Abstract
Type 2 diabetes mellitus (T2D) is a disorder of glucose metabolism. It is a complex process involving the regulation of insulin secretion, insulin sensitivity, gluconeogenesis, and glucose uptake at the cellular level. Diabetic peripheral neuropathy (DPN) is one of the debilitating complications that is present in approximately 50% of diabetic patients. It is the primary cause of diabetes-related hospital admissions and nontraumatic foot amputations. The pathogenesis of diabetic neuropathy is a complex process that involves hyperglycemia-induced oxidative stress and altered polyol metabolism that changes the nerve microvasculature, altered growth factor support, and deregulated lipid metabolism. Recent literature has reported that there are several heterogeneous groups of susceptible genetic loci which clearly contribute to the development of DPN. Several studies have reported that some patients with prediabetes develop neuropathic complications, whereas others demonstrated little evidence of neuropathy even after long-standing diabetes. There is emerging evidence that genetic factors may contribute to the development of DPN. This paper aims to provide an up-to-date review of the susceptible and prognostic genetic factors associated with DPN. An extensive survey of the scientific literature published in PubMed using the search terms “Diabetic peripheral neuropathy/genetics” and “genome-wide association study” was carried out, and the most recent and relevant literature were included in this review.
1. Introduction
Diabetes mellitus is nowadays one of the foremost noncommunicable diseases affecting more than 387 million people worldwide [1]. Type 2 diabetes mellitus (T2D) is a disorder of glucose metabolism. It is a complex process involving the regulation of insulin secretion, insulin sensitivity, gluconeogenesis, and glucose uptake at the cellular level. Dysregulation of one or more of these processes due to environmental or genetic factors can lead to altered glucose metabolism causing diabetes mellitus [2, 3]. More than 90% of cases of T2D show higher incidence of insulin resistance. This phenomenon is acquired due to sedentary lifestyle in combination with multifactorial genetic susceptibility. T2D is associated with increased morbidity and mortality due to its debilitating and progressive nature and associated complications. The condition usually leads to multiorgan failure due to macrovascular and microvascular involvement (Figure 1) [2–5].
Figure 1.
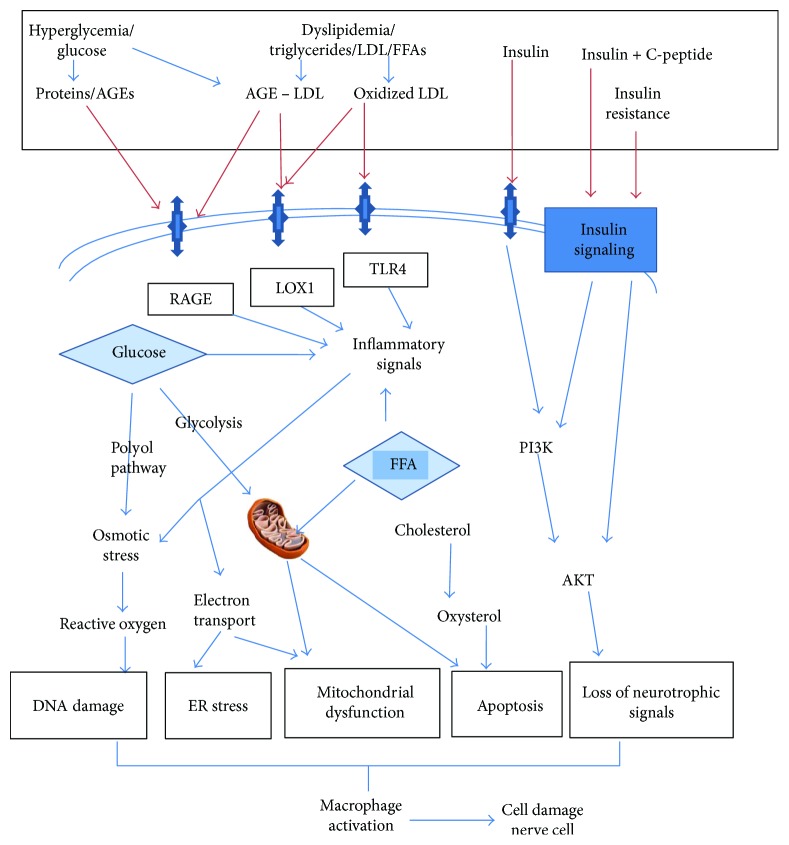
Mechanisms of diabetic neuropathy. Aetiological factors of diabetes initiate a cascade of events leading to DNA damage, endoplasmic reticulum stress, mitochondrial complex dysfunction, apoptosis, and loss of neurotrophic signaling. Ultimate activation of macrophages will cause cell damage in neurons, glial cells, and vascular endothelial cells, all of which can result in nerve dysfunction and neuropathy. AGE = advanced glycation end-products; LDL = low-density lipoprotein; FFA = free fatty acids; ER = endoplasmic reticulum; PI3K = phosphatidylinositol-3-kinase; LOX1 = oxidized LDL receptor 1; RAGE = receptor for advanced glycation end-products; TLR4 = Toll-like receptor 4.
Uncontrolled T2D can complicate pregnancy outcomes. Different kinds of birth defects are more commonly seen in babies born to women with diabetes [3]. Twin studies can estimate the multifactorial genetic involvement in T2D more precisely and have reported high degree of heritability of diabetes-related conditions such as disorders of first phase insulin response and basal and insulin-stimulated glucose uptake [6]. There are different methods for mapping the genetic susceptibility loci in the pathogenesis of T2D. Candidate gene studies and genome-wide studies are commonly used to identify the association of susceptible genetic loci of T2D. The latter includes both genome-wide linkage studies (GWL) and genome-wide association studies (GWAS) [6]. With the advent of recent molecular genetic techniques and rapid screening methods, the method of investigation has shifted to the use of molecular genetic markers for understanding the genetic aetiology of T2D and its complications [6, 7]. The common susceptible genetic variants are known to have a prominent effect on the risk of T2D across the world in multiple ethnic groups [8, 9]. Some variants appear to exert more pronounced genetic effects in specific ethnic groups. Most loci associated with T2D map to regulatory or intronic regions of the genome [9].
