Abstract
The Cre recombinase (Cre) from bacteriophage P1 is an important tool for genetic engineering in mammalian cells. We constructed lentiviral vectors that efficiently deliver Cre in vitro and in vivo. Surprisingly, we found a significant reduction in proliferation and an accumulation in the G2/M phase of Cre-expressing cells. To minimize the toxic effect of Cre, we designed a lentiviral vector that integrates into the host genome, expresses Cre in the target cell, and is subsequently deleted from the genome in a Cre-dependent manner. Thus, the activity of Cre terminates its own expression (self-deleting). We showed efficient modification of target genes in vitro and in the brain after transduction with the self-deleting vectors. In contrast to sustained Cre expression, transient expression of Cre from the self-deleting vector induced significantly less cytotoxicity. Such a self-deleting Cre vector is a promising tool for the induction of conditional gene modifications with minimal Cre toxicity in vivo.
Keywords: gene transfer, regulated gene expression, Cre toxicity
The site-specific recombinase of the bacteriophage P1, Cre, catalyzes the recombination of two 34-bp sequences called loxP, which consist of two 13-bp palindromic sequences flanking an 8-bp core sequence (1, 2). Because of the high efficiency of the Cre/loxP system in mammalian cells (3, 4), this system is now widely used for genetic engineering in vitro and in vivo (5, 6). Transfer of Cre in vivo can be achieved by the use of transgenic mice or by viral delivery to the target organ(s). Thus far, the transfer of Cre by using nonviral delivery vehicles such as liposomes has only been described for use in vitro (7).
We have generated lentiviral vectors that carry the Cre recombinase and have analyzed their ability to mediate loxP-specific recombination. Lentiviruses are enveloped RNA viruses that belong to the family of complex retroviruses (for review see refs. 8 and 9). In contrast to prototypic retroviruses, such as murine leukemia virus, lentiviruses are able to transduce both dividing and nondividing cells. Lentiviral vectors pseudotyped with the G protein of the vesicular stomatitis virus can transduce a wide range of cells and have great potential for human gene therapy (10–13).
The Cre lentiviral vectors efficiently transferred Cre to the target cells and induced Cre-mediated recombination of loxP sites in vitro and in vivo. The Cre-expressing cells unexpectedly exhibited a reduction in proliferation and accumulated in the G2/M phase of the cell cycle. To circumvent this toxic effect of Cre, we designed a Cre lentivirus vector that is itself subject to Cre-mediated excision (self-deleting or LV-Cre-SD). In this way, Cre expression is limited to only the time necessary for recombination. Our studies show that this approach works efficiently in vitro and in vivo.
Materials and Methods
Animals.
Homozygous R26R mice (12–20 weeks of age) were anesthetized by i.p. injection of a ketamine (150 mg/kg)/xylazine (15 mg/kg) mixture. Virus (3 μl) was injected either stereotaxically into the brain (coordinates from Bregma in millimeters: for the striatum, 0.5 anteroposterior, 2.0 mediolateral, 2.5 dorsoventral; for the hippocampus, −2.5 anteroposterior, 2.0 mediolateral, and 1.5 dorsoventral) or into the liver with a 26-gauge Hamilton syringe. All procedures were performed in accordance with the standards of the Salk Institute.
Virus Production.
To make the Cre-lentivirus (LV-Cre), a MluI fragment containing the nuclear localization signal (nls) of the simian virus 40 large T antigen and the cDNA for Cre recombinase (nlsCre) was excised from pMC-Cre (14). The nlsCre was first cloned into the EcoRV site of a pBluescript II KS vector, and then inserted into the XbaI–SalI sites of a pRRL-derived lentiviral vector that contains a central polypurine tract (ppt) and the posttranscriptional element of the woodchuck hepatitis virus (15), resulting in LV-Cre. In addition, a vector (LV-CIG) was designed that contains an internal ribosomal entry site-green fluorescent protein (GFP) cassette downstream of Cre to achieve bicistronic expression of Cre and GFP. The lentiviral vector carrying the lacZ gene (LV-Lac) was constructed by replacing the Cre cassette with the lacZ gene. For the construction of the LV-Cre-SD, one loxP site was cloned into the BspEI site of HIV-CS (16). The nlsCre cassette was then inserted into the EcoRI–XhoI sites of the vector. In addition, a cytomegalovirus (CMV) promoter, a ppt, and a woodchuck hepatitis virus were cloned into this vector. A third-generation, Tat-free packaging system (17) was used to produce recombinant lentivirus, as described (18, 19).
Analysis of Cre-Mediated Recombination in CV-1 Cells.
CV-1 cells (20) were fixed for 5 min at 4°C in PBS containing 2% formaldehyde and 0.2% glutaraldehyde. For β-galactosidase (β-gal) staining, fixed cells were incubated for 4 h at 37°C in staining solution (5 mM ferricyanide/5 mM ferrocyanide/2 mM MgCl, dissolved in PBS) containing 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; Sigma/Aldrich).
