Abstract
An effective method for extraction of intact genomic DNA from the extremely AT-rich polycentric anaerobic fungus Orpinomyces sp. strain PC-2 has been developed. This procedure involves removal of glycogen-like storage polysaccharides using hexadecyltrimethylammonium bromide (CTAB) and high salt washes. The DNA was digested with various restriction enzymes and was suitable for use as a PCR template, for Southern blotting, and for genomic library construction. Genomic DNA analysis of three representative genes (celE, bgl1, and xynA) encoding (hemi-) cellulolytic enzymes of the fungus revealed multiplicity of family 5 endocellulase genes (celE-like), and family 1 β-glucosidase genes (bgl1-like), but only a single copy of family 11 xylanase gene (xynA).
Obligately anaerobic fungi are part of the natural microflora of the alimentary tracts of herbivores and they might account for up to 20% of the total microbial biomass in the rumen [4, 17]. They have been isolated from digesta and feces of many herbivorous animals [17]. According to the number of sporangia developed from the thallus, they have been divided into monocentric and polycentric fungi [5]. Anaerobic fungi can colonize and penetrate plant tissues as a result of filamentous growth, which appears to degrade lignified tissue that is not degraded by other microorganisms in the rumen [13]. These fungi are active in the degradation of plant cell wall polysaccharides and represent a potential source of enzymes against cellulose and hemicelluloses. The importance of the anaerobic fungi is further supported by the synergism with rumen bacteria to enhance degradation of carbohydrates [13]. In addition, the anaerobic fungi may have the potential to contribute substantially to drug metabolism in the alimentary tract of host animals due to the ability to produce a wide range of enzymes with hydrolytic capacity [4]. Molecular evidence shows that hydrolytic enzymes of anaerobic fungi are either associated with fungal cellulosomes attached to the fungal cell wall or secreted outside of the mycelia as individual free enzymes [8, 13, 19]. Many genes encoding hydrolytic enzymes have been cloned and sequenced from the monocentric fungi Neocallimastix patriciarum, N. frontalis, and Piromyces sp. and from the polycentric fungi Orpinomyces sp. strain PC-2 and O. joyonii, mainly through screening cDNA libraries [13]. Genomic DNA analysis of genes encoding β-glucosidases, cellulases, and mannanases of Piromyces sp. E2 revealed a multiplicity of these genes encoding enzymes belong to the same enzyme family in the genome [10–12]. However, little information is available on the genome structures of the (hemi-) cellulolytic enzyme genes from the polycentric anaerobic fungi [8].
High-throughput DNA sequencing and the advent of the proteomics disciplines now offer the potential to obtain a blueprint for the lifestyle of a specific microbe, and to access its genetic potential in a comparative and functional manner. The genome sequences of several rumen bacteria with relevance to fiber degradation, such as Fibrobacter succinogenes, Ruminococcus albus, and Prevotella ruminicola strains 23, are available [13]. Due to the highly AT-rich (80–85 mol%) genomes of anaerobic fungi and to the extent that these genomes test the limits of the physiochemical methods utilized in their analyses, relatively little information is available regarding the genomes of these fungi. Further hampering research are the low yields of DNA and the high carbohydrate contents normally associated with current isolation techniques. For example, the DNA content of 3- to 6-day-old cultures of Neocallimastix sp. is only 2.7–3.2 μg/mg dry weight or 0.32% by weight as determined by spectrofluorometric method on sonicated samples [1, 5]. Subsequently, the biggest obstacle of genomic research among these fungi is the isolation of sufficient amounts of pure and intact genomic DNA [6, 20]. By modifying Rozman and Komel’s method [18], we developed a new procedure for successfully isolating high purity intact genomic DNA from an anaerobic polycentric fungus, Orpinomyces sp. PC-2 [4]. Genome organization of several genes encoding (hemi-) cellulolytic enzymes from this strain was analyzed using the isolated DNA.