Diabetic peripheral neuropathy (DPN) is one of the debilitating microvascular complications of diabetes that is present in approximately 50% of patients. It is the primary cause of diabetes-related hospital admissions and nontraumatic foot amputations [4, 5, 10]. The molecular mechanisms involved in the development of DPN is a complex process that includes activation of the polyol pathway, exaggerated oxidative stress, overactivity of protein kinase C and increased formation of advanced glycation end-products in the presence of hyperglycemia. In addition, there is increasing evidence that genetic factors could also contribute to the development of DPN [10, 11]. The consequences of diabetic neuropathy include neurogenic pain, numbness, lack of coordination of voluntary movements, and a susceptibility to foot ulceration that leads to infections and toe or foot amputations. The rate of toe or foot amputations is 15 times greater in diabetic patients compared with individuals without diabetes. To date, approximately 80 T2D susceptibility genetic loci have been reported in different ethnic groups worldwide [12–14]. Majority of studies on the prevalence and associated aetiological factors of DPN have been conducted in Western countries. There is very limited data currently available for South Asian populations [15].
The objective of this paper is to provide an up-to-date review of the published scientific literature on the susceptible and prognostic genetic variants associated with DPN.
2. Methodology
This is a comprehensive review of the published literature on the susceptible and prognostic genetic variants associated with DPN. These variants were identified by an extensive survey of the scientific literature using the criteria described below. The most recent and relevant papers published in the last 15 years from January 2002 to July 2017 were searched in the PubMed database using the search terms “Diabetic peripheral neuropathy/genetics” and “genome-wide association study.” Altogether, sixty studies describing single nucleotide variants (SNVs) in genes associated with the susceptibility and prognosis of DPN which were published as full text articles in English during the defined period of study were included in this review. Epigenetic modifications which regulate gene expression mainly at the tissue/cellular level were excluded as it was outside the scope of this review.
3. Diabetic Peripheral Neuropathy
According to the Toronto Consensus Panel on Diabetic Neuropathy, DPN is defined as a symmetrical, length-dependent sensorimotor polyneuropathy that develops on a background of longstanding hyperglycemia, associated derangements, and cardiovascular risk factors [16]. The mechanisms underlying the pathogenesis of DPN are different between type 1 and type 2 diabetes mellitus [17]. Recent literature has reported that there are different groups of susceptible genetic loci which are clearly involved in the development of DPN. Different studies reported that some patients with prediabetes develop neuropathic complications, whereas others reported little evidence of neuropathy even after long-standing diabetes. This observation confirms the involvement of genetic aetiological factors associated with the development of DPN [18]. The data from different studies suggest that T2D and its complications may have shared genetic risk factors [12, 18].
4. Genetic Aetiology and Pathogenesis of DPN
The pathogenesis of DPN is a complex process and is involved with hyperglycemia-induced oxidative stress and altered polyol metabolism that changes the nerve microvasculature, growth factor support, and lipid metabolism [4]. It is important to identify these factors alone or in combination to arrange effective DPN treatment, as better understanding of the mechanisms underlying the onset and progression of DPN is of prime importance in the process of management [19]. Different groups of cell types in diabetic complication-prone tissues are targets of damage due to uncontrolled hyperglycemia. Schwann cells are the prime target of hyperglycemia which results in cell damage leading to altered axon integrity and defective growth factor signaling [20, 21]. Defective inflammatory pathways including advanced glycation end-product/receptor (AGE/RAGE) signaling in axons and Schwann cells have been reported in experimental animals with diabetic neuropathy which contributed to nerve damage [22].
Lu et al. in China studied SNVs from previously identified ten genetic loci and analyzed the association of these loci with peripheral nerve function in patients with T2D. They found that rs5219 of KCNJ11 gene polymorphism (E23K, G>A) was associated with peripheral nerve function. The results obtained from nerve conduction studies (NCS) showed that the allele “A” had a protective effect on peripheral nerve function. They also reported that SNVs rs7756992 of CDKAL1 and rs7903146 of TCF7L2 were associated with DPN in the Chinese T2D population [23].
Yigit et al. identified 230 unrelated patients with DPN at the outpatient clinics of the Physical Therapy and Rehabilitation Department of Gaziosmanpasa University, Tokat, in Turkey. They investigated the distributions of the genotype and allele frequencies of the MTHFR gene C677T variant among patients with DPN and a matched control group. A statistically significant difference of MTHFR gene C677T polymorphism between the patients with DPN and the control group was identified [24].
Decreased levels of peroxisome proliferator-activated receptor alpha (PPARA) in chromosome 22 and lipid metabolism-related gene apolipoprotein E (APOE) in chromosome 19 have been identified confirming the findings that altered lipid metabolism may play a role in the progression of DPN [25]. Monastiriotis et al. reviewed the literature to identify the association between APOE polymorphism and DPN and found that the ε4 allele of the apolipoprotein E gene is significantly associated with the pathogenesis of DPN [25].
The alpha2B adrenergic receptor encoded by ADRA2B gene located on chromosome 2 is associated with an array of functions. A polymorphism (12Glu9) resulting in the insertion/deletion of three glutamic acid residues in the third intracellular loop has been described frequently in the literature [26]. In the nervous system, this polymorphism has been reported to be linked with autonomic nervous dysfunction. This is particularly increased with sympathetic nervous system activity, and Papanas et al. found a significant association in this indel allele distribution of alpha2B adrenoceptor gene among T2D patients with DPN in comparison with matched T2D patients without neuropathy [26].
