (451.8KB, txt) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Emmanuel Beaudoing and Chloé Jollivet for technical help regarding the transcriptome assembly and the culture maintenance, respectively. We also thank Élodie Ramond and Samuel Rommelaere for constructive discussions and Christoph Vorburger and Aurélien Vigneron for critically reviewing the manuscript.
This work was funded by ERC advanced grant 339970 and the SNF Sinergia grant CRSII3_154396.
Footnotes
Citation Masson F, Calderon Copete S, Schüpfer F, Garcia-Arraez G, Lemaitre B. 2018. In vitro culture of the insect endosymbiont Spiroplasma poulsonii highlights bacterial genes involved in host-symbiont interaction. mBio 9:e00024-18. https://doi.org/10.1128/mBio.00024-18.
REFERENCES
- 1.Douglas AE. 2016. How multi-partner endosymbioses function. Nat Rev Microbiol 14:731–743. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 2.Welburn SC, Maudlin I, Ellis DS. 1987. In vitro cultivation of rickettsia-like-organisms from Glossina spp. Ann Trop Med Parasitol 81:331–335. doi: 10.1080/00034983.1987.11812127. [DOI] [PubMed] [Google Scholar]
- 3.Dale C, Beeton M, Harbison C, Jones T, Pontes M. 2006. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl Environ Microbiol 72:2997–3004. doi: 10.1128/AEM.72.4.2997-3004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabri A, Leroy P, Haubruge E, Hance T, Frère I, Destain J, Thonart P. 2011. Isolation, pure culture and characterization of Serratia symbiotica sp. nov., the R-type of secondary endosymbiont of the black bean aphid Aphis fabeae. Int J Syst Evol Microbiol 61:2081–2088. doi: 10.1099/ijs.0.024133-0. [DOI] [PubMed] [Google Scholar]
- 5.Brandt JW, Chevignon G, Oliver KM, Strand MR. 2017. Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proc Biol Sci 284:20171925. doi: 10.1098/rspb.2017.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuchi Y. 2009. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ 24:195–204. doi: 10.1264/jsme2.ME09140S. [DOI] [PubMed] [Google Scholar]
- 7.Gasparich GE. 2002. Spiroplasmas: evolution, adaptation and diversity. Front Biosci 7:d619–d640. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Wen B, Gasparich GE, Zhu N, Rong L, Chen J, Xu Z. 2004. A Spiroplasma associated with tremor disease in the Chinese mitten crab (Eriocheir sinensis). Microbiology 150:3035–3040. doi: 10.1099/mic.0.26664-0. [DOI] [PubMed] [Google Scholar]
- 9.Clark TB, Whitcomb RF, Tully JG, Mouches C, Saillard C, Bove JM, Wroblewski H, Carle P, Rose DL, Henegar RB, Williamson DL. 1985. Spiroplasma melliferum, a new species from the honeybee (Apis mellifera). Int J Syst Bacteriol 35:296–308. doi: 10.1099/00207713-35-3-296. [DOI] [Google Scholar]
- 10.Saglio P, Lhospital M, Tully JG, Freundt EA. 1973. Spiroplasma citri gen. and sp. n.: a Mycoplasma-like organism associated with “stubborn” disease of citrus. Int J Syst Evol Microbiol 23:191–204. doi: 10.1099/00207713-23-3-191. [DOI] [Google Scholar]
- 11.Whitcomb RF, Chen TA, Williamson DL, Liao C, Tully JG, Bove JM, Mouches C, Rose DL, Coan ME, Clark TB. 1986. Spiroplasma kunkelii sp. nov.: characterization of the etiological agent of corn stunt disease. Int J Syst Bacteriol 36:170–178. doi: 10.1099/00207713-36-2-170. [DOI] [Google Scholar]
- 12.Saillard C, Vignault JC, Bove JM, Raie A, Tully JG, Williamson DL, Fos A, Garnier M, Gadeau A, Carle P, Whitcomb RF. 1987. Spiroplasma phoeniceum sp. nov., a new plant-pathogenic species from Syria. Int J Syst Bacteriol 37:106–115. doi: 10.1099/00207713-37-2-106. [DOI] [Google Scholar]
- 13.Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. 2006. Heritable endosymbionts of Drosophila. Genetics 174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie J, Tiner B, Vilchez I, Mateos M. 2011. Effect of the Drosophila endosymbiont Spiroplasma on parasitoid wasp development and on the reproductive fitness of wasp-attacked fly survivors. Evol Ecol 53:1065–1079. doi: 10.1007/s10682-010-9453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paredes JC, Herren JK, Schüpfer F, Lemaitre B. 2016. The role of lipid competition for endosymbiont-mediated protection against parasitoid wasps in Drosophila. mBio 7:e1006-16. doi: 10.1128/mBio.01006-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herren JK, Paredes JC, Schüpfer F, Lemaitre B. 2013. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. mBio 4:e00532-12. doi: 10.1128/mBio.