Abstract
The CaaX motif directs C-terminal protein modifications that include isoprenylation, proteolysis and carboxylmethylation. Proteolysis is generally believed to require either Rce1p or Ste24p. While investigating the substrate specificity of these proteases, using the yeast a-factor mating pheromone as a reporter, we observed Rce1p-and Ste24p-independent mating (RSM) when the CKQQ CaaX motif was used in lieu of the natural a-factor CVIA motif. Uncharged or negatively charged amino acid substitutions at the a1 position of the CKQQ motif prevented RSM. Alanine substitutions at the a2 and X positions enhanced RSM. Random mutagenesis of the CaaX motif provided evidence that RSM occurs with approximately 1% of all possible CaaX motif permutations. Combined mutational and genetic data indicate that RSM-promoting motifs have a positively charged amino acid at the a1 position. Two of nine naturally occurring yeast CaaX motifs conforming to this pattern promoted RSM. The activity of the isoprenylcysteine carboxyl methyltransferase Ste14p was required for RSM, indicating that RSM-promoting CaaX motifs are indeed proteolysed. RSM was enhanced by the overexpression of Axl1p or Ste23p, suggesting a role for these M16A subfamily metalloproteases in this process. We have also determined that an N-terminal extension of the a-factor precursor, which is typically removed by the yeast M16A enzymes, is required for optimal RSM. These observations suggest a model that involves targeting of the a-factor precursor to the peptidosome cavity of M16A enzymes where subsequent interactions between RSM-promoting CaaX motifs and the active site of the M16A enzyme lead to proteolytic cleavage.
Keywords: yeast, a-factor, mating pheromone, CaaX, Rce1p, Ste24p, Axl1p, Ste23p
Introduction
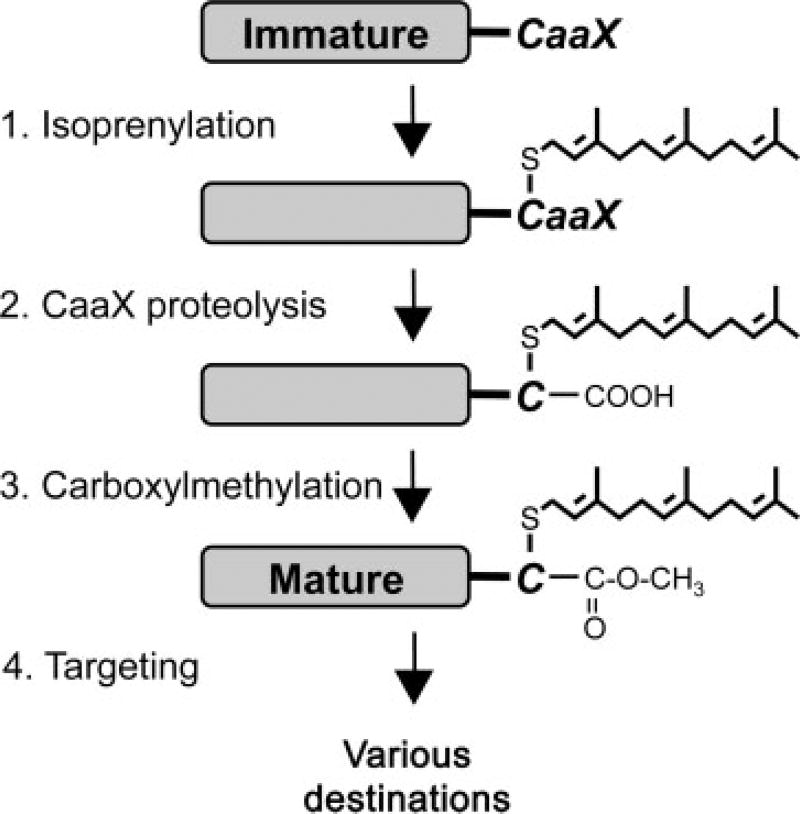
The CaaX motif is a C-terminal tetrapeptide sequence generally described as having an invariant cysteine (C), two aliphatic amino acids (a1 and a2) and one of several amino acids in the terminal position (X). Eukaryotic proteins having a CaaX motif (CaaX proteins) typically undergo three ordered post-translational modifications (reviewed in Spence and Casey, 2001; Young et al., 2001) (Figure 1). The first is isoprenylation of the cysteine by either the C15 farnesyl transferase (FTase) or the C20 geranylgeranyl transferase I (GGTase I). The context of the CaaX motif can dictate which isoprenoid is attached, with geranylgeranylated proteins often having Leu and Phe at the X position. Isoprenylation is followed by an endoproteolytic cleavage that removes the last three amino acids of the motif (i.e. aaX). Two proteases, Rce1p and Ste24p, have been identified that can perform CaaX proteolysis (Boyartchuk et al., 1997b; Tam et al., 2001). CaaX proteolysis is followed by carboxylmethylation of the farnesylated cysteine by an isoprenylcysteine carboxyl methyltransferase (ICMT). Collectively, these modifications modulate the activity, membrane partitioning, subcellular localization, stability and/or protein–protein interaction properties of the modified protein (Bergo et al., 2002a, 2000; Boyartchuk et al., 1997b; Hrycyna et al., 1991; Kim et al., 1999; Marcus et al., 1991; Sapperstein et al., 1994).
Figure 1.
CaaX proteins are extensively modified post-translationally. The C-terminal tetrapeptide CaaX motif directs three ordered post-translational modifications, including isoprenylation, proteolysis and carboxylmethylation. Interfering with these steps can disrupt the activity and/or localization of the protein being modified
CaaX proteins have diverse, biologically important functions. Pertinent examples of CaaX proteins include signalling molecules (e.g. Ras and RhoB), nuclear proteins (e.g. CENP-E, CENP-F and nuclear lamins), Hsp40 chaperones (e.g. Ydj1p and DNJ3) and fungal mating pheromones (e.g. Saccharomyces cerevisiae a-factor). Because of the prominence of CaaX proteins in association with disease (e.g. Ras and cancer), it is generally hypothesized that interfering with CaaX modifications could be incorporated into disease intervention strategies. This hypothesis has led to the development of FTase inhibitors (FTIs) that are currently being investigated for the treatment of cancer and progeroid syndromes (Basso et al., 2006; Young et al., 2005). Inhibitors of the CaaX proteases and ICMT hold similar therapeutic potential and are being investigated (Blum et al., 2008; Manandhar et al., 2007; Wang et al., 2008; Winter-Vann and Casey, 2005). A problematic issue in this research area is the ability of CaaX proteins to be processed by partially redundant activities. For example, several proteins are known to be isoprenylated by GGTase I in the presence of FTIs (Fiordalisi et al., 2003; Sebti and Hamilton, 2000). Likewise, it is possible that targeted inhibition of Rce1p can lead to alternative processing by Ste24p, and vice versa. This issue is less of a concern for targeted inhibition of the ICMT because there appears to be no alternative enzyme that can perform the carboxyl methylation of CaaX proteins.