5. Network of Genes Associated with Common Variants of DPN
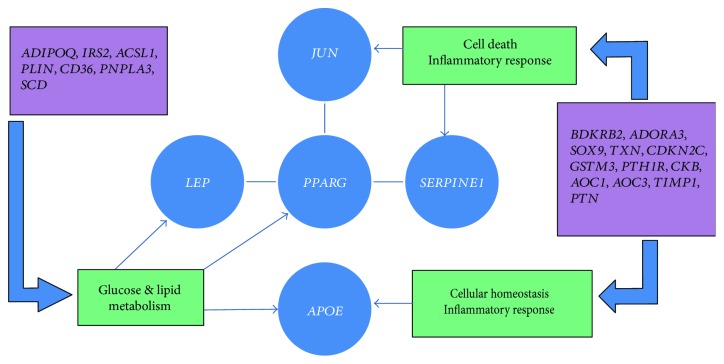
Hur et al. examined two groups of DPN patients. A network of transcription factors jun (JUN), leptin (LEP), serpin peptidase inhibitor E type 1 (SERPINE1), apolipoprotein E (APOE), and peroxisome proliferator-activated receptor gamma (PPARG) was examined to identify their potential relationship (Figure 2). Further subsets of genes related to defense response, inflammatory response, regulation of lipid metabolic processes, and PPAR signaling pathways were then analyzed to identify the association of gene expression and development of DPN (Figure 2) [27]. They demonstrated that increased glucose metabolism due to hyperglycemia resulted in increased oxidative stress, mitochondrial dysfunction, and cell death in both in vitro and in vivo models of diabetic neuropathy [27].
Figure 2.
Five main genes (in blue circles) associated with diabetic peripheral neuropathy: JUN, PPARG, LEP, SERPINE1, and APOE and their relationship with defense response, inflammatory response, glucose, and lipid metabolism pathways (in green-coloured cages) are represented in the figure. Additional genes involved with DPN in relation to abovementioned metabolic pathways are indicated in purple-coloured cages. ADIPOQ = adiponectin, C1Q and collagen domain containing; IRS2 = insulin receptor substrate 2; ACSL1 = acyl-CoA synthetase long chain family member 1; PLIN = lipid storage droplet 2-like; CD36 = CD36 molecule; PNPLA3 = patatin-like phospholipase domain containing 3; SCD = stearoyl-CoA desaturase; BDKRB2 = bradykinin receptor B2; ADORA3 = adenosine A3 receptor; SOX9 = sex determining region Y-box 9; TXN = thioredoxin; CDKN2C = cyclin-dependent kinase inhibitor 2C; GSTM3 = glutathione S-transferase mu 3; PTH1R = parathyroid hormone 1 receptor; CKB = creatine kinase B; AOC1 = amine oxidase, copper containing 1; AOC3 = amine oxidase, copper containing 3; TIMP1 = TIMP metallopeptidase inhibitor 1; PTN = pleiotrophin.
6. Variants Associated with Defense Response and Inflammatory Response in the Pathogenesis of DPN
Hur et al. reported that the molecules which are involved with the process of inflammation such as chemotactic agents and cytokines are involved with the development and progression of DPN as well as diabetic nephropathy [27, 28]. Kakoki et al. identified that the bradykinin receptor B2 (BDKRB2) is of particular interest in disease progression of DPN. BDKRB2 gene was found to be involved in progressive glomerulosclerosis and also susceptibility to DPN [29].
Membrane-associated adenosine A3 receptor (ADORA3) is also involved in the pathogenesis of DPN [30]. Variants of BDKRB2 and ADORA3 were found to be involved in enhanced inflammation and dysregulated defense responses, thus contributing to more substantial nerve damage in patients with progressive DPN [27, 30]. Gene variants of TXN, CDKN2C, GSTM3, PTH1R, CKB, AOC1, AOC3, TIMP1, and PTN have been identified as other additional genes associated with defense response and inflammatory response in the pathogenesis of DPN (Figure 2) [27].
7. Variants Associated with Glucose Metabolic Processes and PPAR Signaling Pathway in the Pathogenesis of DPN
According to Hur et al., PPARG, which encodes a nuclear receptor for glitazone, plays a key role in regulating glucose and lipid metabolism [27, 31]. Agonists of PPARG are effective in treatment of DPN and nephropathy in experimental animal models [27, 32]. Another key gene is APOE, encoding an apolipoprotein, which regulates the normal catabolism of triglycerides and cholesterol. A polymorphism of this gene is linked to the progression of DPN [33]. Gene variants of ADIPOQ, IRS2, ACSL1, PLIN, CD36, PNPLA3, and SCD were identified as other additional gene variants associated with glucose metabolic processes and PPAR signaling pathway in the pathogenesis of DPN (Figure 2) [27].
8. Genetic Variants Involved in Different Phenotypes of DPN
According to Cheng et al., in an experiment involving both human and animal models, sensory neurodegeneration in the chronic stage of diabetes was found to be associated with early damage to the distal axons of both upper and lower limb neurons showing a pattern that accounts for the distribution of “glove-and-stocking” loss of sensation characteristically seen in DPN. These changes accompany widespread abnormalities involving electrophysiology and alterations in gene expression that indicate a degenerative phenotype. However, existing knowledge on the development of DPN which includes oxidative and nitrergic stress, polyol accumulation, microangiopathy, inappropriate AGE-RAGE signaling, and/or mitochondrial dysfunction account for diverse mRNA changes that alter miRNA expression patterns resulting in diverse DPN phenotypes [34].
9. Genetic Variants Involved in Gender Dimorphism of DPN
Significant gender dimorphisms in the responsiveness of patients to antidiabetic drugs have been reported in the literature [35–37]. These observations highlighted the importance of understanding the gender-specific differences in manifestation of diabetes mellitus and its complications such as DPN.