Materials and Methods
Isolation of genomic DNA
Orpinomyces sp. PC-2 was as described by Borneman et al. [4]. The fungus was grown anaerobically at 39°C for 2.5 days in a semidefined medium supplemented with 0.3% (w/v) cellobiose [8]. A method modified from Rozman and Komel [18] was used for genomic DNA isolation. Harvested fresh mycelium was rinsed with deionized water. Excess water was removed by vacuum filtration onto Whatman No. 1 filter paper. The mycelium was rapidly frozen in liquid nitrogen and ground into a fine powder with a mortar and pestle. Ground mycelium (1.5 g wet weight) was suspended in a 50-ml polypropylene screw cap tube containing 10 ml extraction buffer (100 mM Tris-HCl, pH 8.0, 100 mM EDTA, 250 mM NaCl, 0.45 mg/ml proteinase K). One milliliter of 10% sodium lauroylsarcosine was added when the cells were completely suspended. Lysis was allowed to proceed overnight with gentle shaking at 50°C. Following lysis, 1.02 ml of 5 M NaCl and 0.81 ml of 10% (w/v) hexadecyltrimethylammonium bromide (CTAB) in 0.7 M NaCl was added for every 6-ml extract and the mixture was incubated at 65°C for 30 min with occasional inverting.
After the samples had cooled to room temperature, an equal volume of chloroform:isoamyl alcohol (24:1) was added and mixed gently by inversion until an even milky white suspension appeared. The mixture was centrifuged for 10 min at 9,700g at 4°C. Sixty percent volume of isopropanol was added to the upper aqueous phase to initiate DNA precipitation. The pellet was spun down and washed with 5 ml of 70% ethanol then resuspended in 500 μl 10 mM Tris-HCl, pH 7.6, 1 mM EDTA (TE). The sample was transferred to a microcentrifuge tube and incubated with 0.1 mg RNase A at 37°C for 1 hour. The supernatant containing DNA was precipitated with 50 μl of 3 M sodium acetate and 550 μl ethanol. The resulting DNA pellet was washed twice with 70% ethanol, drained, and resuspended in 200 μl TE. Determination of the purified genomic DNA concentration was performed with the PicoGreen dsDNA quantification system, using calf thymus DNA as a standard, according to the manufacturer’s protocol (Molecular Probes, Eugene, OR).
Primer Design, PCR, and Cloning
DNA probes from the genomic DNA of Orpinomyces sp. PC-2 were synthesized by PCR. Primers are listed in Table 1. Probe pCELE recognizes a 467-bp segment of a family 5 cellulase catalytic domain; probe pBGL1 recognizes a 637-bp segment of a family 1 β-glucosidase catalytic domain; and probe pXYNA recognizes a 376-bp segment of a family 11 xylanase catalytic domain. PCR was performed in a Mastercycler gradient (Eppendorf). The amplification conditions were one cycle of 95°C for 3 min, 30 cycles with each cycle including 30 sec of melting at 95°C, 30 sec of annealing at 50°C, and 60 sec of extension at 72°C, and one final extension cycle at 72°C for 5 min. The PCR products were cloned directly into the pCR2.1-TOPO vector (Invitrogen), sequenced for verification, and examined by 2% agarose gel electrophoresis.
Table 1.
Primers used in amplifying DNA probes
| Primer name | GenBank no. | Sequence from 5′ to 3′ | Position | |
|---|---|---|---|---|
|
| ||||
| Begin | End | |||
| CelE-F | U97153 | atccaaaggctactccag | 197 | 214 |
| CelE-R | U97153 | tggaatcataaggtgacg | 646 | 663 |
| Bgll-F | AF016864 | atgtcgaatctggtgacagatcc | 338 | 360 |
| Bgll-R | AF016864 | agaatattgtgggaacaccagac | 952 | 974 |
| XynA-F | U57819 | accttcaaggctgaatggag | 207 | 226 |
| XynA-R | U57819 | gacggacactgaagtattgc | 563 | 582 |
Southern Blot Analysis
The genomic DNA (3 μg) was digested overnight with 3 units of the following restriction enzymes: Hind III or Acc I and EcoR I or Xba I overnight. The digested DNA was subjected to electrophoresis on a 0.7% (w/v) agarose gel. The fractionated DNA was transferred onto an IMMOBILON nylon membrane (Millipore). The DNA was cross-linked to the membrane and hybridized with the DNA probes. Labeling of the probes and membrane visualization was performed with the DIG Easy Hyb labeling and detection kit (Roche Applied Science) as described in detailed here. The membrane was pre-hybridized with 15 ml pre-warmed DIG Easy Hyb at 65°C for 1 hour. The DIG labeled PCR probe was denatured by boiling at 95°C for 5 min and rapidly cooled on ice. The denatured probe was mixed with 10 ml of pre-warmed DIG Easy Hyb (40°C). Ten milliliters of the probe/hybridization solution was added to the membrane and it was incubated overnight at 40°C. The membrane was washed twice with 2 × SSC/0.1% SDS and twice with 0.5 × SSC/0.1% SDS at room temperature for 5 min each. The membrane was blocked by incubation in blocking buffer for 1 hour at room temperature. Next the membrane was incubated for 30 min in antibody solution (blocking buffer/anti-DIG-AP; 1:10000), then it was washed twice in Washing Buffer for 15 min at room temperature. The membrane was equilibrated in Detection Buffer for 5 min. The membrane was incubated in 1 ml ready-to-use CSPD substrate solution. After removing the excess solution, the membrane was sealed in a plastic bag and exposed to film until achieving desired result.