The two CaaX proteases are both ER-localized membrane proteins but are otherwise unrelated by primary sequence (Schmidt et al., 1998). Ste24p is a zinc-dependent metalloprotease that has been purified and demonstrated to possess in vitro CaaX proteolytic activity (Tam et al., 2001). The mechanism of Rce1p remains undefined. Several lines of evidence support the function of Rce1p as a CaaX protease, including genetic and overexpression studies (Bergo et al., 2002a; Boyartchuk et al., 1997b; Otto et al., 1999; Tam et al., 2001). Bioinformatic and inhibitor profiles suggest that it is a metalloprotease (Manandhar et al., 2007; Pei and Grishin, 2001).
Rce1p and Ste24p have partially overlapping substrate specificity, meaning that each enzyme has specific substrates and also shared ones. For example, Rce1p specifically modifies Ras GTPases, Ste24p specifically modifies prelamin A and both enzymes modify the yeast a-factor precursor (Bergo et al., 2002b; Boyartchuk et al., 1997b; Pendas et al., 2002). Yeast a-factor has been a convenient reporter for investigating CaaX modifications, because defects in any of the three post-translational events results in a sterile mating phenotype and because it can be used to readily monitor either Rce1p or Ste24p activity (Boyartchuk et al., 1997b). The yeast system is also useful for the evaluation of CaaX proteases from other eukaryotic species, because they all have the ability to recognize the yeast a-factor precursor as a substrate (Bracha et al., 2002; Cadiñanos et al., 2003a, 2003b; Plummer et al., 2006).
While investigating the substrate specificities of the yeast CaaX proteases using a-factor as a reporter, we observed the ability of certain CaaX motifs to promote yeast mating in the absence of Rce1p and Ste24p. This study compares Rce1p and Ste24p-independent mating (RSM) with mating promoted by the established CaaX proteases and provides evidence that a substantial number of CaaX motifs, including naturally occurring yeast motifs, can promote RSM. Moreover, we provide evidence that the yeast M16A metalloproteases Axl1p and Ste23p, which normally cleave an N-terminal extension found on the a-factor precursor, can enhance RSM, suggesting that these enzymes may be responsible for this activity.
Materials and methods
Yeast strains
The yeast strains used in this study are listed in Table 1. yWS829 was created by disrupting the STE14 gene in yWS164, using the BamHI–ClaI fragment from pSM284 (Sapperstein et al., 1994). The disruption was specific, as confirmed by PCR using appropriate primers flanking the sites of integration and Southern analysis using the URA3 cassette to probe a BamHI digest of genomic DNA prepared from the candidate disruption strain. Yeast strains were routinely grown at 30 °C on rich media (YEPD) or appropriate synthetic dropout media (SC–) when propagating plasmid-transformed strains (Michaelis and Herskowitz, 1988). Yeast DNA transformations were carried out according to published methods (Elble, 1992).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| IH1783 | MATa trp1 leu2 ura3 his4 | ATCC 204278 |
| IH1793 | MATα lys1 | ATCC 204279 |
| RC757 | MATα sst2-1 rme his6 met1 can1 cyh2 | Powers et al. (1986) |
| SM2331 | MATa trp1 leu2 ura3 his4 mfa1Δ mfa2Δ | Chen et al. (1997) |
| yWS164 | MATa trp1 leu2 ura3 his4 mfa1Δ mfa2Δ rce1::TRP1 ste24::KANR | Cadiñanos et al. (2003b) |
| yWS829 | MATa trp1 leu2 ura3 his4 mfa1Δ mfa2Δ rce1::TRP1 ste24::KANR ste14::URA3 | This study |
Yeast plasmids
The yeast plasmids used in this study are listed in Table 2. pWS610 and pWS612 were constructed by subcloning the appropriate NotI–XhoI fragment encoding a-factor from pSM1605 and pWS196, respectively, into pRS415. pWS817 was created similarly but with pWS654 and pRS315. pWS196 and all other a-factor-encoding plasmids bearing altered CaaX motifs were created by PCR-directed plasmid-based recombination (Oldenburg et al., 1997). The parent plasmid (i.e. pSM1605, pWS438 or pWS610) was treated with MluI and, in most instances, SphI to generate a gap in the 3′ untranslated region (UTR) of the MFA1 gene, very near the 3′ end of the open reading frame (ORF). The digested plasmid was co-transformed into yeast with a PCR product having sequence homology to the plasmid in regions flanking the restriction site(s) to allow for gap repair. The PCR product was generated using a mutagenic forward oligo that contained 39 bases of homology to the parent plasmid, nine bases encoding the desired aaX sequence and an 18–21 base extension for annealing to a template (i.e. pSM1605, pWS438 or pWS610). The reverse oligo was complementary to the vector outside the polylinker into which the MFA1-encoding fragment was subcloned; its use generates homology to the plasmid on the SphI side of the digested plasmid. Following co-transformation of the digested plasmid and PCR product, individual yeast colonies surviving appropriate selection (SC –Ura or SC –Leu) were screened for those containing a plasmid encoding the altered MFA1 gene, as determined by restriction enzyme mapping and subsequent sequencing of isolated plasmids; a silent site (e.g. SphI or PstI) was typically incorporated along with the desired mutation. All plasmids derived from pWS438 were converted to low-copy plasmids by subcloning the NotI–XhoI MFA1-encoding fragment into pRS315 at the same sites. pWS196 was the only plasmid derived from pSM1605 and was manipulated as described above.
Table 2.