O'Brien et al. reported the first instance of a female T2D mouse model presenting with a neuropathic phenotype including decreased intraepidermal nerve fiber density, impaired motor and sensory nerve conduction velocities, and thermal hypoalgesia [38]. A GWAS involving 961 diabetic neuropathic pain cases and 3260 diabetic controls in the Genetics of Diabetes Audit and Research Tayside by Meng et al. found that a cluster in the 1p35.1 region, the zinc finger and SCAN domain containing 20 (ZSCAN20) with a lowest p value of a variant at rs71647933 in females, and a cluster in the 8p23.1 region next to HMGB1P46 with a lowest p value of a variant at rs6986153 in males were significantly associated with DPN. This GWAS on diabetic neuropathic pain provides evidence for the sex-specific involvement of 1p35.1 region (ZSCAN20) and 8p23.1 region (HMGB1P46) [39].
10. Other Gene Loci Involved in the Pathogenesis and Prognosis of DPN
A fibrinolysis-regulating gene, SERPINE1, which encodes for plasminogen activator inhibitor 1 (PAI-1) has been identified in association with higher incidences of diabetic complications such as diabetic neuropathy and nephropathy in knockout PAI-1 mice [40, 41]. The cell cycle controlling JUN is also involved in the progression of DPN and is associated with inflammation and insulin resistance which is activated in multiple tissues including the peripheral sensory nerves of patients with types 1 and 2 diabetes [42].
Three other subsets of important gene variants were documented in literature associated with “cell projection and axonogenesis” involving nerve growth factor receptor (NGFR) and “cellular homeostasis and inflammatory response” involving thioredoxin, and “cytoskeletal protein binding” with stathmin 1 (STMN1) genes. NGFR exhibits protection against nerve cell and axonal damage, and the expression of nerve growth factor receptor protein in plasma correlates with DPN progression in diabetic rat models [43]. Thioredoxin, which regulates cellular oxidative stress with its antioxidant activity, also plays an important role in associated diabetes. Thioredoxin's antioxidant activity is significantly inhibited by hyperglycemic states in the blood. It complicates diabetes by playing an important role by deregulating vascular oxidative stress and inflammation in diabetic patients [44].
A study in North Catalonia, Spain, by Jurado et al. identified the protective effect of a single angiotensin-converting enzyme (ACE) gene polymorphism on the development of DPN in T2D patients. Despite ACE gene variants which are associated with diabetic renal disease and/or diabetic retinopathy, the heterozygous genotype stands as a protective factor against the development of DPN [45]. Heterozygous (D/I) ACE gene polymorphism reported a statistically significantly reduced risk of developing DPN whereas homozygous (D/D) ACE gene polymorphism reported an increased risk [45].
Mitochondrial transcription factor A (TFAM) is located in mitochondria, and its level regulates mitochondrial DNA (mtDNA) copy number. Chandrasekaran et al. showed that TFAM over expression prevented a decrease in mtDNA copy number in diabetic dorsal root ganglia (DRG) neurons, helped prevent DPN, and protected DRG neurons from oxidative stress in experimental mouse models [46].
Aldo-keto reductase family 1 member B (AKR1B1) in chromosome 7 encodes a member of the aldo-keto reductase superfamily, which consists of more than 40 known enzymes and proteins. This catalyzes the reduction of a number of aldehydes and is thereby implicated in the development of diabetic complications by catalyzing the reduction of glucose to sorbitol. Saraswathy et al. identified significant association of AKR1B1 gene mutations in painful diabetic neuropathy [47].
There is increasing evidence that microRNAs (miRNAs) act as regulators of gene expression in multiple biological processes and associated complications [48]. Ciccacci et al. looked for an association between variants in miRNA genes and DPN. The results of this study identified a role for MIR146a and MIR128a SNVs in the susceptibility to DPN and were shown to have a significant association [49]. The rs2910164 (G>C) in MIR146a is associated with lower risk, and rs11888095 (C>T) in MIR128a is associated with higher risk of susceptibility to DPN [49].
Nitric oxide (NO) production and local release in the tissues significantly contributed to endothelial dysfunction. The process takes place by the modulation of the nitric oxide synthase (NOS) enzymes responsible for NO synthesis. Endothelium-derived NO plays a key role in the regulation of vascular tone and has vasoprotective effects by removing superoxide radicals and suppressing platelet aggregation, leukocyte adhesion, and smooth muscle cell proliferation. However, dysfunctional endothelial nitric-oxide synthase (eNOS) might play a critical role in the pathogenic pathway leading to diabetic vascular complications including DNP. Therefore, eNOS is considered as a candidate for the progression of DPN [50, 51].
In the early stages of DPN, abnormalities in the vasa nervorum and loss of nerve fibers can be seen in association with hyperglycemia. Damage to the nervous tissue results in increased intravascular endothelial growth factor (VEGF) plasma levels in diabetic animal models [52]. The ischemia and hypoxia in the nerves of patients with T2D due to microangiopathy of vasa nervorum have always been observed and may be a key pathogenic mechanism of DPN [53]. An association study by Ghisleni et al. showed a clear association between diabetic polyneuropathy and the C936T polymorphism of the VEGF gene and the C242T polymorphism of the p22phox allele of CYBA gene [52].
Functional GRP78 variants in heat shock protein family A (Hsp70) member 5 (HSPA5) genes are likely to have some influence on the gene expression, which results in the dysfunction of peripheral nerves and neuropathy. According to Jia et al., functional GRP78 rs391957 variants, which are located in the promoter region, 57168556T>C, are known to cause abnormal promoter activities significantly associated with DPN [54].