Results and Discussion
Isolation of the Fungal Genomic DNA
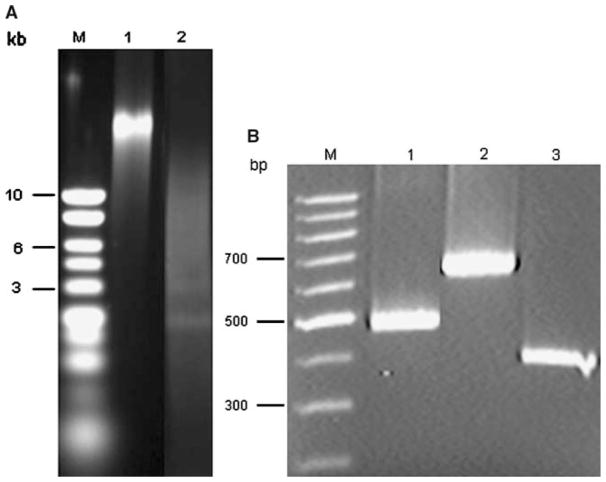
To obtain large amounts of healthy mycelium, the Orpinomyces sp. PC-2 was cultured in a medium containing glucose, xylose, or cellobiose. Cellobiose was found to be a better carbon source than the other two sugars for growth (data not shown). The yield of DNA was 73.5 μg from approximately 1.5 g (wet weight) of mycelium. The procedure was scaled up fivefold by starting with 5 × 1.5 g wet weight of mycelium simultaneously. The preparation was free of contaminating RNA on an agarose gel (Fig. 1A) and had an A260/A280 of about 1.81. The quality and purity of the DNA was further examined by restriction enzyme digestion (Fig. 1A). To construct a cosmid DNA library for the genomic sequence, both quantity and quality of DNA are extremely important. There are also some DNA quality criteria needed to perform genome sequencing. The successful enzymatic digestion of the DNA confirmed its quality. Although it has been known that the DNA contents of anaerobic fungi are lower than those of aerobic fungi, little information (except that it is low) is available regarding yield of DNA isolated from the anaerobic fungi [5]. Using the current method, it was estimated that the yield of the anaerobic fungal DNA was about 25–50% of DNA isolated from aerobic fungi [3, 18]. For comparison, we employed this method to isolate genomic DNA from the monocentric anaerobic fungus N. frontalis EB188 [13] as well and similar results were obtained.
Fig. 1.
Isolated Orpinomyces sp. PC-2 genomic DNA. (A) Before and after digestion with restriction enzymes. Lane M, DNA molecular standards. Lane 1, genomic DNA before restriction digestion. Lane 2, genomic DNA after restriction digestion with EcoR I and Hind III. (B) Detection of the carbohydrate-degrading enzyme genes by PCR. Lane M, DNA molecular standards. Lane 1, celE. Lane 2, bgl1. Lane 3, xynA.
It is crucial to start with fresh mycelium for DNA isolation. Cells were kept frozen after the mycelium was disrupted in liquid nitrogen and immediately transferred to lysis buffer to avoid DNA shearing. Established protocols for DNA purification were examined for comparison and yielded low amounts of truncated DNA (data not shown). For example, only about 15 μg of truncated genomic DNA was obtained from about 1.5 g wet weight of mycelium by a method using a cell-wall digesting enzyme, Triton X-100, and guanidine-HCl as lysis buffer, compared to a 73.5 μg yield by our protocol.