Isolation of Bone Marrow Progenitors.
Isolation of lineage negative (Lin−) cells.
Bone marrow cells were flushed from the femur and tibia of R26R mice. Mononuclear cells were isolated by Ficoll/Paque (Amersham Pharmacia) density separation and enriched for progenitor cells by antibody depletion for lineage-positive cells by using the StemSep enrichment antibody mixture (StemCell Technologies, Vancouver). Lin− cells were transduced with LV-CIG vectors [multiplicity of infection (moi) = 200] for 2 h at 37°C.
Colony-forming assay.
Lin− cells were incubated in methylcellulose medium (MethoCult, StemCell Technologies) with recombinant cytokines (50 ng/ml stem cell factor/10 ng/ml IL-3/10 ng/ml IL-6/3 units/ml erythropoietin) at a cell density of 1,000 cells per plate (20 cm) (2). At day 12, colonies were stained with X-gal.
Suspension culture.
To differentiate bone marrow progenitors, Lin− cells were incubated for 10 days in serum-free medium (StemSpan, StemCell Technologies) in the presence of cytokines (100 ng/ml stem cell factor/20 ng/ml IL-3/20 ng/ml IL-6/50 ng/ml thrombopoietin). Either cell suspensions were stained with X-gal, or immunofluorescence was performed on cytospins by using an antibody-detecting CD11b (PharMingen), a myeloid marker, or by using an antibody against CD41 (PharMingen), a marker for megakaryocytes.
Cell-Cycle Analysis.
Cell-cycle analysis was done as described (21). In brief, cells were harvested at 48, 96, and 140 h after infection and resuspended in PBS containing 1% FBS. Cells (1 × 106 to 3 × 106) were fixed with 95% ethanol and washed twice before incubating for 15 min at 37°C with propidium iodide (PI)/RNase A solution (5 μg/ml PI/250 μg/ml RNase A in PBS/1% FBS). Incorporation of PI was quantified by FACScan (Becton Dickinson). Cellular DNA contents were calculated by using the MULTICYCLE program (Phoenix Flow Systems, San Diego).
Histological Procedures.
Animals were anesthetized and then transcardially perfused with either 4.0% paraformaldehyde (for immunohistochemistry) or 1.5% glutaraldehyde (for X-gal staining). Dissected tissues were postfixed overnight at 4°C and then saturated with 30% sucrose for at least 2 days at 4°C. Brains were sliced at 40 μm on a sliding microtome, stored at −20°C in a cryoprotectant, and stained as free floating sections. Livers were cut on a cryostat at 12 μm and stained on slides. After blocking (1 h, room temperature) with 0.1% Triton X-100 and 3% normal donkey serum (PBS++), the tissue was incubated with primary antibodies in PBS++ overnight at 4°C, followed by incubation with secondary antibodies in PBS++ for 2 h at room temperature. The primary antibodies used were the monoclonal anti-Cre (MAB3120, Chemicon) at 1:1,000, the polyclonal anti-Cre (no. 69050-3, Novagen) at 1:1,000, anti-β-gal (5′->3′) at 1:1,000, anti-NeuN (clone A60 kindly provided by R. Mullen, University of Utah, Salt Lake City) at 1:20, and anti-glial fibrillary acidic protein (GFAP; Advanced ImmunoChemical, Long Beach, CA) at 1:250. Secondary antibodies (conjugated to FITC, Cy3, Cy5, or biotin) were all generated in donkey (The Jackson Laboratory) and used at 1:250 dilution. The anti-β-gal signal was enhanced with biotinylated secondary and streptavidin-conjugated fluorophore. Fluorescence was viewed with either a Nikon inverted microscope or a Bio-Rad confocal microscope. X-gal staining of tissues was performed for 4 h at 37°C followed by counterstaining with nuclear fast red.
Results
Analysis of Cre-Mediated Recombination in a Reporter Cell Line.
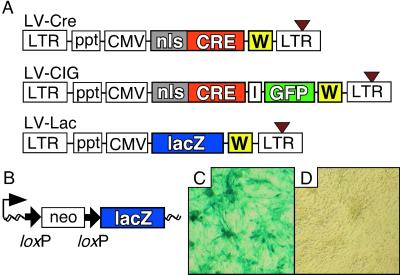
The latest generation of HIV-based lentiviral vectors (15, 16) was used to generate the vectors carrying the Cre recombinase under the control of an internal CMV promoter (LV-Cre) (Fig. 1A). A bicistronic vector (LV-CIG) was constructed by cloning Cre and an internal ribosomal entry site upstream of an expression cassette for the enhanced GFP (Fig. 1A Middle). In addition, we designed a vector that carries the lacZ cDNA (Fig. 1A Bottom). Viral particles were produced by using a third-generation, split-genome packaging system (17) and pseudotyped with the G protein of the vesicular stomatitis virus (18). Initially, CV-1 reporter cells (20) were used to determine the activity of Cre delivered by a lentiviral vector. These cells carry a switching unit that reports Cre/loxP recombination by expression of β-gal (Fig. 1B). LV-Cre efficiently transduced CV-1 cells (Fig. 1C).