Adiponectin gene (ADPN) serves as a protective factor in preventing diabetes progression by suppressing inflammatory responses and increasing insulin sensitivity [55]. SNVs of ADPN may influence T2DM, but ADPN variants SNV45 (45T/G, rs2241766) and SNV276 (276G/T, rs1501299) are the two most prominent variants influencing the disease progression, especially pathogenesis of DPN [56, 57]. A case-control study conducted by Ji et al. to evaluate the association between ADPN gene variants and pathogenesis of DPN in T2D patients indicated an increased risk of DPN in T2D patients, by downregulating ADPN expression which resulted in significantly reduced circulating ADPN plasma levels. Furthermore, they reported that the polymorphism frequencies of GG and GT haplotypes in the DPN group were significantly lower than those in the matched control group, while the frequency of the TG haplotype in the DPN group was markedly higher than that in the control group, showing a clear association between ADPN gene variants and the risk of DPN [58].
11. Genetic Variants of DPN in Different Ethnic Populations
Up to now, only few ethnic groups have described population-specific genetic variants associated with DPN. In a study conducted by Lu et al., 10 SNVs associated with pathogenesis of T2DM were studied. They reported that rs5219 on KCNJ11 (E23K) gene is significantly associated with peripheral nerve function in a Chinese population with T2D [23]. Jia et al. studied the significance of functional GRP78 gene variants in predicting the onset of type 2 DPN in the Chinese population. They suggested that the GRP78 rs391957 promoter polymorphism is a potential risk factor for type 2 DPN in this population [54]. Prasad et al. studied forty-two patients with T2D from the Institute of Diabetology, Madras Medical College, and Rajiv Gandhi Government General Hospital in Chennai, Tamil Nadu, India. In this study, the extent of DNA damage in patients suffering from T2D, both with and without neuropathy, was analyzed. No genetic variants were evaluated in this study. The data demonstrated that the frequency of DNA damage was significantly higher in the T2D patients with DPN than in the controls [59]. Stoian et al. conducted a study in the University Center of Tırgu Mures, Romania. In their case-control study, which included a total of 182 participants, including 84 unrelated patients with T2D and an age-matched control group consisting of 98 unrelated individuals without T2D, they evaluated the influence of GSTM1, GSTT1, and GSTP1 variants on T2D and DPN risk. Their data suggested that GSTM1, GSTT1, and GSTP1 gene variants were not associated with individual susceptibility to developing DPN in patients with T2D in the Romanian population [60].
An association study of C936T polymorphism of the VEGF gene and the C242T polymorphism of the p22phox gene with T2D and DPN in a population of Caucasian ethnicity was studied by Ghisleni et al. According to their results, the C936T polymorphism of the VEGF gene and C242T polymorphism of the p22phox gene did not correlate with the risk of developing diabetes mellitus or neuropathic signs and symptoms. When considering the results of other studies, a substantial heterogeneity in the findings is observed, which demonstrates a complex link between the risk factors of DM and genetic predisposition to DPN [52]. Common susceptible and prognostic genetic factors associated with DPN in T2D are diverse in different pathophysiological pathways, and it is difficult to separate each genetic variant from the other as most of the variants are interrelated with each other (Table 1).
Table 1.
Genetic variants associated with DPN.
| Gene | Chromosomal location | Variants | Associated risk/remarks | Reference |
|---|---|---|---|---|
| Advanced glycation end receptor (AGER) | 6p21.32 | rs1800624 | Higher risk/defective inflammatory pathways | [22] |
| Peroxisome proliferator-activated receptor alpha (PPARA) | 3p25.2 | rs1801282 | Higher risk/defective inflammatory pathways | [27] |
| Bradykinin receptor B2 (BDKRB2) | 14q32.2 | rs1799722 | Higher risk/defective inflammatory pathways/African-Americans | [29] |
| Potassium voltage-gated channel subfamily J member 11 (KCNJ11) | 11p15.1 | E23K, G>A rs5219 | Higher risk/Chinese population/altered signaling pathways | [23] |
| CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) | 6p22.3 | rs7756992 | Higher risk/Chinese population | [23] |
| Transcription factor 7-like 2 (TCF7L2) | 10q25.2-q25.3 | rs7903146 | Higher risk/Chinese population | [23] |
| Methylenetetrahydro folate reductase (MTHFR) | 1p36.22 | C677T rs1801133 | Higher risk/altered folate metabolism | [24] |
| Apolipoprotein E (APOE) | 19q13.32 | ε4 allele- rs429358 rs7412 | Higher risk/altered lipid metabolism | [25] |
| Adrenoceptor alpha 2B (ADRA2B) | 2q11.2 | 12Glu9 rs879255577 | Higher risk/defects in regulation of neurotransmitter release from sympathetic nerves | [26] |
| microRNA 146a (MIR146a) | 5q33.3 | rs2910164 (G>C) | Lower risk | [49] |
| microRNA128a (MIR128a) | 2q21.3 | rs11888095 (C>T) | Higher risk | [49] |
| High mobility group box 1 pseudogene 46 (HMGB1P46) | 8q23.1 | rs6986153 | Males/higher risk | [39] |
| Zinc finger and SCAN domain containing 20 (ZSCAN20) | 1p35.1 | rs71647933 | Females/higher risk | [39] |
| Serpin family E member 1 (SERPINE1) | 7q22.1 | rs1799768 | Progressive type of DPN | [40] |
| Nerve growth factor receptor (NGFR) | 17q21.33 | rs734194 | Progressive type of DPN | [43] |
| Angiotensin-converting enzyme (ACE) | 17q23.3 | rs1799752 diallelic polymorphism: presence/absence of 287bp in intron 16 Heterozygous D/I Homozygous D/D |
Japanese population Lower risk Higher risk |