Genomic Organization of celE, bgl1, and xynA From Orpinomyces sp. PC-2
The genomic DNA preparation was used to confirm the presence of celE, bgl1, and xynA genes by PCR. There was a clear band for each gene fragment on a 1.5 % (w/v) agarose gel, indicating that the DNA obtained by this method was of good quality (Fig. 1B).
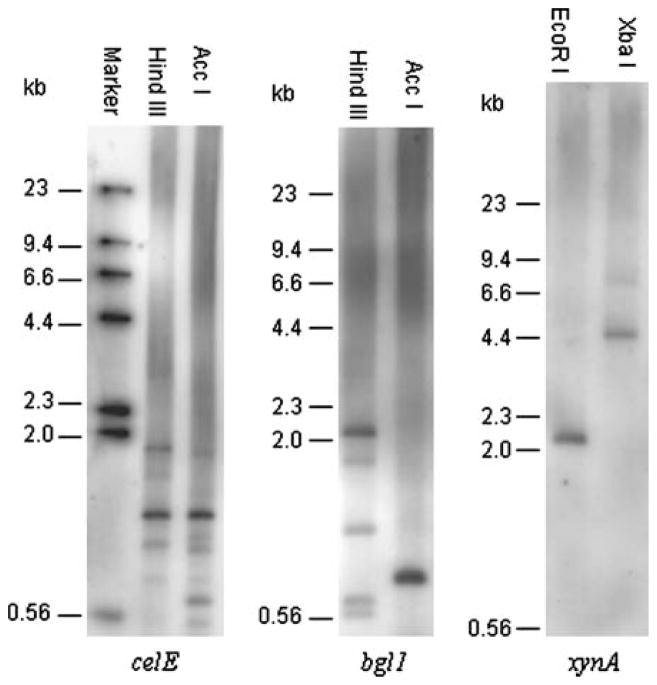
CelE belongs to the glycosyl hydrolase family 5 enzymes [7]. It randomly hydrolyzes carboxymethylcellu-lose and cello-oligosaccharides in the pattern of endoglucanases. CelE contains 477 amino acid residues and is highly homologous (72.3%) to CelB of Orpinomyces sp. PC-2 [7] and to several endocellulases from other anaerobic fungi [9, 16, 21]. It has a non-catalytic repeated peptide domain at the C-terminal end and is a component of the Orpinomyces sp. PC-2 cellulosome. The celE gene was devoid of introns, which may be originally transferred from rumen bacteria and subsequently duplicated in the anaerobic fungus [7]. Although several similar genes encoding family 5 enzymes have been found among species of both monocentric and polycentric anaerobic fungi [9, 21], no information is available regarding genomic organization. A PCR product of pCELE corresponding to the 5′ catalytic region of celE was used to prepare a probe. Southern hybridization analysis revealed at least eight genes most likely coding for homologous enzymes of CelE in Orpinomyces sp. PC-2 (Fig. 2A). The autoradiograph showed one strong and more than seven weak signals in both of the lanes.
Fig. 2.
Genomic Southern blot analysis of Orpinomyces sp. PC-2 carbohydrate-degrading enzyme genes. (A) Probed with a PCR product from celE. (B) Probed with a PCR product from bgl1. (C) Probed with a PCR product from xynA.
Bgl1 is a glycosyl hydrolase family 1 β-glucosidase, which converts oligosaccharides to glucose. The enzyme contains 657 amino acid residues and was homologous to bacterial β-glucosidases [15]. It lacks a dockerin and is not a component of the Orpinomyces sp. strain PC-2 cellulosome. Bgl1 had a significant level of identity with a β-glucosidase from Piromyces E2 (Cel1A, 72%), which is carried as multiple copies in Piromyces E2 [12]. A PCR product of pBGL1 corresponding to the 3′ catalytic region of bgl1 was used to prepare a probe. Southern hybridization analysis revealed at least five genes most likely coding for homologous enzymes of Bgl1 in Orpinomyces sp. PC-2 (Fig. 2B), which is similar to the cel1A gene from Piromyces E2. An intensive band in lane Hind III matched the predicted 2068-bp fragment of the cDNA sequence of bgl1 cut by the enzyme, the other intensive band in lane Acc I matched the predicted 796-bp fragment of the cDNA sequence of bgl1 cut by the enzyme, which indicate they represent bgl1 in Orpinomyces sp. PC-2 [15].