Plasmids used in this study
| Plasmid | Genotype | Reference |
|---|---|---|
| P80 | CEN URA3 AXL1 | Adames et al. (1995) |
| pRS315 | CEN LEU2 | Sikorski and Hieter (1989) |
| pRS316 | CEN URA3 | Sikorski and Hieter (1989) |
| pRS415 | CEN LEU2 | Sikorski and Hieter (1989) |
| pRS424 | 2µ TRP1 | Sikorski and Hieter (1989) |
| pSM284 | integrating ste14:URA3 | Sapperstein et al. (1994) |
| pSM703 | 2µ URA3 PPGK | Zhang et al. (2001) |
| pSM1107 | CEN URA3 HA::STE24 | Fujimura-Kamada et al. (1997) |
| pSM1153 | CEN TRP1 AXL1 | Schmidt et al. (1998) |
| pSM1366 | CEN URA3 UBI-MFA1 M | Tam et al. (1998) |
| pSM1368 | CEN URA3 UBI-MFA1 P1 | Tam et al. (1998) |
| pSM1369 | CEN URA3 UBI-MFA1 P2 | Tam et al. (1998) |
| pSM1605 | 2µ URA3 MFA1 | Schmidt et al. (2000) |
| pSM1314 | CEN URA3 RCE1::HAc | Schmidt et al. (1998) |
| pWS196 | 2µ URA3 MFA1-CASQ | This study |
| pWS438 | 2µ LEU2 MFA1 | Cadiñanos et al. (2003b) |
| pWS601 | 2µ URA3 PPGK-AXL1 | This study |
| pWS602 | 2µ URA3 PPGK-STE23-BglII | This study |
| pWS610 | CEN LEU2 MFA1 | This study |
| pWS612 | CEN LEU2 MFA1-CASQ | This study |
| pWS654 | 2µ LEU2 MFA1-CALQ | Plummer et al. (2006) |
| pWS718 | 2µ LEU MFA1-CKQQ | This study |
| pWS727 | CEN LEU2 MFA1-CKQQ | This study |
| pWS737 | CEN LEU2 MFA1-CAQQ | This study |
| pWS738 | CEN LEU2 MFA1-CKAQ | This study |
| pWS739 | CEN LEU2 MFA1-CKQA | This study |
| pWS817 | CEN LEU2 MFA1-CALQ | This study |
| pWS844 | CEN LEU2 MFA1-CRQQ | This study |
| pWS845 | CEN LEU2 MFA1-CKIS | This study |
| pWS846 | CEN LEU2 MFA1-CKQS | This study |
| pWS847 | CEN LEU2 MFA1-CKGE | This study |
| pWS848 | CEN LEU2 MFA1-CKCI | This study |
| pWS849 | CEN LEU2 MFA1-CKYI | This study |
| pWS850 | CEN LEU2 MFA1-CKCT | This study |
| pWS851 | CEN LEU2 MFA1-CEQQ | This study |
| pWS852 | CEN LEU2 MFA1-CHQQ | This study |
| pWS853 | CEN LEU2 MFA1-CDQQ | This study |
| pWS854 | CEN LEU2 MFA1-CRVK | This study |
| pWS855 | CEN LEU2 MFA1-CRNR | This study |
| pWS856 | CEN LEU2 MFA1-CRMV | This study |
| pWS883 | 2µ LEU2 MFA1-CKVA | This study |
| pWS884 | 2µ LEU2 MFA1-CRVA | This study |
| pWS885 | 2µ LEU2 MFA1-CRMS | This study |
| pWS886 | 2µ LEU2 MFA1-CRVN | This study |
| pWS887 | 2µ LEU2 MFA1-CKMT | This study |
| pWS891 | 2µ LEU2 MFA1-CKIT | This study |
| pWS892 | CEN URA3 UBI-MFA1 P1 (CKQQ) | This study |
| pWS893 | CEN URA3 UBI-MFA1 P2 (CKQQ) | This study |
| pWS894 | CEN URA3 UBI-MFA1 M (CKQQ) | This study |
| pWS912 | CEN LEU2 MFA1-CRME | This study |
pWS601 and pWS602 were also created by PCR-directed plasmid-based recombination. These plasmids encode AXL1 and STE23, respectively, behind the constitutive phosphoglycerate kinase (PGK) promoter. pSM703 was the recipient vector used in the construction of these plasmids, which was gapped within its polylinker prior to use.
Constructs encoding ubiquitin fusions were created by PCR-directed plasmid-based recombination, essentially as described above for the creation of a-factor CaaX motif mutants. A PCR fragment encoding the CKQQ motif was derived from pWS718 and recombined into MluI-linearized pSM1368 and pSM1369 to create pWS892 and pWS893, respectively. To create pWS894, a PCR fragment also derived from pWS718 was produced that would incorporate the DNA sequence encoding mature a-factor upon recombination into MluI-linearized pWS892.
Serial dilution mating assay
The ability of the various CaaX motifs to promote a-factor maturation was judged using a genetic assay that scores diploid formation resulting from the mating of haploid mating partners. The MATa haploid strain used (yWS164) lacks both CaaX protease-encoding genes and a-factor-encoding genes (Cadiñanos et al., 2003b). Mating competence was restored in this strain by co-transformation with plasmids encoding an a-factor species and a CaaX protease. Transformation with the latter was not necessary in the case of certain a-factor CaaX variants.
In brief, the serial dilution mating assay involves the mixing of MATa and MATα cell suspensions on a medium selective for diploid growth (Plummer et al., 2006). The cultures are prepared by first growing the MATa yeast in selective medium and the MATα yeast in non-selective YEPD for 24 h, then normalizing the cultures to a cell density of A600 = 1.00 ± 0.05 with appropriate sterile medium. A portion of each normalized MATa culture was diluted 10-fold with a normalized MATα culture, such that the final volume of the mating mixture was 100 µl. This primary mixture was subjected to several additional 10-fold dilutions, using normalized MATa cells as the diluent, until a set of five samples was prepared. A portion of each serially diluted mixture (5 µl) was spotted onto solid SD medium. Growth of diploid cells on SD medium was scored after 3–4 days growth at 30 °C. The results of the mating test were digitally recorded by scanning the plates, using a standard flat bed scanner. Unless otherwise noted, images are representative of mating results observed within one experiment where the indicated yeast strains were evaluated as a set to facilitate better assessment of relative mating efficiencies.
Quantitative mating assay
Assays were performed essentially as previously described (Michaelis and Herskowitz, 1988). In brief, MATa yeast were cultured for 36 h to saturation in SC –Ura –Leu medium; MATα yeast were cultured for 24 h in YEPD liquid. Ten-fold serial dilutions of MATa yeast were prepared in SC –Ura –Leu liquid in triplicate, and a portion (100 µl) of an empirically determined dilution was mixed with an equal volume (100 µl) of undiluted MATα yeast. The mixtures were spread onto SD solid minimal medium. In parallel, a portion (100 µl) of the MATa 10 dilution was spread onto SC –Ura –Leu solid medium to derive the titre of viable cells in the sample. The number of colonies observed on the SD plate after 3 days of growth was recorded, adjusted for the dilution factor and normalized for the titre of viable cells. Normalized values were used to determine mating efficiencies relative to a wild-type strain (IH1783 containing pRS315 and pRS316) that was defined as having 100% mating efficiency. The MATa dilutions were chosen such that the resultant number of diploid colonies was typically in the range 10–100/plate, or in the case of non-maters an undiluted sample was used.