[45] |
| Aldo-keto reductase family 1 member B (AKR1B1) | 7q33 | rs5053 rs759853 | Higher risk/altered glucose metabolism | [47] |
| Vascular endothelial growth factor (VEGF) | 6p21.1 | C936T/rs3025039 rs2010963 rs699947 | Higher risk | [52] |
| Cytochrome b-245 alpha chain (CYBA) | 16q24.2 | C242T rs4673 | Higher risk | [52] |
| Heat shock protein family A (Hsp70) member 5 (HSPA5) | 9q33.3 | Promoter region 57168556T>C rs391957 | Higher risk | [54] |
| Adiponectin (ADIPOQ) | 3q27.3 | 45T/G rs2241766 | Higher risk | [56, 57] |
| 276G/T rs1501299 | Higher risk |
12. Conclusions
Although targeted gene sequencing is still a method of choice to identify rare functional mutations in monogenic disorders, exome sequencing becomes an attractive and cost-effective alternative when other disease-mapping strategies provide few or ambiguous results. This review has attempted to identify the common susceptibility and prognostic genetic factors associated with DPN in T2D. Knowledge about these factors is vital as DPN is one of the debilitating complications associated with T2D and identification of the common genetic variants would be valuable for the future development of gene panels targeted for the early detection and prognosis of DPN. Together with these gene panels, further gene expression studies will need to be conducted to modulate effective targeted therapies for DPN in these patients.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.International Diabetes Federation. IDF Diabetes Atlas. 6th. Brussels, Belgium: International Diabetes Federation; 2013. [PubMed] [Google Scholar]
- 2.Wolfs M. G. M., Hofker M. H., Wijmenga C., van Haeften T. W. Type 2 diabetes mellitus: new genetic insights will lead to new therapeutics. Current Genomics. 2009;10(2):110–118. doi: 10.2174/138920209787847023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh S. The genetics of type 2 diabetes mellitus: a review. Journal of Scientific Research. 2011;55:35–48. Banaras Hindu University, Varanasi. [Google Scholar]
- 4.Edwards J. L., Vincent A. M., Cheng H. T., Feldman E. L. Diabetic neuropathy: mechanisms to management. Pharmacology & Therapeutics. 2008;120(1):1–34. doi: 10.1016/j.pharmthera.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. Atlanta, GA, USA: US Department of Health and Human Services; 2014. [Google Scholar]
- 6.Guja C., Gagniuc P., Ionescu-tîrgovişte C. Genetic factors involved in the pathogenesis of type 2 diabetes. The Proceedings of the Romanian Academy, Series B. 2012;1:44–61. [Google Scholar]
- 7.Kommoju U. J., Reddy B. M. Genetic etiology of type 2 diabetes mellitus: a review. International Journal of Diabetes in Developing Countries. 2011;31(2):51–64. doi: 10.1007/s13410-011-0020-8. [DOI] [Google Scholar]
- 8.Kota S. K., Meher L. K., Jammula S., Kota S. K., Modi K. D. Genetics of type 2 diabetes mellitus and other specific types of diabetes; its role in treatment modalities. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2012;6(1):54–58. doi: 10.1016/j.dsx.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Kato N. Insights into the genetic basis of type 2 diabetes. Journal of Diabetes Investigation. 2013;4(3):233–244. doi: 10.1111/jdi.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent A. M., Russell J. W., Low P., Feldman E. L. Oxidative stress in the pathogenesis of diabetic neuropathy. Endocrine Reviews. 2004;25(4):612–628. doi: 10.1210/er.2003-0019. [DOI] [PubMed] [Google Scholar]
- 11.Yagihashi S., Mizukami H., Sugimoto K. Mechanism of diabetic neuropathy: where are we now and where to go? Journal of Diabetes Investigation. 2011;2(1):18–32. doi: 10.1111/j.2040-1124.2010.00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura M., Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [review] Endocrine Journal. 2011;58(9):723–739. doi: 10.1507/endocrj.EJ11-0113. [DOI] [PubMed] [Google Scholar]
- 13.Zeggini E., Scott L. J., Saxena R., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ropper A. H., Gorson K. C., Gooch C. L., et al. Vascular endothelial growth factor gene transfer for diabetic polyneuropathy: a randomized, double-blinded trial. Annals of Neurology. 2009;65(4):386–393. doi: 10.1002/ana.21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal D., Gudala K., Muthyala H., Esam H. P., Nayakallu R., Bhansali A. Prevalence and risk factors of development of peripheral diabetic neuropathy in type 2 diabetes mellitus in a tertiary care setting. Journal of Diabetes Investigation. 2014;5(6):714–721. doi: 10.1111/jdi.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesfaye S., Boulton A. J. M., Dyck P. J., et al. Diabetic neuropathies: update on definitions, diagnostic criteria, estimation of severity, and treatments. Diabetes Care. 2010;33(10):2285–2293. doi: 10.2337/dc10-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callaghan B. C., Cheng H. T., Stables C. L., Smith A. L., Feldman E. L. Diabetic neuropathy: one disease or two? Current Opinion in Neurology. 2012;25(5):536–541. doi: 10.1097/WCO.0b013e328357a797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papanas N., Vinik A. I., Ziegler D. Neuropathy in prediabetes: does the clock start ticking early? Nature Reviews Endocrinology. 2011;7(11):682–690. doi: 10.1038/nrendo.2011.113. [DOI] [PubMed] [Google Scholar]