XynA is a major xylanase found in Orpinomyces sp. PC-2 [14]. It randomly hydrolyzes xylan in the pattern of endoxylanase. XynA contains 362 amino acid residues and has a glycosyl hydrolase family 11 enzyme catalytic domain at the N-terminal part and a noncatalytic repeated peptide domain at its C-terminal part [14]. Similar enzymes homologous to XynA were found in several other anaerobic fungi [13]. A PCR product of pXYNA corresponding to the 5′ catalytic region of xynA was used to prepare a probe. Southern hybridization analysis revealed only one gene encoding the glycosyl hydrolase family 11 xylanase in Orpinomyces sp. PC-2 (Fig. 2C). A family 10 xylanase gene (xynB) of N. patriciarum, which does not exhibit any significant homology with the Orpinomyces sp. PC-2 xynA, was expressed in the fungus [2]. In order to elucidate the expression profile of the xynB-like gene of Orpinomyces sp. PC-2, a cDNA library, constructed in λZAPII, of Orpinomyces sp. PC-2 was intensively screened for xylanase-expressing clones. Only the xynA gene was identified from screening over 200 positive clones by sequencing or PCR verification. This revealed that the xynB-like gene may not be efficiently expressed and is not a major component of the cellulase-hemicellulase enzyme system in Orpinomyces sp. PC-2.
We have developed an effective and reliable genomic DNA isolation method for anaerobic fungi. The isolated DNA is of high quality with good yield and shows no apparent degradation, making it suitable for a variety of purposes, including Southern blot hybridization analysis, PCR, and library construction for genome sequencing. Although there are morphological differences between monocentric and polycentric anaerobic fungi, their genes encoding hydrolytic enzymes have similar structures in their genomes and more importantly have the same origin. There are many signs of evidence to demonstrate that horizontal gene transfer events occurred between anaerobic fungi and, subsequently, the transferred genes were duplicated in individual fungi [3]. It seems that gene duplications happen more frequently in genes encoding the hydrolytic enzymes responsible for degrading less accessible carbohydrates, such as cellulose, than in those genes responsible for degrading easier accessible carbohydrates, such as xylan and lichenin. A future successful genome sequence for this important group of fungi will facilitate gene finding, expression, structure, and function studies.
Acknowledgments
We thank Drs. John B. Sutherland and Seong-Jae Kim for their critical review of the manuscript.
Literature Cited
- 1.Billon-Grand G, Fiol JB, Breton A, Bruyere A, Oulhaj Z. DNA of some anaerobic rumen fungi: G + C content determination. FEMS Microbiol Lett. 1991;82:267–270. doi: 10.1016/0378-1097(91)90272-c. [DOI] [PubMed] [Google Scholar]
- 2.Black GW, Hazlewood GP, Xue G-P, Orpin CG, Gilbert HJ. Xylanase B from Neocallimastix patriciarum contains a non-catalytic 455-residue linker sequence comprised of 57 repeats of an octapeptide. Biochem J. 1994;299:381–387. doi: 10.1042/bj2990381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgia PT, Eagleton LE, Miao Y. A preparations from Aspergillus and other filamentous fungi. BioTechniques. 1994;17:431–432. [PubMed] [Google Scholar]
- 4.Borneman WS, Akin DE, Ljungdahl LG. Fermentation products and plant cell wall degrading enzymes produced by monocentric and polycentric anaerobic rumen fungi. Appl Environ Microbiol. 1989;55:1066–1073. doi: 10.1128/aem.55.5.1066-1073.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownlee AG. Remarkably AT-rich genomic DNA from the anaerobic fungus Neocallimastix. Nucleic Acids Res. 1989;17:1327–1335. doi: 10.1093/nar/17.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownlee AG. A rapid DNA isolation procedure applicable to many refractory filamentous fungi. Fungal Genet Newslett. 1988;35:8–9. [Google Scholar]