Genetic screen to identify CaaX motifs that permit RSM
A library of plasmids encoding all possible permutations of the CaaX motif appended to yeast a-factor was created in yWS164. The individual plasmid-bearing colonies were assessed by replica methods for the ability to produce a-factor. Both mating and halo assays were used (Nijbroek and Michaelis, 1998). In brief, the population of transformants was replica-plated onto separate lawns of IH1793 and RC757. The lawns were prepared by scraping freshly grown strains from a YEPD plate (i.e. 48 h growth at 30 °C), diluting the cells into liquid YEPD, adjusting the density to A600 = 1.00 ±0.05, pouring the cell suspension onto a plate of SD (IH1793) or YEPD (RC757) (~3–5 ml/plate), immediately decanting the majority of the surface liquid, and allowing the residual liquid to absorb for 30 min at room temperature. The replica-printed plates were incubated at 30 °C for 120 h (IH1793 lawn) or 16 h (RC757 lawn) to allow for the growth of diploids and the formation of halos, respectively. Plasmids were isolated from colonies exhibiting mating competence and the ability to growth arrest RC757 cells (Robzyk and Kassir, 1992). The plasmids were retransformed into yWS164, the phenotypes reconfirmed and plasmids sequenced.
The plasmid library was created by plasmid-based PCR-directed recombination. pWS654 was gapped with PstI, which cuts within the sequence encoding the aaX portion of the MFA1 gene, and MluI, which cuts 3′ of the MFA1 ORF. The forward oligo used to generate the PCR fragment had 39 bases of homology to the MFA1 gene (5′ to PstI cut site), a nine-base sequence that was randomized for every possible nucleotide combination (i.e. the randomized aaX sequence), and 24 bases for annealing of the primer to the pWS438 plasmid used for target amplification; the first codon of the 24 base sequence encoded a stop codon. The reverse primer was homologous to DNA just outside the polylinker of pWS438 into which the MFA1 gene was subcloned. This sequence is also present on pWS654. The plasmid-derived DNA fragments and the PCR-generated DNA fragments were co-transformed into yWS164 to facilitate recombination events that formed plasmids allowing for selective growth of the yeast on SC –Leu medium.
Results
a-factor-CKQQ promotes Rce1p and Ste24p-independent mating (RSM)
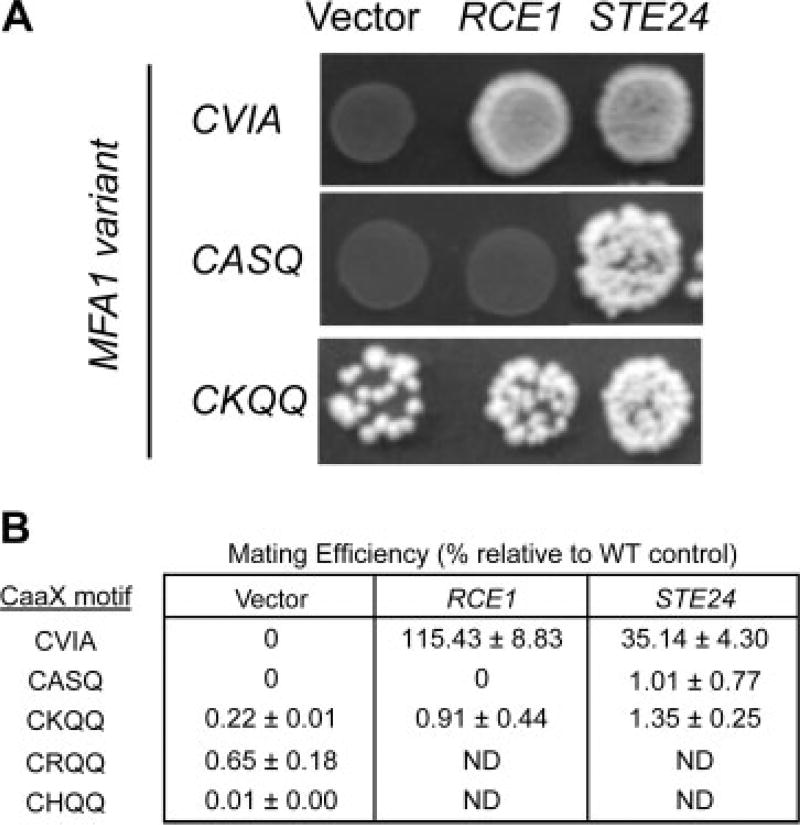
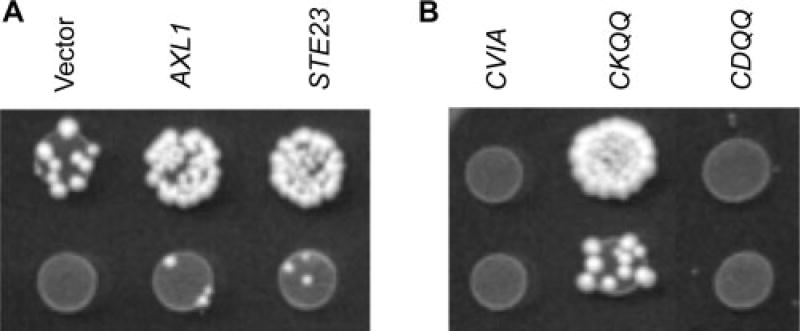
The specificities of the yeast Rce1p and Ste24p CaaX proteases can be monitored using the yeast a-factor mating pheromone as a reporter molecule. During such an investigation, we observed mating by a strain expressing the a-factor-CKQQ variant in the absence of the established CaaX proteases (Figure 2A). The CKQQ motif was derived from Pex19p and is also present on the mammalian Ser/Thr kinase Lkb1, a known tumour suppressor. Both proteins are known to be isoprenylated and thus substrates for CaaX proteolysis (Collins et al., 2000; Gotte et al., 1998). RSM was not observed when either wild-type a-factor (CVIA) or a variant known to be Ste24p-specific (CASQ) was expressed.
Figure 2.
Evidence for Rce1p and Ste24p–independent mating (RSM). (A) The a-factor-CKQQ variant promotes mating in a yeast strain lacking endogenous copies of the CaaX proteases and a-factor genes (yWS164). This phenotype is not associated when a-factor is appended with its natural CaaX motif (CVIA) or one that is Ste24p–specific (CASQ). When co-expressed with Ste24p, but not Rce1p, the CKQQ variant promotes more efficient mating. The plasmids used were pRS316, pSM1107, pSM1314, pWS610, pWS612 and pWS727. (B) Quantitative mating tests were conducted to quantifiably compare the amount of mating promoted by a-factor CaaX motif variants. Mating efficiencies are reported relative to a wild-type control (IH1783) containing both CaaX protease genes, both a-factor genes and empty vectors (pRS315 and pRS316) to maintain the same plasmid markers as the tested strains. The plasmids used were pSM1314 (RCE1), pSM1107 (STE24), pWS610 (CVIA), pWS612 (CASQ), pWS727 (CKQQ), pWS844 (CRQQ) and pWS852 (CHQQ), which were evaluated alone and in combination. In instances where plasmids were evaluated alone, an appropriate empty vector was included to maintain markers. ND, not determined
Through close inspection of diploid colony densities, we predicted that Ste24p, and to a lesser extent Rce1p, could enhance CKQQ-dependent RSM. This prediction was confirmed through quantitative mating tests (Figure 2B) (Michaelis and Herskowitz, 1988). We also confirmed that the CVIA motif was readily cleaved by Rce1p and to a lesser extent by Ste24p in a manner consistent with the reported properties of these enzymes (Boyartchuk et al., 1997a; Trueblood et al., 2000). We also determined that the Ste24p/CKQQ and Ste24p/CASQ pairings had approximately equal mating efficiencies. Overall, our analysis revealed that RSM mating efficiency was low (<1%), but within an order of magnitude of that observed for CASQ and CKQQ motifs in the presence of CaaX proteases.