- 19.Figueroa-Romero C., Sadidi M., Feldman E. L. Mechanisms of disease: the oxidative stress theory of diabetic neuropathy. Reviews in Endocrine and Metabolic Disorders. 2008;9(4):301–314. doi: 10.1007/s11154-008-9104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C., Rouen S., Dobrowsky R. T. Hyperglycemia and downregulation of caveolin-1 enhance neuregulin-induced demyelination. Glia. 2008;56(8):877–887. doi: 10.1002/glia.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGuire J. F., Rouen S., Siegfreid E., Wright D. E., Dobrowsky R. T. Caveolin-1 and altered neuregulin signaling contribute to the pathophysiological progression of diabetic peripheral neuropathy. Diabetes. 2009;58(11):2677–2686. doi: 10.2337/db09-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukic I. K., Humpert P. M., Nawroth P. P., Bierhaus A. The RAGE pathway: activation and perpetuation in the pathogenesis of diabetic neuropathy. Annals of the New York Academy Sciences. 2008;1126(1):76–80. doi: 10.1196/annals.1433.059. [DOI] [PubMed] [Google Scholar]
- 23.Lu J., Luo Y., Wang J., et al. Association of type 2 diabetes susceptibility loci with peripheral nerve function in a Chinese population with diabetes. Journal of Diabetes Investigation. 2017;8(1):115–120. doi: 10.1111/jdi.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yigit S., Karakus N., Inanir A. Association of MTHFR gene C677T mutation with diabetic peripheral neuropathy and diabetic retinopathy. Molecular Vision. 2013;19:1626–1630. [PMC free article] [PubMed] [Google Scholar]
- 25.Monastiriotis C., Papanas N., Trypsianis G. The ε4 allele of the APOE gene is associated with more severe peripheral neuropathy in type 2 diabetic patients. Angiology. 2013;64(6):451–455. doi: 10.1177/0003319712453645. [DOI] [PubMed] [Google Scholar]
- 26.Papanas N., Papatheodorou K., Papazoglou D., Kotsiou S., Christakidis D., Maltezos E. An insertion/deletion polymorphism in the alpha2B adrenoceptor gene is associated with peripheral neuropathy in patients with type 2 diabetes mellitus. Experimental and Clinical Endocrinology & Diabetes. 2007;115(05):327–330. doi: 10.1055/s-2007-967084. [DOI] [PubMed] [Google Scholar]
- 27.Hur J., Sullivan K., Pande M., et al. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain. 2011;134(11):3222–3235. doi: 10.1093/brain/awr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivero A., Mora C., Muros M., Garcia J., Herrera H., Navarro-Gonzalez J. F. Pathogenic perspectives for the role of inflammation in diabetic nephropathy. Clinical Science. 2009;116(6):479–492. doi: 10.1042/CS20080394. [DOI] [PubMed] [Google Scholar]
- 29.Kakoki M., Sullivan K. A., Backus C., et al. Lack of both bradykinin B1 and B2 receptors enhances nephropathy, neuropathy, and bone mineral loss in Akita diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10190–10195. doi: 10.1073/pnas.1005144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawelczyk T., Grden M., Rzepko R., Sakowicz M., Szutowicz A. Region-specific alterations of adenosine receptors expression level in kidney of diabetic rat. The American Journal of Pathology. 2005;167(2):315–325. doi: 10.1016/S0002-9440(10)62977-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan S. Z., Usher M. G., Mortensen R. M. PPARs: the vasculature, inflammation and hypertension. Current Opinion in Nephrology and Hypertension. 2009;18(2):128–133. doi: 10.1097/MNH.0b013e328325803b. [DOI] [PubMed] [Google Scholar]
- 32.Yamagishi S., Ogasawara S., Mizukami H., et al. Correction of protein kinase C activity and macrophage migration in peripheral nerve by pioglitazone, peroxisome proliferator activated-γ-ligand, in insulin-deficient diabetic rats. Journal of Neurochemistry. 2008;104(2):491–499. doi: 10.1111/j.1471-4159.2007.05050.x. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Tang K., Zhang Z., et al. Genetic diversity of the apolipoprotein E gene and diabetic nephropathy: a meta-analysis. Molecular Biology Reports. 2011;38(5):3243–3252. doi: 10.1007/s11033-010-9999-z. [DOI] [PubMed] [Google Scholar]
- 34.Cheng C., Kobayashi M., Martinez J. A., et al. Evidence for epigenetic regulation of gene expression and function in chronic experimental diabetic neuropathy. Journal of Neuropathology & Experimental Neurology. 2015;74(8):804–817. doi: 10.1097/NEN.0000000000000219. [DOI] [PubMed] [Google Scholar]
- 35.Donnelly L. A., Doney A. S., Hattersley A. T., Morris A. D., Pearson E. R. The effect of obesity on glycaemic response to metformin or sulphonylureas in type 2 diabetes. Diabetic Medicine. 2006;23(2):128–133. doi: 10.1111/j.1464-5491.2005.01755.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y. M., Cha B. S., Kim D. J., et al. Predictive clinical parameters for therapeutic efficacy of rosiglitazone in Korean type 2 diabetes mellitus. Diabetes Research and Clinical Practice. 2005;67(1):43–52. doi: 10.1016/j.diabres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Osterbrand M., Fahlen M., Oden A., Eliasson B. A. A method to predict the metabolic effects of changes in insulin treatment in subgroups of a large population based patient cohort. European Journal of Epidemiology. 2007;22(3):151–157. doi: 10.1007/s10654-007-9107-4. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien P. D., Hur J., Robell N. J., Hayes J. M., Sakowski S. A., Feldman E. L. Gender-specific differences in diabetic neuropathy in BTBR ob/ob mice. Journal of Diabetes and its Complications. 2016;30(1):30–37. doi: 10.1016/j.jdiacomp.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng W., Deshmukh H. A., Donnelly L. A., et al. A genome-wide association study provides evidence of sex-specific involvement of Chr1p35.1 (ZSCAN20-TLR12P) and Chr8p23.1 (HMGB1P46) with diabetic neuropathic pain. EBioMedicine. 2015;4(2)(10):1386–1393. doi: 10.1016/j.ebiom.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Festa A., D’Agostino R., Jr., Tracy R. P., Haffner S. M. Elevated levels of acute-phase proteins and plasminogen activator inhibitor-1 predict the development of type 2 diabetes: the insulin resistance atherosclerosis study. Diabetes. 2002;51(4):1131–1137. doi: 10.2337/diabetes.51.4.1131. [DOI] [PubMed] [Google Scholar]