Figure 1.
Lentiviral vectors and LV-Cre-mediated recombination in vitro and in vivo. (A) Schematic representation of the lentiviral vectors used in this study. (Top) The Cre lentiviral vector (LV-Cre) contains a Cre expression cassette with an nls. Expression of Cre is driven by the CMV promoter. (Middle) LV-CIG contains an internal ribosomal entry site (I) and a GFP cassette downstream of Cre. (Bottom) LV-Lac carries the lacZ gene instead of Cre. All vectors contain the Rous sarcoma virus promoter in the 5′ long-terminal repeat (LTR) (for efficient virus production in the context of a Tat-free packaging system), and self-inactivating mutations in the 3′ LTR (brown triangle). To enhance transgene expression in the target cells, a central ppt of HIV-1 pol (ppt) and a posttranscriptional element (W) from the woodchuck hepatitis virus were included. (B) Structure of the Cre-responsive lacZ reporter gene unit of CV-1 cells. Before Cre-mediated recombination, transcription from the CMV-β-actin promoter (thin black arrow) stops at the polyadenylation site of the neomycin resistance (neo) cassette. After recombination of the loxP sites (bold black arrows), this cassette is excised and the lacZ reporter gene is transcribed. Wavy line, chromosomal DNA. (C and D) X-gal stain of the CV-1 cells 72 h after infection with LV-Cre (moi ≈ 2) (C) and of uninfected CV-1 cells (D).
Analysis of Cre Recombination in Vivo.
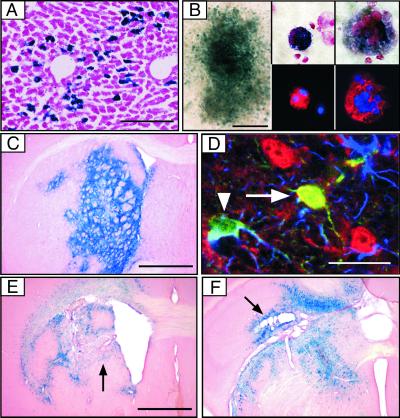
ROSA26 Cre reporter (R26R) mice were used to analyze the ability of lentiviral vectors to express active Cre in vivo (22). R26R mice carry a Cre-responsive reporter cassette that is similar to the one used in CV-1 cells (Fig. 1B). At 3 weeks after intraparenchymal injection of LV-Cre [1.2 ×108 infectious units (IU)] into the liver, we observed strong expression of β-gal not only at the site of injection (Fig. 2A) but also in uninjected liver lobes (not shown). Close to the site of injection, more than two-thirds of the β-gal-positive (β-gal+) cells were identified as hepatocytes, whereas more distal to the site of injection, the ratio of transduced hepatocytes to nonhepatocytes was ≈1:3.
Figure 2.
Lentiviral transfer of Cre into tissues and cells of R26R reporter mice. (A) Expression of β-gal 3 weeks after intraparenchymal injection of 6 × 107 IU into the liver. (Scale bar = 200 μm.) (B) Analysis of β-gal expression in colonies formed by progenitor bone marrow cells (Left; scale bar = 400 μm) and in in vitro differentiated progenitor cells (Center and Right). Identification of differentiated progenitors by staining for the myeloid marker CD11b (Center Lower, red) and for the megakaryocyte marker CD41 (Right Lower, red); 4′,6-diamidino-2-phenylindole is shown in blue. Original magnification ×300 (Center) and ×1000 (Right). (C--F) Analysis of Cre expression and recombination in the brain. (C) X-gal stain of coronal section 1 week after injection of 6 × 107 IU LV-Cre into the striatum. (Scale bar = 1 mm.) (D) Identification of transduced cells in the cortex of R26R mice by staining for β-gal (green), the astrocyte marker GFAP (blue), and neuronal marker NeuN (red) (arrowhead, β-gal- and GFAP-positive cell; arrow, β-gal- and NeuN-positive cell). (Scale bar = 30 μm.) (E and F) X-gal stain of coronal section 3 weeks after injection of 6 × 107 IU LV-Cre into the striatum (E) and hippocampus (F). Black arrows indicate tissue damage. (Scale bar = 1 mm.)
To analyze the recombination efficiency of the LV-Cre in hematopoietic progenitors, Lin− bone marrow cells were isolated from R26R mice and incubated in methylcellulose and cytokines (23). At 12 days after transduction with LV-CIG, Cre recombinase activity was assessed by X-gal staining of the colony-forming cell progenitors. Approximately 40% of the colonies were β-gal+ 2 weeks after infection with LV-CIG (moi 200) (Fig. 2B). Next, transduced Lin− cells were incubated for 10 days in the presence of stem cell factor/IL-2/IL-6/thrombopoietin to induce differentiation of granulocytes, monocytes, and megakaryocytes. The differentiated cells were then analyzed by X-gal staining and immunofluorescence by using antibodies against CD11b (myeloid cells) and CD41 (megakaryocytes) (Fig. 2B Center and Right). Robust β-gal expression was observed in polymorphonuclear cells and megakaryocytes (Fig. 2B), demonstrating that lentiviral vectors can transfer Cre to early, Lin− hematopoietic progenitors that can differentiate into several hematopoietic lineages.