Other CaaX motifs also support RSM
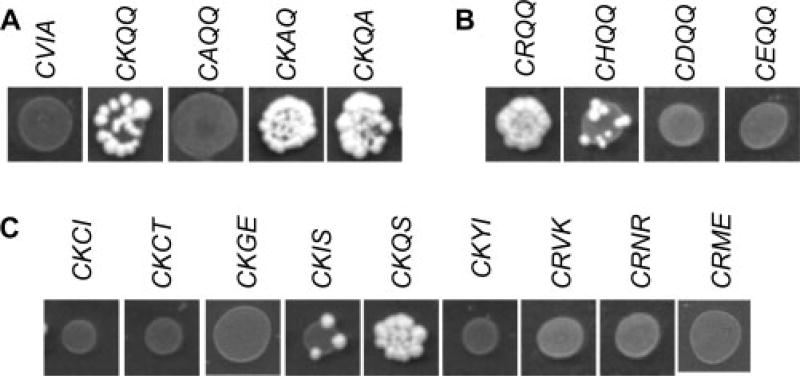
To investigate the extent of motifs that support RSM, the a1, a2 and X positions of the CKQQ motif were independently altered to Ala. This analysis revealed that Lys at the a1 position was a critical determinant for RSM (Figure 3A). Alterations at the a2 and X position did not abolish RSM. Both CKAQ and CKQA appeared to support more efficient mating than the CKQQ motif. The possibility of a charge requirement at the a1 position was investigated in more detail by substituting various polar amino acids (Figure 3B). Of the motifs evaluated, CRQQ and CHQQ promoted RSM while CDQQ and CEQQ did not. The CRQQ motif appeared to support more efficient mating than CKQQ, while the CHQQ motif appeared to support less efficient mating. This observation was confirmed by quantitative mating tests (Figure 2B).
Figure 3.
Multiple CaaX motifs promote RSM. Serial dilution mating tests were conducted as described in Figure 2, using yWS164 and plasmids encoding the indicated a-factor CaaX variants. Only the first dilution spot is shown for each mating test. (A) Ala substitutions at the a1, a2 and X positions of the CKQQ motif. The plasmids used were pWS610, pWS727, pWS737, pWS738 and pWS739. (B) Charged amino acid substitutions at the a1 position. The plasmids used were pWS610, pWS727, pWS844, pWS851, pWS852 and pWS853. (C) Naturally occurring motifs that correspond to the consensus C(K/R/H)aX. See Table 3 for the source gene of the natural CaaX motifs. The plasmids used were pWS845, pWS846, pWS847, pWS848, pWS849, pWS850, pWS854, pWS855 and pWS912
Given our findings, we hypothesized that the C(K/R/H)aX motif might be a good predictor of RSM substrates. To test this hypothesis, we examined additional natural yeast CaaX motifs and several synthetic sequences (i.e. not occurring in yeast) corresponding to this consensus. Only a subset of these motifs promoted mating when appended to a-factor (Figure 3C, Table 3). These observations indicate that C(K/R/H)aX can be used to identify candidate substrates for RSM, but that this consensus sequence is not an absolute predictor of RSM substrates. Our result was somewhat expected, since the consensus-matching CKIA motif has been previously identified as not promoting mating activity (Trueblood et al., 2000).
Table 3.
Summary of ability of CaaX motifs to promote Rce1p and Ste24p-independent mating (RSM)
| Motif | RSM Observed | Sourcea |
|---|---|---|
| CALM | Nob | Synthetic |
| CALQ | Nob | Synthetic |
| CAMQ | Nob | Synthetic |
| CAQQ | No | Synthetic |
| CASQ | No | YDJ1 |
| CDQQ | No | Synthetic |
| CEQQ | No | Synthetic |
| CHQQ | Yes | Synthetic |
| CKAQ | Yes | Synthetic |
| CKCI | No | POP8 |
| CKCT | No | YBL049w/MOH1 |
| CKGE | No | YMR197c/VTI1 |
| CKIS | Yes | YPR092wc |
| CKIT | Yes | RSM screen |
| CKMT | Yes | RSM screen |
| CKQA | Yes | Synthetic |
| CKQQ | Yes | PEX19, LKB1, NAP1 |
| CKQS | Yes | NAP1 |
| CKVA | Yes | RSM screen |
| CKYI | No | YMR060c/SAM37/TOM37 |
| CRME | No | YJL059W/YHC3 |
| CRMS | Yes | RSM screen |
| CRMV | Yesb | Synthetic |
| CRNR | No | YML041C/VPS71 |
| CRQQ | Yes | Synthetic |
| CRVA | Yes | RSM screen |
| CRVK | No | YMR158W/MRPS8 |
| CRVN | Yes | RSM screen |
| CVIA | No | MFA1, MFA2 |
All the indicated genes are those of S. cerevisiae except for LKB1 and NAP1, which are human.
Krishnankutty and Schmidt, unpublished observation; see also Plummer et al. (2006).
Reported as a dubious ORF in the Saccharomyces Genome Database (www.yeastgenome.org).
A relatively large number of CaaX motifs can support RSM
To broadly investigate the propensity of CaaX motifs to promote mating in the absence of Rce1p and Ste24p, we set up a genetic screen to identify motifs capable of producing biologically active a-factor in an rce1 ste24 null background. For the screen, a degenerate PCR oligonucleotide was used to create a population of plasmids encoding yeast a-factor with randomly appended aaX sequences. Theoretically, 8000 aaX permutations were possible. The plasmid library was created in yeast through recombination-mediated methods. Evaluation of over 3000 yeast colonies by replica-based mating tests revealed a substantial number having the ability to mate and induce growth arrest of MATα sst2-1 yeast. RSM was observed at a rate of 0.93% ± 0.61%, suggesting that approximately 75 CaaX motifs can promote RSM. Six plasmids capable of promoting RSM were recovered and sequenced. This limited analysis revealed sequences having either Lys or Arg at the a1 position but no consistent pattern at the a2 and X positions (Table 3). Future investigations to identify additional RSM motifs will be required to fully define the RSM consensus.