- 41.Nicholas S. B., Aguiniga E., Ren Y., Kim J., Wong J., Govindarajan N. Plasminogen activator inhibitor-1 deficiency retards diabetic nephropathy. Kidney International. 2005;67(4):1297–1307. doi: 10.1111/j.1523-1755.2005.00207.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang R., Trevillyan J. M. c-Jun N-terminal kinase pathways in diabetes. The International Journal of Biochemistry & Cell Biology. 2008;40(12):2702–2706. doi: 10.1016/j.biocel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Chilton L., Middlemas A., Gardiner N., Tomlinson D. R. The p75 neurotrophin receptor appears in plasma in diabetic rats—characterisation of a potential early test for neuropathy. Diabetologia. 2004;47(11):1924–1930. doi: 10.1007/s00125-004-1550-0. [DOI] [PubMed] [Google Scholar]
- 44.Schulze P. C., Yoshioka J., Takahashi T., He Z., King G. L., Lee R. T. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. Journal of Biological Chemistry. 2004;279(29):30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- 45.Jurado J., Ybarra J., Romeo J. H., Garcia M., Zabaleta-del-Olmo E. Angiotensin-converting enzyme gene single polymorphism as a genetic biomarker of diabetic peripheral neuropathy: longitudinal prospective study. Journal of Diabetes and its Complications. 2012;26(2):77–82. doi: 10.1016/j.jdiacomp.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Chandrasekaran K., Anjaneyulu M., Inoue T., et al. Mitochondrial transcription factor A regulation of mitochondrial degeneration in experimental diabetic neuropathy. American Journal of Physiology Endocrinology and Metabolism. 2015;309(2):E132–E141. doi: 10.1152/ajpendo.00620.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraswathy R., Anand S., Kunnumpurath S. K., Kurian R. J., Kaye A. D., Vadivelu N. Chromosomal aberrations and exon 1 mutation in the AKR1B1 gene in patients with diabetic neuropathy. The Ochsner Journal. 2014;14(3):339–342. [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenzen J., Kumarswamy R., Dangwal S., Thum T. MicroRNAs in diabetes and diabetes-associated complications. RNA Biology. 2012;9(6):820–827. doi: 10.4161/rna.20162. [DOI] [PubMed] [Google Scholar]
- 49.Ciccacci C., Morganti R., Di Fusco D., et al. Common variants in MIR146a, MIR128a and MIR27a genes contribute to neuropathy susceptibility in type 2 diabetes. Acta Diabetologica. 2014;51(4):663–671. doi: 10.1007/s00592-014-0582-2. [DOI] [PubMed] [Google Scholar]
- 50.Prabhakar S. S. Role of nitric oxide in diabetic nephropathy. Seminars in Nephrology. 2004;24(4):333–344. doi: 10.1016/j.semnephrol.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Nakagawa T., Sato W., Glushakova O., et al. Diabetic endothelial nitric oxide synthase knockout mice develop advanced diabetic nephropathy. Journal of the American Society of Nephrology. 2007;18(2):539–550. doi: 10.1681/ASN.2006050459. [DOI] [PubMed] [Google Scholar]
- 52.Ghisleni M. M., Biolchi V., Jordon B. C., Rempel C., Genro J. P., Pozzobon A. Association study of C936T polymorphism of the VEGF gene and the C242T polymorphism of the p22phox gene with diabetes mellitus type 2 and distal diabetic polyneuropathy. Molecular Medicine Reports. 2015;12(3):4626–4633. doi: 10.3892/mmr.2015.3988. [DOI] [PubMed] [Google Scholar]
- 53.Wirostko B., Wong T. Y., Simó R. Vascular endothelial growth factor and diabetic complications. Progress in Retinal and Eye Research. 2008;27(6):608–621. doi: 10.1016/j.preteyeres.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Jia Y., Tong Y., Min L. Significance of functional GRP78 polymorphisms in predicting the onset of type 2 diabetic peripheral neuropathy in Chinese population. Neurological Research. 2015;37(8):683–687. doi: 10.1179/1743132815Y.0000000054. [DOI] [PubMed] [Google Scholar]
- 55.Abdelgadir M., Karlsson A. F., Berglund L., Berne C. Low serum adiponectin concentrations are associated with insulin sensitivity independent of obesity in Sudanese subjects with type 2 diabetes mellitus. Diabetology & Metabolic Syndrome. 2013;5(1):p. 15. doi: 10.1186/1758-5996-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han L. Y., Wu Q. H., Jiao M. L., et al. Associations between single-nucleotide polymorphisms (+45T>G, +276G>T, −11377C>G, −11391G>A) of adiponectin gene and type 2 diabetes mellitus: a systematic review and meta-analysis. Diabetologia. 2011;54(9):2303–2314. doi: 10.1007/s00125-011-2202-9. [DOI] [PubMed] [Google Scholar]
- 57.Sandy An S., Palmer N. D., Hanley A. J., et al. Genetic analysis of adiponectin variation and its association with type 2 diabetes in African Americans. Obesity. 2013;21(12):E721–E729. doi: 10.1002/oby.20419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji Z. Y., Li H. F., Lei Y., et al. Association of adiponectin gene polymorphisms with an elevated risk of diabetic peripheral neuropathy in type 2 diabetes patients. Journal of Diabetes and its Complications. 2015;29(7):887–892. doi: 10.1016/j.jdiacomp.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 59.Prasad M., Bronson S. C., Warrier T., et al. Evaluation of DNA damage in type 2 diabetes mellitus patients with and without peripheral neuropathy: a study in south Indian population. Journal of Natural Science, Biology and Medicine. 2015;6(1):80–84. doi: 10.4103/0976-9668.149096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoian A., Bănescu C., Bălaşa R. I., et al. Influence of GSTM1, GSTT1, and GSTP1 polymorphisms on type 2 diabetes mellitus and diabetic sensorimotor peripheral neuropathy risk. Disease Markers. 2015;2015:10. doi: 10.1155/2015/638693.638693 [DOI] [PMC free article] [PubMed] [Google Scholar]