- 7.Chen H, Li X-L, Blum DL, Ljungdahl LG. Two genes of the anaerobic fungus Orpinomyces sp. strain PC-2 encoding cellulases with endoglucanase activities may have arisen by gene duplication. FEMS Microbiol Lett. 1998;159:63–68. doi: 10.1111/j.1574-6968.1998.tb12842.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Li X-L, Blum DL, Ximenes EA, Ljungdahl LG. celF of Orpinomyces PC-2 has an intron and encodes a cellulase (CelF) containing a carbohydrate-binding module. Appl Biochem Biotech. 2003;105–108:775–785. doi: 10.1385/abab:108:1-3:775. [DOI] [PubMed] [Google Scholar]
- 9.Eberhardt RY, Gilbert HJ, Hazlewood GP. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology. 2000;146:1999–2008. doi: 10.1099/00221287-146-8-1999. [DOI] [PubMed] [Google Scholar]
- 10.Harhangi HR, Akhmanova A, Steenbakkers PJM, Jetten MSM, van der Drift C, den Op Camp JM. Genomic DNA analysis of genes encoding (hemi-) cellulolytic enzymes of the anaerobic fungus Piromyces sp. E2. Gene. 2003;314:73–80. doi: 10.1016/s0378-1119(03)00705-4. [DOI] [PubMed] [Google Scholar]
- 11.Harhangi HR, Freelove ACJ, Ubhayasekera W, van Dinther M, Steenbakkers PJM, Akhmanova A, van der Drift C, Jetten MSM, Mowbray SL, Gilbert HJ, den Op Camp JM. Cel6A, a major exocellulase from the cellulosome of the anaerobic fungi Piromyces sp. E2 and Piromyces equi. Biochem Biophys Acta. 2003;1628:30–39. doi: 10.1016/s0167-4781(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 12.Harhangi HR, Steenbakkers PJM, Akhmanova A, Jetten MSM, van der Drift C, den Op Camp JM. A highly expressed family 1 β-glucosidase with transglycosylation capacity from the anaerobic fungus Piromyces sp. E2. Biochem Biophys Acta. 2002;1574:293–303. doi: 10.1016/s0167-4781(01)00380-3. [DOI] [PubMed] [Google Scholar]
- 13.Krause DO, Deman SE, Mackie RI, Morrison M, Rae AL, Attwood GT, McSweeney CS. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev. 2003;27:663–693. doi: 10.1016/S0168-6445(03)00072-X. [DOI] [PubMed] [Google Scholar]
- 14.Li X-L, Chen H, Ljungdahl LG. Monocentric and polycentric anaerobic fungi produce structurally related cellulases and xylanases. Appl Environ Microbiol. 1997;63:628–635. doi: 10.1128/aem.63.2.628-635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X-L, Ljungdahl LG, Ximenes EA, Chen H, Felix CR, Cotta MA, Dien BS. Properties of a recombinant beta-glucosidase from polycentric anaerobic fungus Orpinomyces PC-2 and its application for cellulose hydrolysis. Appl Biochem Biotechnol. 2004;113–116:233–250. doi: 10.1385/abab:113:1-3:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu X, Selinger B, Yanke L-J, Cheng K-J. Isolation and analysis of two cellulose cDNAs from Orpinomyces joyonii. Gene. 2000;245:119–126. doi: 10.1016/s0378-1119(00)00028-7. [DOI] [PubMed] [Google Scholar]
- 17.Rezaeian M, Beakes GW, Parker DS. Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol Res. 2004;108:1227–1233. doi: 10.1017/s0953756204000929. [DOI] [PubMed] [Google Scholar]
- 18.Rozman D, Komel R. Isolation of genomic DNA from filamentous fungi with high glucan level. BioTechniques. 1994;16:382–383. [PubMed] [Google Scholar]
- 19.Steenbakkers PJM, Li X-L, Ximenes EA, Arts JG, Chen H, Ljungdahl LG, den Op Camp JM. Noncatalytic docking domains of cellulosomes of anaerobic fungi. J Bacteriol. 2001;183:5325–5333. doi: 10.1128/JB.183.18.5325-5333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor JW, Natvig DO. Isolation of fungal DNA. In: Fuller MS, Jaworski A, editors. Zoosporic fungi in teaching and research. Athens, GA: Southeastern Publishing Corp; 1987. pp. 252–258. [Google Scholar]
- 21.Zhou L, Xue G-P, Orpin CG, Black GW, Gilbert HJ, Hazlewood GP. Intronless celB from the anaerobic fungus Neocallimastix patriciarum encodes a modular family A endoglucanase. Biochem J. 1994;297:359–364. doi: 10.1042/bj2970359. [DOI] [PMC free article] [PubMed] [Google Scholar]