RSM is dependent on Ste14p
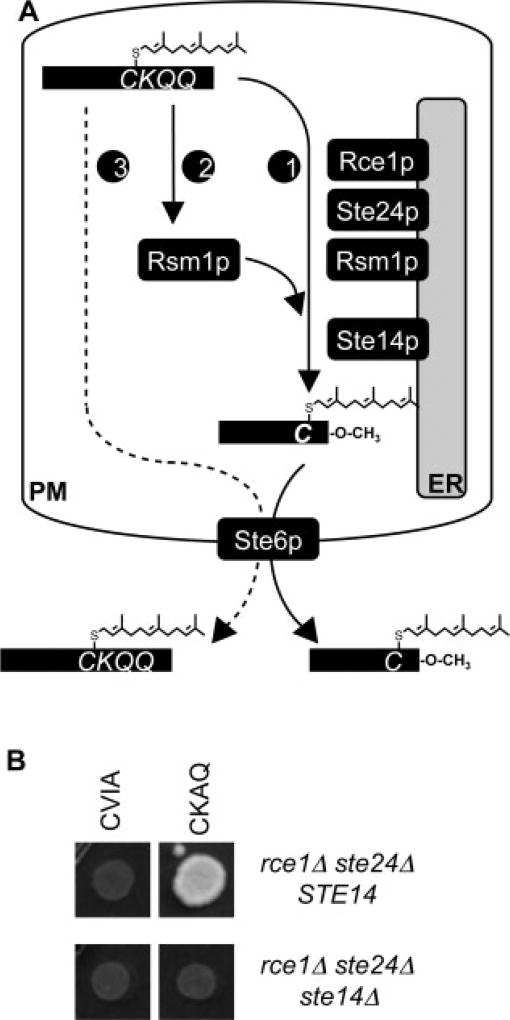
Two hypotheses were developed to explain our observations for RSM. The most straightforward was that a third CaaX proteolytic activity is responsible for RSM (Figure 4A). Alternatively, it was possible that certain RSM-promoting motifs were uncleaved, and that the uncleaved motifs somehow mimicked the biophysical properties of a carboxyl-methylated C-terminus, such that cellular export and receptor binding by the pheromone were now possible. To distinguish between these possibilities, we predicted that a proteolytic-dependent mechanism would require the isoprenylcysteine carboxyl methyltransferase (ICMT) for activation of the biological activity of a-factor, whereas a carboxyl-methyl mimic would not. We thus evaluated the dependence of RSM on the Ste14p ICMT. Using a-factor-CKAQ as a reporter, we observed that RSM was indeed dependent on Ste14p (Figure 4B). This observation strongly implicates involvement of a proteolytic activity in promoting RSM.
Figure 4.
RSM requires carboxylmethylation. (A) Models for RSM. In the absence of Rce1p and Ste24p, CaaX proteolysis of a-factor-CKQQ is mediated by either an ER (1) or a non-ER-localized (2) protease (Rsm1p). If the latter, the dependence of RSM on Ste14p indicates that a trafficking step is required to return proteolysed a-factor to the Ste14p ICMT that resides at the ER. Alternatively, non-proteolysed a-factor could promote mating (3). In this scenario, RSM would be independent of Ste14p. ER, endoplasmic reticulum; PM, plasma membrane. (B) Loss of Ste14p in a CaaX protease-deficient strain prevents RSM. The plasmids used were pWS610 and pWS738, which were separately transformed into yWS164 and yWS829. yWS164-derived strains were additionally transformed with pRS316 to provide a comparable set of auxotrophic markers to that of the yWS829-derived strains
However, our interpretation is subject to the concern that another CaaX protein might have impaired function in the absence of STE14, and that this impairment contributes to the negative mating phenotype observed. Unlike a-factor, however, no other CaaX protein has been identified whose function is fully impaired in the absence of carboxylmethylation. Nevertheless, it remains formally possible that the function of some CaaX protein, perhaps one involved in cell fitness (e.g. Ras2p) or the mating response (e.g. Ste18p), is partially compromised in the absence of STE14, such that the weak mating observed with a-factor-CKQQ is now below the detection threshold of our methods.
RSM is enhanced by the yeast M16A proteases Axl1p and Ste23p
To further advance the hypothesis that RSM is promoted by a proteolytic activity, we sought to identify protease gene(s) involved. Using a candidate approach, we first examined other proteases associated with a-factor maturation, specifically the M16A subfamily proteases Axl1p and Ste23p. These proteases independently cleave an N-terminal extension found on the a-factor precursor during a-factor biogenesis, with Axl1p being responsible for the majority of this activity (Kim et al., 2005). When overexpressed, each protease was capable of enhancing RSM associated with a-factor-CKQQ (Figure 5A). Protease overexpression did not promote RSM in the presence of wild-type a-factor or a charge switch mutant (CVIA and CDQQ, respectively) (Figure 5B). This observation suggests that the effect of protease overexpression is linked to recognition of a specific subset of CaaX motifs. Our results are consistent with Axl1p and Ste23p contributing to the proteolytic activity that promotes RSM, but do not exclude the possibility of the M16A proteases activating a secondary protease having this role.
Figure 5.
RSM is enhanced by M16A proteases. (A) Axl1p and Ste23p were independently overexpressed in yWS164 in the presence of the a-factor CKQQ variant and the strains were subjected to a serial dilution mating test, as described in Figure 2. The first two dilution spots are shown. The plasmids used were pRS316, pWS601, pWS602 and pWS727. (B) The ability of overexpressed Ste23p to enhance RSM was evaluated for CaaX motifs that were previously identified as either not promoting (e.g. CVIA and CDQQ) or promoting RSM (e.g. CKQQ). The plasmids used were pWS602, pWS610, pWS727 and pWS853. The experiments shown in (A, B) were performed on separate days and a slight variation in mating efficiency is apparent for the same strain, which is labelled STE23 in (A) and CKQQ in (B)
The N-terminal extension of a-factor is important for RSM
Given the possible and likely involvement of M16A enzymes in cleaving RSM-promoting CaaX motifs, we hypothesized that the N-terminal extension of a-factor would somehow be involved in regulating RSM. The first third of this N-terminal extension is removed by Ste24p to yield a partial extension, which is subsequently fully removed by the activity of a yeast M16A enzyme (Tam et al., 1998). By analogy to substrates of other M16A enzymes, the partial N-terminal extension presumably binds an exosite on the M16A enzyme (Shen et al., 2006).