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herren JK, Paredes JC, Schüpfer F, Arafah K, Bulet P, Lemaitre B. 2014. Insect endosymbiont proliferation is limited by lipid availability. Elife 3:e02964. doi: 10.7554/eLife.02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harumoto T, Anbutsu H, Fukatsu T. 2014. Male-killing Spiroplasma induces sex-specific cell death via host apoptotic pathway. PLoS Pathog 10:e1003956. doi: 10.1371/journal.ppat.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harumoto T, Anbutsu H, Lemaitre B, Fukatsu T. 2016. Male-killing symbiont damages host’s dosage-compensated sex chromosome to induce embryonic apoptosis. Nat Commun 7:12781. doi: 10.1038/ncomms12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paredes JC, Herren JK, Schüpfer F, Marin R, Claverol S, Kuo C-H, Lemaitre B, Béven L. 2015. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. mBio 6:e02437-14. doi: 10.1128/mBio.02437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack RJ, Telford SR, Spielman A. 1993. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol 31:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breton M, Duret S, Arricau-Bouvery N, Béven L, Renaudin J. 2008. Characterizing the replication and stability regions of Spiroplasma citri plasmids identifies a novel replication protein and expands the genetic toolbox for plant-pathogenic spiroplasmas. Microbiology 154:3232–3244. doi: 10.1099/mic.0.2008/019562-0. [DOI] [PubMed] [Google Scholar]
- 23.Saillard C, Carle P, Duret-Nurbel S, Henri R, Killiny N, Carrère S, Gouzy J, Bové J-M, Renaudin J, Foissac X. 2008. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics 9:195. doi: 10.1186/1471-2164-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiles BG, Barth G, Barth H, Popoff MR. 2013. Clostridium perfringens epsilon toxin: a malevolent molecule for animals and man? Toxins 5:2138–2160. doi: 10.3390/toxins5112138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton PT, Peng F, Boulanger MJ, Perlman SJ. 2016. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc Natl Acad Sci U S A 113:350–355. doi: 10.1073/pnas.1518648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Ankyrin-repeat containing proteins of microbes: a conserved structure with functional diversity. Trends Microbiol 18:132–139. doi: 10.1016/j.tim.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siozios S, Ioannidis P, Klasson L, Andersson SGE, Braig HR, Bourtzis K. 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8:e55390. doi: 10.1371/journal.pone.0055390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herren JK, Lemaitre B. 2011. Spiroplasma and host immunity: activation of humoral immune responses increases endosymbiont load and susceptibility to certain Gram-negative bacterial pathogens in Drosophila melanogaster. Cell Microbiol 13:1385–1396. doi: 10.1111/j.1462-5822.2011.01627.x. [DOI] [PubMed] [Google Scholar]
- 29.Hahn J, Inamine G, Kozlov Y, Dubnau D. 1993. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol Microbiol 10:99–111. doi: 10.1111/j.1365-2958.1993.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 30.Hackett KJ, Lynn DE, Williamson DL, Ginsberg AS, Whitcomb RF. 1986. Cultivation of the Drosophila sex-ratio spiroplasma. Science 232:1253–1255. [DOI] [PubMed] [Google Scholar]
- 31.Williamson DL, Steiner T, McGarrity GJ. 1983. Spiroplasma taxonomy and identification of the sex ratio organisms: can they be cultivated? Yale J Biol Med 56:583–592. [PMC free article] [PubMed] [Google Scholar]
- 32.Killiny N, Castroviejo M, Saillard C. 2005. Spiroplasma citri spiralin acts in vitro as a lectin binding to glycoproteins from its insect vector Circulifer haematoceps. Phytopathology 95:541–548. doi: 10.1094/PHYTO-95-0541. [DOI] [PubMed] [Google Scholar]
- 33.Duret S, Batailler B, Dubrana M-P, Saillard C, Renaudin J, Béven L, Arricau-Bouvery N. 2014. Invasion of insect cells by Spiroplasma citri involves spiralin relocalization and lectin/glycoconjugate-type interactions. Cell Microbiol 16:1119–1132. doi: 10.1111/cmi.12265. [DOI] [PubMed] [Google Scholar]
- 34.Dubrana M-P, Béven L, Arricau-Bouvery N, Duret S, Claverol S, Renaudin J, Saillard C. 2016. Differential expression of Spiroplasma citri surface protein genes in the plant and insect hosts. BMC Microbiol 16:53. doi: 10.1186/s12866-016-0666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berho N, Duret S, Renaudin J. 2006. Absence of plasmids encoding adhesion-related proteins in non-insect-transmissible strains of Spiroplasma citri. Microbiology 152:873–886. doi: 10.1099/mic.0.28541-0. [DOI] [PubMed] [Google Scholar]