Lentiviral Delivery of Cre into the Brain.
Adult R26R mice were injected stereotaxically with LV-Cre into various regions of the brain. At 1 week after a single injection (6 × 107 IU) into the striatum, most of the cells within a volume of ≈1- to 1.5-mm radius were β-gal+ (Fig. 2C). Similar results were obtained after transduction of hippocampus, thalamus, septum, and cerebellum (data not shown). The lentiviral vector delivered Cre to neurons and astrocytes, as demonstrated by immunofluorescence staining with antibodies against the astrocyte marker GFAP, and the neuronal marker NeuN (Fig. 2D). Histological analyses performed at later times (3–18 weeks after injection) unexpectedly revealed the damage of brain structures and the presence of abnormal cavities in the injected striatum (Fig. 2E) or hippocampus (Fig. 2F). Because we have not observed tissue damage after injection of lentiviral vectors carrying GFP into the striatum and hippocampus (refs. 18 and 24; data not shown), these results indicated that sustained expression of Cre might be detrimental to the transduced cells.
Cre Expression Reduces Cell Proliferation.
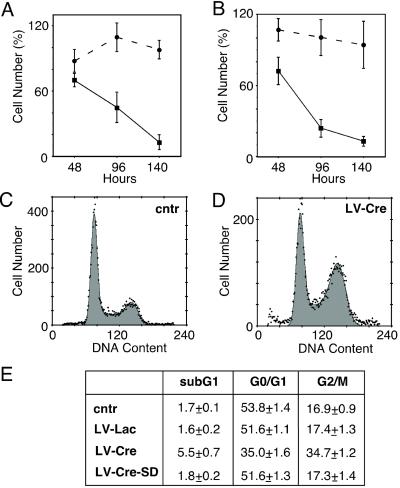
To address the question of a potential toxic effect of Cre, we analyzed the effects of Cre expression on cell growth and viability. Transduction of CV-1 cells with LV-Lac (moi 2) resulted in the transduction of ≈90% of the cells but did not significantly affect the number of cells, as compared with uninfected cells (Fig. 3A). Thus, neither lentiviral infection nor β-gal expression per se significantly affects cell proliferation. In contrast, transduction of CV-1 cells with LV-Cre (moi 2) caused a significant (P < 0.01) decrease in cell number to 70 ± 6%, 45 ± 6%, and 13 ± 7% of the uninfected control cells at 48, 96, and 140 h, respectively (Fig. 3A). To study whether the Cre-induced inhibition of cell proliferation requires the presence of loxP sites, we tested the effect of Cre in COS cells. Expression of Cre caused an almost identical reduction of cell growth in CV-1 and COS cells (Fig. 3B), indicating that this effect is not caused by recombination of loxP sites. In addition, we observed a 6-fold increase in the number of detached and floating cells 96 h after transduction with LV-Cre (moi 2), as compared with the uninfected COS cells (data not shown).
Figure 3.
Effect of Cre expression on cell number and cell cycle. (A and B) Quantification of the number of cells attached to the substrate 48, 96, and 140 h after infection with LV-Lac (circles) or LV-Cre (squares). The number of CV-1 (A) and COS (B) cells expressed as percentage of the uninfected control cells (mean ± SEM, n = 4 experiments). (C–E) Analysis of propidium iodine incorporation into cellular DNA by flow cytometry. Representative cell-cycle profiles of uninfected (cntr) COS cells (C) and COS cells infected with LV-Cre (D). (E) Statistical analysis of cell-cycle profiles (n = 3 experiments).
The negative effect of Cre on cell viability and proliferation was further analyzed by propidium iodide incorporation into cellular DNA followed by FACScan analysis. At 48 hours after transduction, 34.7 ± 1.2% of the Cre-expressing cells had accumulated in the G2/M phase as compared with 17.4 ± 1.3% and 16.9 ± 0.9% in LV-Lac-transduced and nontransduced cells, respectively (Fig. 3 C–E). Furthermore, a 3-fold increase in the sub-G1 population was observed in the LV-Cre-infected cells, as compared with LV-Lac or uninfected cells, indicating an increase in apoptosis in the Cre-expressing cells (Fig. 3E).
Transient Cre Expression with LV-Cre-SD.