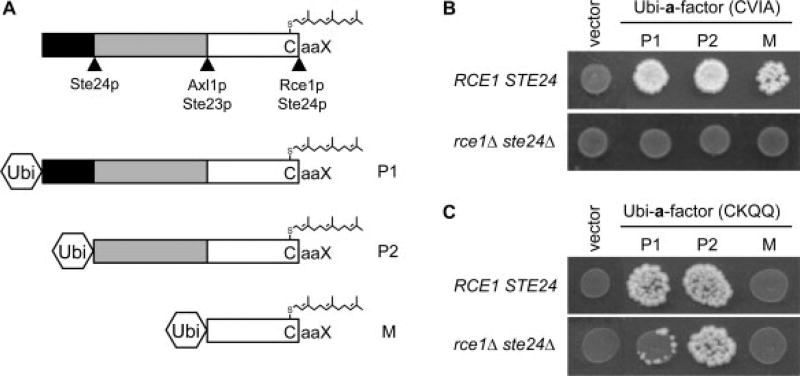
First, we examined whether the N-terminal extension shields the natural a-factor CaaX motif but not RSM-promoting CaaX motifs from M16A recognition. To test this possibility, a-factor was expressed with and without its N-terminal extension, using a ubiquitin fusion approach that allows expression of very short peptides (Boyartchuk and Rine, 1998; Tam et al., 1998). The fusions incorporated the full-length a-factor precursor sequence (Ubi-P1), a truncated sequence reflecting partial loss of the N-terminal extension (Ubi-P2) or the mature sequence of a-factor (Ubi-M) (Figure 6A). The fusions had an associated CaaX motif, either CVIA or CKQQ. None of the fusions containing the wild-type CaaX motif CVIA were capable of promoting RSM despite encoding functional a-factor products (Figure 6B), indicating that the N-terminal extension does not simply shield CaaX motifs from M16A recognition. Of note, Ubi-M appeared less effective at promoting mating in the presence of CaaX proteases relative to its longer counterparts, which is consistent with its rapid turnover in cells (Tam et al., 1998).
Figure 6.
The N-terminal extension of a-factor modulates RSM. (A) Diagram depicting the proteolytic cleavage sites in the a-factor precursor and ubiquitin fusions created to bypass certain proteolytic steps associated with a-factor biogenesis. See text for additional details on the ubiquitin fusions. (B) Ubiquitin fusions to various lengths of the a-factor precursor were expressed in either SM2331 or yWS164 and the strains were subjected to a serial dilution mating test, as described in Figure 2. The plasmids used were pRS316, pSM1366, pSM1368 and pSM1369. (C) A mating test was performed as described in (B), using ubiquitin–a-factor fusions appended with the CKQQ motif. The plasmids used were pRS316, pWS892, pWS893 and pWS894
We next examined whether the N-terminal extension is needed to recruit the a-factor precursor to the ‘peptidosome’ cavity of the M16A enzyme, where CaaX motifs with appropriate biophysical properties (i.e. RSM promoting motifs) would be cleaved. To test this possibility, ubiquitin-a-factor fusions having a CKQQ motif were evaluated (Figure 6C). In the presence of CaaX proteolytic activity, all of the ubiquitin fusions had reduced ability to promote mating relative to their CVIA counterparts, with Ubi-M(CKQQ) being completely incapable of promoting mating. RSM was observed with Ubi-P1(CKQQ) and Ubi-P2(CKQQ) but not with Ubi-M(CKQQ). The lack of recognition of Ubi-M(CKQQ) by both Rce1p and Ste24p suggests that the N-terminal extension is a recognition and/or a targeting determinant that may facilitate interaction with these CaaX proteases. The fact that Ubi-P2(CKQQ) promotes mating somewhat better than Ubi-P1(CKQQ) is consistent with our assertion that the N-terminal extension of a-factor, specifically the partial extension present on the P2 intermediate, is important for RSM. However, the fact that Ubi-M(CKQQ) cannot promote mating in the presence of CaaX proteases precludes our ability to conclusively demonstrate an essential recruitment role for the N-terminal extension in RSM.
Discussion
We have identified multiple CaaX motifs that, when used in lieu of the natural a-factor CVIA motif, can promote yeast mating in the absence of the established CaaX proteases Rce1p and Ste24p. Rce1p-and Ste24p-independent mating (RSM) can be promoted by several CaaX motifs naturally present in the yeast genome as well as multiple synthetic sequences (Table 3). Our genetic analysis allows us to project that approximately 75 motifs can promote RSM. These motifs represent approximately 1% of all possible CaaX permutations. The fact that RSM-promoting motifs have been previously overlooked is probably not surprising, given that only about 1% of CaaX motifs have previously been evaluated in the context of the a-factor reporter. (Boyartchuk et al., 1997b; Plummer et al., 2006; Trueblood et al., 2000). Moreover, with the exception of CKIA, none of the previously evaluated motifs were matches for the consensus sequence C(K/R/H)aX, which we have derived as a good but not absolute predictor of an RSM-promoting motif. Sequencing of all the RSM-promoting motifs identified by our unbiased genetic screen may provide additional insight into whether there is an RSM consensus motif. Evaluation of existing motifs suggests that a charged residue at the X position may not be compatible with RSM.
Our observations are consistent with RSM involving proteolysis of the susceptible motifs and identify the yeast M16A metalloproteases Axl1p and Ste23p as having involvement in this process. We do not know whether these proteases act indirectly or directly. Indirect action could be as simple as M16A enzymes activating a distinct protease having RSM activity, or as complex as M16A enzymes serving as scaffolds to help recruit such an activity or to properly present substrates to this activity. Determining whether a direct or indirect scenario is more likely will require purification of the yeast M16A enzymes and the synthesis of a compatible substrate. The hypothesis that yeast M16A enzymes can directly cleave CaaX motifs, however, is supported by additional observations. Three yeast activities, one membrane-associated and two soluble, have been previously identified that are able to cleave the a-factor CaaX motif in vitro (Ashby et al., 1992; Hrycyna and Clarke, 1992). The membrane-associated activity is likely a combination of Rce1p and Ste24p activities, and neither can be responsible for RSM due to their absence in our test strain. One of the soluble activities is a PEP4-dependent carboxypeptidase, most likely carboxypeptidase Y. This enzyme is a compartmentalized vacuolar protease and is not expected to come in contact with a-factor intermediates, which are hypothesized to be modified by enzymes having cytosol-orientated active sites; mature a-factor is exported directly from the cytosol and across the plasma membrane by the Ste6p ABC-type transporter (Kuchler et al., 1989). The second soluble in vitro activity is associated with an undefined 110 kDa enzyme and is phenanthroline-sensitive. Axl1p and Ste23p are approximately this size and are both predicted to be phenanthroline-sensitive, based on their functional homology to other M16A proteases, such as the human insulin-degrading enzyme (IDE) (Kim et al., 2005). Thus, they are likely responsible for the in vitro activity reported. However, one major inconsistency remains between our in vivo observations and the reported 100 kDa in vitro activity that cleaves CaaX motifs; we do not observe cleavage of the CVIA CaaX motif in vivo, whereas this is observed in vitro. A major difference between the two types of experiments is, respectively, the use of a full-length biologically active reporter vs. a shorter peptide-based biologically inactive reporter, and this could underlie the specificity differences observed.