- 36.Béven L, Duret S, Batailler B, Dubrana M-P, Saillard C, Renaudin J, Arricau-Bouvery N. 2012. The repetitive domain of ScARP3d triggers entry of Spiroplasma citri into cultured cells of the vector Circulifer haematoceps. PLoS One 7:e48606. doi: 10.1371/journal.pone.0048606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breton M, Duret S, Danet J-L, Dubrana M-P, Renaudin J. 2010. Sequences essential for transmission of Spiroplasma citri by its leafhopper vector, Circulifer haematoceps, revealed by plasmid curing and replacement based on oncompatibility. Appl Environ Microbiol 76:3198–3205. doi: 10.1128/AEM.00181-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaenike J, Polak M, Fiskin A, Helou M, Minhas M. 2007. Interspecific transmission of endosymbiotic Spiroplasma by mites. Biol Lett 3:23–25. doi: 10.1098/rsbl.2006.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haselkorn TS, Markow TA, Moran NA. 2009. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol Ecol 18:1294–1305. doi: 10.1111/j.1365-294X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 40.Ballinger MJ, Perlman SJ. 2017. Generality of toxins in defensive symbiosis: ribosome-inactivating proteins and defense against parasitic wasps in Drosophila. PLoS Pathog 13:e1006431. doi: 10.1371/journal.ppat.1006431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCammon SL, Dally EL, Davis RE. 1990. Electroporation and DNA methylation effects on the transfection of Spiroplasma, p 60–65. In Stanek G, Tully J, Whitcomb R (ed), Recent advances in mycoplasmology: proceedings of the 7th Congress of the International Organization for Mycoplasmology, Baden near Vienna, 1988. G. Fischer, Stuttgart, Germany. [Google Scholar]
- 42.Lartigue C, Duret S, Garnier M, Renaudin J. 2002. New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48:149–159. doi: 10.1016/S0147-619X(02)00121-X. [DOI] [PubMed] [Google Scholar]
- 43.Pool JE, Wong A, Aquadro CF. 2006. Finding of male-killing Spiroplasma infecting Drosophila melanogaster in Africa implies transatlantic migration of this endosymbiont. Heredity (Edinb) 97:27–32. doi: 10.1038/sj.hdy.6800830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taguchi G. 1991. Taguchi methods: research and development. American Supplier Institute, Dearborn, MI. [Google Scholar]
- 45.Rao RS, Kumar CG, Prakasham RS, Hobbs PJ. 2008. The Taguchi methodology as a statistical tool for biotechnological applications: a critical appraisal. Biotechnol J 3:510–523. doi: 10.1002/biot.200700201. [DOI] [PubMed] [Google Scholar]
- 46.Williamson DL, Sakaguchi B, Hackett KJ, Whitcomb RF, Tully JG, Carle P, Bové JM, Adams JR, Konai M, Henegar RB. 1999. Spiroplasma poulsonii sp. nov., a new species associated with male-lethality in Drosophila willistoni, a neotropical species of fruit fly. Int J Syst Bacteriol 49:611–618. doi: 10.1099/00207713-49-2-611. [DOI] [PubMed] [Google Scholar]
- 47.Anbutsu H, Fukatsu T. 2003. Population dynamics of male-killing and non-male-killing spiroplasmas in Drosophila melanogaster. Appl Environ Microbiol 69:1428–1434. doi: 10.1128/AEM.69.3.1428-1434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 52.Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. 2015. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wang S, Li W. 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 56.R Core Team 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 57.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29–R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tully JG, Whitcomb RF, Clark HF, Williamson DL. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892–894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. poulsonii titer in BSK-H-spiro medium diluted weekly with fresh medium. Each bar represents the average titer in three independent replicate experiments. Error bars represent standard deviations. S. poulsonii was not detected by qPCR from passage 5 on when the culture was diluted by 10. Download FIG S1, TIF file, 0.4 MB (469KB, tif) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple alignment of contig 7 with reference plasmid sequences of S. citri and S. kunkelii using the MAUVE progressive algorithm. Conserved synteny blocks (red arrows) contain an ARP operon (green block) and a traE operon (blue block). Download FIG S2, PDF file, 0.7 MB (777.8KB, pdf) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recipes for fly extract, lipid mix, and BSK-H-spiro medium. Download TEXT S1, PDF file, 0.05 MB (52.4KB, pdf) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed differential expression analysis of S. poulsonii transcriptome in the host versus in vitro. Download DATA SET S1, TXT file, 0.4 MB (451.8KB, txt) .
Copyright © 2018 Masson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.