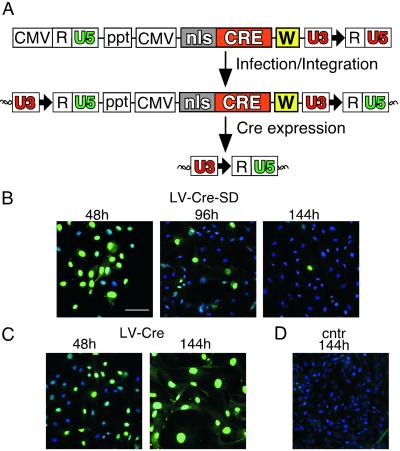
To minimize the cytotoxic effects of Cre, we designed a regulated LV-Cre. Ideally, the system should efficiently express enough Cre protein to achieve rapidly the recombination of the loxP sites in the target gene, after which the Cre expression should be terminated quickly. To this end, we introduced into the lentiviral vector a Cre-responsive deletion unit, which consists of a single loxP site incorporated into the U3 region of the 3′ LTR (LV-Cre-SD; Fig. 4A). As with other retroviruses, the U3 region of the lentiviral 3′ LTR is duplicated and transferred to the 5′ LTR during reverse transcription of the viral RNA (for details see refs. 9, 25, and 26). Thus, once integrated into the host genome, the LV-Cre-SD provirus contains one loxP site in the 5′ LTR and one loxP site in the 3′ LTR. Cre-mediated recombination of these two loxP sites should lead to the deletion of the lentiviral vector and termination of Cre expression.
Figure 4.
Development of LV-Cre-SD for transient Cre expression. (A) Strategy for regulating Cre expression by using LV-Cre-SD (Top). Self-deletion is achieved by incorporation of a single loxP site (bold black arrow) into the U3 region of the 3′ LTR. Because of the duplication of the U3 region during reverse transcription, the integrated provirus contains two U3 regions with loxP sites (Middle). The internal CMV promoter drives expression of Cre, which in turn will catalyze the recombination of the loxP sites in the U3 regions, resulting in the excision of almost the complete provirus including the Cre cassette (Bottom). In this way, Cre terminates its own expression. (B–D) Immunofluorescence analysis of Cre expression (green nuclei) in CV-1 cells transduced with LV-Cre-SD (B), LV-Cre (C), or uninfected (cntr) cells (D) at the indicated times after infection. Representative merged images (green channel, anti-Cre; blue color, 4′,6-diamidino-2-phenylindole). (Scale bar = 90 μm.)
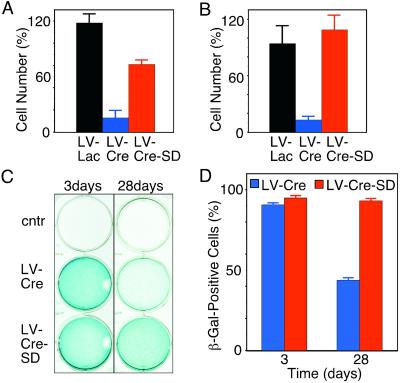
Transduction of CV-1 cells with LV-Cre-SD (moi ≈ 2) resulted in the expression of Cre in more than 90% of the cells within 48 h (Fig. 4B)—a transduction efficacy similar to that observed for the conventional LV-Cre (Fig. 4C). During the next 48 h, Cre expression levels decreased in the LV-Cre-SD-transduced cell (Fig. 4B) but not in the cells infected with LV-Cre (data not shown). At 144 h, only 5–10% of the LV-Cre-SD transduced cells expressed Cre, whereas Cre expression was observed in >90% of the LV-Cre-transduced cells (Fig. 4 B and C). Western blot analyses confirmed the down-regulation of Cre within 140 h after infection with LV-Cre-SD (data not shown). Cell counts performed on CV-1 and COS cells 140 h after infection with LV-Cre-SD revealed a significantly (P < 0.01) greater number of cells than the LV-Cre-infected cells (Fig. 5 A and B). Although the number of CV-1 cells observed at 140 h (Fig. 5A) after treatment with LV-Cre-SD was significantly lower than the number of those transduced with LV-Lac, no significant differences in cell number were detected at 280 h (data not shown). No significant decrease in the cell number was observed in the COS cells 140 h after infection with LV-Cre-SD in comparison with uninfected cells (Fig. 5B). In addition, cell-cycle analysis revealed no significant changes as compared with the untreated or LV-Lac-treated cells (Fig. 3E).
Figure 5.
Effect of the conventional and the LV-Cre-SD on cell number. CV-1 (A) and COS (B) cells were counted 140 h after infection with LV-Lac (black columns), LV-Cre (blue), or LV-Cre-SD (red). The number of cells is depicted as percentage of the uninfected (cntr) control cells (mean ± SEM, n = 3 experiments). (C) Analysis of reporter gene (β-gal) activation by the LV-Cre-SD. CV-1 cells were infected with two particles per cell of LV-Cre (Middle) or LV-Cre-SD (Bottom). At 3 days (Left) or 28 days (Right) after infection, cells were stained with X-Gal. (D) Quantification of β-gal+ cells 3 days or 28 days after infection with LV-Cre (blue) or LV-Cre-SD (red) (mean ± SEM, n = 3 experiments).