Another issue that remains to be resolved is the physiological impact of M16A enzymes on the maturation of CaaX proteins having RSM-promoting motifs. We believe that M16A enzymes cleave RSM-promoting motifs only in the specific context of the yeast a-factor reporter. This conclusion is based on the observation that the a-factor CKQQ variant, when produced without its N-terminal extension, is an unsuitable substrate for RSM. The N-terminal extension, by analogy to other M16A substrates, is likely involved in binding to the exosite of M16A enzymes. This extension helps anchor the substrate in the so-called ‘peptidosome’ cavity of the M16A enzyme. We thus hypothesize that certain CaaX motifs, once drawn into the cavity of an M16A enzyme, have the propensity to compete against the ideal M16A cleavage sequence for binding to the active site, perhaps by their ability to form extended β-sheet interactions with the M16A enzyme. The ability of other substrates to behave similarly would be limited to those having an exosite binding sequence and being small enough to fit within the M16A enzyme cavity, which is predicted to hold proteins <50 amino acids in size (Shen et al., 2006). None of the naturally occurring yeast proteins having RSM-promoting motifs are small enough to fit in an M16A cavity. Hence, we believe that these proteins are not modified to any appreciable extent by M16A enzymes. However, we cannot discount the possibility that small CaaX proteins with RSM-promoting motifs exist in other systems that would be suitable M16A substrates in those systems. If such candidates exist in humans, IDE might be able to help mature these proteins under conditions where CaaX protease inhibition is a therapeutic strategy.
Acknowledgments
We are grateful to members of the Schmidt and Sabatini Laboratories, University of Georgia, for technical advice and critical discussions. This work was supported in part through a Georgia Cancer Coalition Distinguished Cancer Clinician/Scientist Scholar Award (to WKS) and a grant from the National Institutes of Health (to WKS; R01 GM67092).
References
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Ashby M, King D, Rine J. Endoproteolytic processing of a farnesylated peptide in vitro. Proc Natl Acad Sci USA. 1992;89:4613–4617. doi: 10.1073/pnas.89.10.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Ambroziak P, Gregory C, et al. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol. 2002a;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino B, Ross J, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA. 2002b;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, et al. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- Blum R, Cox AD, Kloog Y. Inhibitors of chronically active ras: potential for treatment of human malignancies. Recent Patents Anticancer Drug Discov. 2008;3:31–47. doi: 10.2174/157489208783478702. [DOI] [PubMed] [Google Scholar]
- Boyartchuk V, Trueblood C, Vik A, Rine J. Yeast Cell Biology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997a. Roles of CAAX proteases in a-factor and Ras maturation. [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997b;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Rine J. Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics. 1998;150:95–101. doi: 10.1093/genetics/150.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha K, Lavy M, Yalovsky S. The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J Biol Chem. 2002;277:29856–29864. doi: 10.1074/jbc.M202916200. [DOI] [PubMed] [Google Scholar]
- Cadin˜anos J, Schmidt WK, Fueyo A, et al. Identification, functional expression and enzymic analysis of two distinct CaaX proteases from Caenorhabditis elegans. Biochem J. 2003a;370:1047–1054. doi: 10.1042/BJ20021514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadin˜anos J, Varela I, Mandel D, et al. AtFACE-2, a prenylated-protein protease from Arabidopsis thaliana related to Ras converting enzymes. J Biol Chem. 2003b;278:42091–42097. doi: 10.1074/jbc.M306700200. [DOI] [PubMed] [Google Scholar]
- Chen P, Sapperstein S, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SP, Reoma JL, Gamm DM, Uhler MD. LKB1, a novel serine/threonine protein kinase and potential tumour suppressor, is phosphorylated by cAMP-dependent protein kinase PKA and prenylated in vivo. Biochem J. 2000;345(3):673–680. [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Fiordalisi JJ, Johnson RL, II, Weinbaum CA, et al. High affinity for farnesyltransferase and alternative prenylation contribute individually to K-Ras4B resistance to farnesyltransferase inhibitors. J Biol Chem. 2003;278:41718–41727. doi: 10.1074/jbc.M305733200. [DOI] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte K, Girzalsky W, Linkert M, et al. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna CA, Clarke S. Maturation of isoprenylated proteins in Saccharomyces cerevisiae. J Biol Chem. 1992;267:10457–10464. [PubMed] [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ambroziak P, Otto JC, et al. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- Kim S, Lapham A, Freedman C, et al. Yeast as a tractable genetic system for functional studies of the insulin-degrading enzyme. J Biol Chem. 2005;280:27481–27490. doi: 10.1074/jbc.M414192200. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar SP, Hildebrandt ER, Schmidt WK. Small-molecule inhibitors of the Rce1p CaaX protease. J Biomol Screen. 2007;12:983–993. doi: 10.1177/1087057107307226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S, Caldwell GA, Miller D, et al. Significance of C-terminal cysteine modifications to the biological activity of the Saccharomyces cerevisiae a-factor mating pheromone. Mol Cell Biol. 1991;11:3603–3612. doi: 10.1128/mcb.11.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijbroek GL, Michaelis S. Functional assays for the analysis of yeast ste6 mutants. Methods Enzymol. 1998;292:193–212. doi: 10.1016/s0076-6879(98)92016-x. [DOI] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JC, Kim E, Young SG, Casey PJ. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- Pendas AM, Zhou Z, Cadinanos J, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Plummer LJ, Hildebrandt ER, Porter SB, et al. Mutational analysis of the ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem. 2006;281:4596–4605. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S, Michaelis S, Broek D, et al. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986;47:413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Kassir Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesyl-cysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Michaelis S. Reconstitution of the Ste24p-dependent N-terminal proteolytic step in yeast a-factor biogenesis. J Biol Chem. 2000;275:6227–6233. doi: 10.1074/jbc.275.9.6227. [DOI] [PubMed] [Google Scholar]
- Sebti SM, Hamilton AD. Farnesyltransferase and geranyl-geranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence R, Casey PJ. Mechanism of catalysis by protein farnesyltransferase. In: Tamanoi F, Sigman D, editors. The Enzymes. Academic Press; New York: 2001. pp. 1–18. [Google Scholar]
- Tam A, Nouvet F, Fujimura-Kamada K, et al. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A, Schmidt WK, Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J Biol Chem. 2001;276:46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Picologlou EA, et al. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tan W, Zhou J, et al. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–18684. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- Young SG, Ambroziak P, Kim E, Clarke S. Postisoprenylation protein processing: CXXX CaaX endoproteases and isoprenylcysteine carboxyl methyltransferase. In: Tamanoi F, Sigman DS, editors. The Enzymes. Academic Press; New York: 2001. pp. 155–213. [Google Scholar]
- Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria — new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, et al. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]