Next, we analyzed the ability of LV-Cre-SD to modify the loxP-flanked target gene, i.e., the neo cassette in the CV-1 reporter cells. At 3 days after infection with LV-Cre-SD and LV-Cre, a similar number of cells were β-gal+ (Fig. 5 C and D). At this time, the ratio of β-gal+ to β-gal-negative (β-gal−) cells was ≈20:1 for both LV-Cre and LV-Cre-SD. However, 4 weeks after transduction with LV-Cre, we observed a significant reduction in the ratio of β-gal+/β-gal− cells to ≈1:1, whereas no significant change in the ratio of β-gal+/β-gal− cells was observed after transduction with LV-Cre-SD (Fig. 5D).
Cre-Mediated Recombination in Vivo with LV-Cre-SD.
To analyze the function of the regulated Cre virus in vivo, we injected 6 × 107 IU of LV-Cre-SD into the brains of R26R mice. First, we analyzed the time course of Cre expression by immunohistochemistry. Although we detected expression of Cre 1 week after injection of LV-Cre-SD in the striatum (Fig. 6A), Cre expression was reduced more than 100-fold 5 weeks after injection (Fig. 6B). At that time, only a small subset of cells expressed detectable amounts of Cre (compare Fig. 6 A and B Insets). Thus, the incorporation of a single loxP site into the U3 region of the 3′ LTR is a feasible way to achieve transient Cre expression from the modified lentiviral vector in vivo. To determine whether the concentration of Cre expressed from the Cre-suicide vector was sufficient to achieve the recombination of the cellular target gene, we assessed the amount of β-gal expression by immunohistochemical staining of consecutive sections 1 or 5 weeks after injection (Fig. 6 C and E).
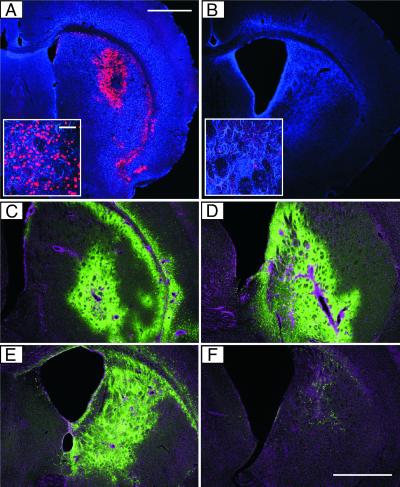
Figure 6.
Transient Cre expression and reporter gene activation by the LV-Cre-SD in vivo. (A and B) Immunofluorescence analysis of Cre expression. The striatum of R26R mice was injected with 6 × 107 IU of LV-Cre-SD. At 1 (A) or 5 weeks (B) later, mice were killed and analyzed for Cre expression (red). GFAP was used as background stain (blue). (A Inset and B Inset) Cre-positive cells (red) in the striatum of LV-Cre-SD-injected mice. (Scale bar = 62.5 μm.) (C–F) Immunohistochemical analysis of β-gal expression. At 1 (C and D) or 5 (E and F) weeks after injection of 6 × 107 IU of LV-Cre-SD (C and E) or LV-Cre (D and F) into the striatum of R26R mice, coronal sections of fixed brains were stained for β-gal expression (green). The nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (magenta). Coronal sections of the same mice are shown in A and C as well as B and E. (Scale bar for A–F = 1 mm.)
At 1 week after striatal injection of either LV-Cre-SD or LV-CIG, the β-gal+ volume was a size similar to that in mice injected with LV-Cre (Fig. 6 C and D; see also Fig. 2C), demonstrating that the LV-Cre-SD induces recombination of a target gene that contains loxP sites in the brain with an efficacy similar to conventional LV-Cre. Furthermore, analysis of β-gal expression 1 week after injection of LV-Cre-SD or LV-Cre (each 1.2 × 108 IU) into the liver revealed almost identical numbers of recombined cells with the two vectors (data not shown).
We repeatedly found a clear difference in the extent of β-gal expression when comparing the conventional and the regulated Cre vector at a later time. All mice (n = 8) were injected on the same day under identical conditions and demonstrated similar β-gal expression patterns 1 week after injection. However, at 5 weeks after injection, there was a clear reduction in the number of β-gal-expressing cells in the LV-Cre-injected mice (Fig. 6F) as compared with the LV-Cre-SD-injected mice (Fig. 6E). Analysis of the number of transduced cells in brains injected with LV-CIG revealed a significant reduction of GFP-positive (i.e., transduced cells) 5 weeks after injection as compared with the mice that were analyzed 1 week after injection (data not shown). These findings indicate that sustained expression of high concentrations of Cre might be deleterious/disadvantageous to the target cells not only in vitro (see Figs. 3 and 5) but also in vivo. The Cre-induced cytotoxic effects can be minimized by transient Cre expression with the LV-Cre-SD, which induces efficient recombination in several different tissues of transgenic mice.
Discussion
The Cre/loxP system is a powerful tool for the manipulation of the mammalian genome, and the interbreeding of Cre-expressing transgenic lines with mice that carry genes flanked by loxP sites has resulted in efficient modification of a variety of genes in vivo. Viral vectors have also been shown to achieve Cre expression in different tissues. Thus far, vectors derived from adenoviruses (27–30) and herpes simplex virus (31) have been described that carry Cre. Adenoviral vectors can be produced at high titers (up to 1012 plaque-forming units/ml) and efficiently transduce susceptible cells. However, their major disadvantage is the host immune response against the viral proteins (for review see ref. 13) that can lead to the destruction of the viral vectors and of the transduced cells. Although Cre-expressing herpes simplex virus vectors have mediated recombination in vivo, the titers of these vectors (in the range of 105–106 plaque-forming units/ml) (31) and their efficacy (only 30% of the infected cells underwent recombination; ref. 32) are relatively low. In the present report, we have described lentiviral vectors for the delivery of Cre recombinase. These vectors induce efficient recombination of loxP sites in a variety of target cells, including neurons, hepatocytes, and hematopoietic progenitors.
Apparently, a potential drawback of the Cre/loxP system is the cytotoxicity of Cre. We observed a decrease in proliferation as well as an increase in apoptosis in Cre-expressing cells. A similar effect of Cre has been described recently by several other groups (33, 34). Our analyses revealed an accumulation of Cre-expressing cells in the G2/M phase of the cell cycle. The fact that the G2/M arrest was found in cells that do not contain loxP sites indicates that it is an intrinsic feature of the Cre recombinase and not a result of loxP-mediated recombination. In addition, we observed tissue damage after expression of Cre for several weeks in the brain. With use of lentiviral vectors that carry other transgenes, we and others have observed long-term expression of the transgenes without obvious indications of tissue damage (18, 24, 35). Cre-induced cytotoxicity might be an explanation for the reduced number of Cre-expressing, β-gal+ cells that we observed in vitro and after prolonged (3–5 weeks) Cre expression in the brain. Shut-off or silencing of the β-gal promoter is unlikely to be the explanation for these results, because we did not observe it after transient Cre expression, and in R26R mice, β-gal is expressed from an ubiquitously active, endogenous promoter (22).
A possible explanation for the toxic effects of Cre expression might be the Cre-mediated recombination of genomic DNA at sites that share homology with the lox sites of the bacteriophage P1. Indeed, such pseudo-lox sites have recently been discovered in the mouse and human genomes and shown to be substrates for Cre-mediated recombination (36). This “illegitimate” (37) recombination results in DNA strand breaks and chromosome rearrangements (33, 34). Endogenous pseudo-lox sites share only a limited homology with loxP sites, and Cre-mediated recombination of pseudo-lox sites is less efficient than it is for bona fide loxP sites (36). Thus, transient expression of Cre should minimize the chances of recombination at the endogenous pseudo-lox sites and/or give the target cell time to repair the DNA damage efficiently.
To optimize the Cre expression system such that Cre is only expressed for the period necessary to achieve recombination of the target loxP sites, we designed a self-deleting lentiviral vector. This approach obviates the use of exogenous ligands or drugs for the regulation of expression, because the transgene (i.e., Cre) delivered by the lentiviral vector regulates its own expression. The LV-Cre-SD carries the Cre cDNA and a single loxP site in its U3 region of the 3′ LTR. Several groups have shown that loxP sites can be incorporated into simple/prototypic and complex retroviruses and that these loxP sites are duplicated in the integrated provirus, making it susceptible to Cre-mediated excision (38–42). Recently, a murine leukemia virus-based self-excising vector was described and shown to achieve transient expression of Cre in vitro (33). We now demonstrate that by incorporation of Cre and loxP into one single lentiviral vector, one can achieve transient expression of Cre in vivo. In contrast to murine leukemia virus-based vectors (for details see ref. 13), LV-Cre can transduce a broad spectrum of terminally differentiated, nondividing cells, and therefore, this regulated Cre delivery system should be a useful tool for genetic engineering in vitro and in vivo. In addition, it is conceivable that the concept of a self-excising LV-Cre could be used for the short-term expression of other toxic genes by incorporating both the toxic gene of interest and Cre together in a bicistronic Cre-self-deleting vector.
Acknowledgments
We thank Nina Tonnu, Kevin Gobeske, and Nushin Sherkat for technical assistance. The ROSA26 Cre reporter mice were kindly provided by Philippe Soriano, Fred Hutchinson Cancer Research Center, Seattle. A.P. received a Heisenberg Scholarship from the Deutsche Forschungsgemeinschaft and is supported by the Fonds der Chemischen Industrie. E.P.B. is supported by the John Douglas French Alzheimer's Foundation. F.H.G. is supported by grants from the National Institutes of Health/National Institute on Aging, Lookout Fund, and Christopher Reeve Paralysis Foundation. I.M.V. is an American Cancer Society Professor of Molecular Biology and is supported by grants from the National Institutes of Health, the March of Dimes, the Wayne and Gladys Valley Foundation, the Lebensfeld Foundation, and the H. N. and Frances C. Berger Foundation.
Abbreviations
- LV-Cre
Cre lentivirus
- nls
nuclear localization signal
- GFP
green fluorescent protein
- LV-CIG
lentiviral vector designed to achieve bicistronic expression of Cre and GFP
- LV-Lac
lentiviral vector carrying the lacZ gene
- LV-Cre-SD
Cre-lentivirus self-deleting
- CMV
cytomegalovirus
- ppt
polypurine tract
- Lin−
lineage negative
- moi
multiplicity of infection
- β-gal
β-galactosidase
- GFAP
glial fibrillary acidic protein
- LTR
long terminal repeat
- IU
infectious units
References
- 1.Sternberg N, Hamilton D, Hoess R. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 3.Sauer B, Henderson N. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 5.Sauer B. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 7.Bugeon L, Syed N, Dallman M J. Transgenic Res. 2000;9:229–232. doi: 10.1023/a:1008990510849. [DOI] [PubMed] [Google Scholar]
- 8.Vogt P K. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 1–25. [PubMed] [Google Scholar]
- 9.Joag S V, Stephens E B, Narayan O. In: in Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 1977–1996. [Google Scholar]
- 10.Trono D. Gene Ther. 2000;7:20–23. doi: 10.1038/sj.gt.3301105. [DOI] [PubMed] [Google Scholar]
- 11.Somia N, Verma I M. Nat Rev Genet. 2001;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 12.Kay M A, Glorioso J C, Naldini L. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer A, Verma I M. Annu Rev Genomics Hum Genet. 2001;2:177–211. doi: 10.1146/annurev.genom.2.1.177. [DOI] [PubMed] [Google Scholar]
- 14.Gu H, Zou Y R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 15.Follenzi A, Ailles L E, Bakovic S, Geuna M, Naldini L. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi H, Blomer U, Takahashi M, Gage F H, Verma I M. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer A, Kessler T, Silletti S, Cheresh D A, Verma I M. Proc Natl Acad Sci USA. 2000;97:12227–12232. doi: 10.1073/pnas.220399597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanegae Y, Takamori K, Sato Y, Lee G, Nakai M, Saito I. Gene. 1996;181:207–212. doi: 10.1016/s0378-1119(96)00516-1. [DOI] [PubMed] [Google Scholar]
- 21.Ruffner H, Verma I M. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 24.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffin J M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 1767–1848. [Google Scholar]
- 26.Rabson A B, Wills J W. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 205–261. [Google Scholar]
- 27.Anton M, Graham F L. J Virol. 1995;69:4600–4606. doi: 10.1128/jvi.69.8.4600-4606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai K, Mitani K, Miyazaki J. Biochem Biophys Res Commun. 1995;217:393–401. doi: 10.1006/bbrc.1995.2789. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Krushel L A, Edelman G M. Proc Natl Acad Sci USA. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohlmann A, Gotthardt M, Willnow T E, Hammer R E, Herz J. Nat Biotechnol. 1996;14:1562–1565. doi: 10.1038/nbt1196-1562. [DOI] [PubMed] [Google Scholar]
- 31.Brooks A I, Muhkerjee B, Panahian N, Cory-Slechta D, Federoff H J. Nat Biotechnol. 1997;15:57–62. doi: 10.1038/nbt0197-57. [DOI] [PubMed] [Google Scholar]
- 32.Brooks A I, Cory-Slechta D A, Federoff H J. Proc Natl Acad Sci USA. 2000;97:13378–13383. doi: 10.1073/pnas.230169397. . (First Published November 14, 2000; 10.1073/pnas.230169397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver D P, Livingston D M. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 34.Loonstra A, Vooijs M, Beverloo H B, Allak B A, van Drunen E, Kanaar R, Berns A, Jonkers J. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consiglio A, Quattrini A, Martino S, Bensadoun J C, Dolcetta D, Trojani A, Benaglia G, Marchesini S, Cestari V, Oliverio A, et al. Nat Med. 2001;7:310–316. doi: 10.1038/85454. [DOI] [PubMed] [Google Scholar]
- 36.Thyagarajan B, Guimaraes M J, Groth A C, Calos M P. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt E E, Taylor D S, Prigge J R, Barnett S, Capecchi M R. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. . (First Published November 21, 2000; 10.1073/pnas.240471297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergemann J, Kuhlcke K, Fehse B, Ratz I, Ostertag W, Lother H. Nucleic Acids Res. 1995;23:4451–4456. doi: 10.1093/nar/23.21.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russ A P, Friedel C, Grez M, von Melchner H. J Virol. 1996;70:4927–4932. doi: 10.1128/jvi.70.8.4927-4932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westerman K A, Leboulch P. Proc Natl Acad Sci USA. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flowers C C, Woffendin C, Petryniak J, Yang S, Nabel G J. J Virol. 1997;71:2685–2692. doi: 10.1128/jvi.71.4.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, Trono D. Mol Ther. 2000;2:404–414. doi: 10.1006/mthe.2000.0141. [DOI] [PubMed] [Google Scholar]