Abstract
Enterococcus faecalis azoreductase (AzoA) is a very active enzyme with a broad spectrum of substrate specificity and is capable of degrading various azo dyes. The enzyme has an absolute requirement for reduced FMN, which delivers a total of four electrons from NADH to the substrate, resulting in the cleavage of the nitrogen double bond. In this study, we report the identification of amino acid residues critical for FMN binding in AzoA. FMN is stabilized by 22 amino acid residues, eight of which, Trp-105, Asn-106, Leu-107, Gly-150, Gly-151, Tyr-153, Asn-121 and Tyr-129, are involved in binding the FMN isoalloxazine ring. In silico analysis of the amino acid residues revealed that the Trp residue at position 105 of AzoA is the most likely significant contributor to the binding of FMN to the enzyme and is involved in FMN stabilization and destabilization. Site-directed mutagenesis analysis of Trp-105 was performed to determine the role of this amino acid residue in FMN binding and azo dye reductive activity. The mutant proteins were overexpressed in Escherichia coli and purified by anion-exchange and size-exclusion chromatography. The replacement of Trp-105 by the small side-chain amino acids Ala and Gly caused complete loss of both affinity for FMN and enzyme activity. Substitution of Tyr for Trp-105 did not significantly decrease the Vmax of the enzyme (22 % reduction). Substitutions with three bulky side-chain amino acids, Gln, Phe and His, produced enzymes with lower Vmax values (decreases of 68.2, 30.6 and 8.2-fold, respectively). However, these mutated enzymes maintained Km values similar to the wild-type enzyme. This study provides an insight into the catalytic properties of AzoA in FMN stabilization and enzyme activity.
INTRODUCTION
Azo dyes are a major group of colorants used in textiles, plastics, pharmaceuticals, cosmetics and food products (Chen, 2006; Stolz, 2001). Many synthetic azo dyes can be converted to potentially carcinogenic aromatic amines after reduction (Platzek et al., 1999). Because of the widespread use of azo dyes, human intestinal micro-organisms are exposed to these compounds (Hanauer, 1996). The human microbiota play a key role in the metabolism of azo dyes, with azo reduction being the most important reaction related to the toxicity and mutagenicity of these compounds (Chung, 1983). The microbial flora of the human gastrointestinal tract comprises at least several thousand species of bacteria (Eckburg et al., 2005). Enterococcus faecalis is one of the predominant intestinal bacteria, and it is able to cometabolize many substrates, including azo dyes (Chen et al., 2004). FMN-dependent azoreductases, key components of the system responsible for biodegradation of azo dyes, catalyse the reduction of the nitrogen double bonds (-N=N-) of azo compounds to generate aromatic amines via hydrazo intermediates. Catalysis involves sequential transfer of four electrons from NAD(P)H to the substrate via FMN. The flavin could carry out this process in two sequential steps by NADH or NADPH reduction with subsequent oxidation by the azo compound via a hydrazo intermediate (Deller et al., 2006). Biochemical and 3D structural information is available for FMN-dependent azoreductases of Escherichia coli, Ent. faecalis, Staphylococcus aureus, Pseudomonas aeruginosa, Bacillus subtilis and Saccharomyces cereviseae (Chen et al., 2004, 2005; Ito et al., 2006; Liu et al., 2007; Wang et al., 2007).
Azoreductase from Ent. faecalis (AzoA) is one of the most active oxygen-tolerant FMN-dependent azoreductases characterized to date, showing a broad spectrum of substrate specificity. Recently, we reported the 3D structure of AzoA determined at 2.07 Å (0.207 nm) resolution (Liu et al., 2007). That study provided direct evidence for a 46 kDa asymmetrical homodimer with one FMN molecule bound to each monomer. Dimeric AzoA has two separate active sites that are located at the interfaces between the two monomers, and the binding site of FMN lies inside each active site, indicating that the dimeric arrangement is required for active holoenzyme. In the active site, the FMN sits inside a positively charged cavity formed by several stretches of residues; the si face of the isoalloxazine ring of FMN is solvent-accessible, whereas its re face is buried in the protein. The structural environment of the active site could be correlated with the mechanical view adopted previously for azoreductase (Ito et al., 2006; Liu et al., 2007; Wang et al., 2007).
AzoA absolutely requires reduced FMN for enzyme activity. Despite the large body of biochemical and 3D structural information available on bacterial FMN-dependent azoreductases (Chen et al., 2004, 2005; Ito et al., 2006; Liu et al., 2007; Wang et al., 2007), little is known about the role of specific amino acid residues involved in the flavin binding and catalytic mechanism. In this study, a computational protein–FMN interaction analysis was conducted, which identified Trp-105 on loop ℓ9 as a potential determinant for FMN stabilization. We utilized site-directed mutagenesis with the equivalents of Trp-105 from related proteins of known 3D structure and then performed kinetic analysis to evaluate the relative contributions of the side-chains on position 105 to the FMN binding and catalysis.
METHODS
Materials
Methyl red (MR), NADH and FMN were obtained from Sigma-Aldrich. All other reagents were reagent grade and high purity. E. coli BL21-Gold(DE3)pLysS (Stratagene) was used for recombinant DNA studies. E. coli strains were cultured at 37 °C in Luria–Bertani (LB) medium with appropriate antibiotics (50 μg ml−1). pET-11a (Stratagene) was used for cloning and expression.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuickChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The mutagenic oligonucleotide primers are listed in Table 1. The template was pAZOA, which consists of the azoA gene cloned into pET-11a (Chen et al., 2004). Complete sequences were determined on a model 3100 Genetic Analyzer (Applied Biosystems) using T7 promoter and terminator primers to confirm that only the intended mutations had been introduced. pAZOA and its mutant plasmids were used for transformation of E. coli BL21-Gold (DE3)pLysS.
Table 1.
Primers used for site-directed mutagenesis of AzoA
Underlined nucleotides indicate mutations incorporated into primers.
| Mutation | Primer sequences (5′→3′) |
|---|---|
| W105Y | GCTAATCCAATGTACAACTTAAACGTAC |
| GTACGTTTAAGTTGTACATTGGATTAGC | |
| W105Q | GTAATTGCTAATCCAATGCAAAACTTAAACGTACCGAC |
| GTCGGTACGTTTAAGTTTTGCATTGGATTAGCAATTAC | |
| W105F | GTAATTGCTAATCCAATGTTCAACTTAAACGTACCGAC |
| GTCGGTACGTTTAAGTTGAACATTGGATTAGCAATTAC | |
| W105H | GTAATTGCTAATCCAATGCACAACTTAAACGTACCG |
| CGGTACGTTTAAGTTGTGCATTGGATTAGCAATTAC | |
| W105A | GTAATTGCTAATCCAATGGCGAACTTAAACGTACCG |
| CGGTACGTTTAAGTTCGCCATTGGATTAGCAATTAC | |
| W105G | GCTAATCCAATGGGAAACTTAAACGTAC |
| GTACGTTTAAGTTTCCCATTGGATTAGC |
Expression and purification of wild-type and mutant proteins
E. coli BL21-Gold(DE3)pLysS cells harbouring pAZOA or pAZOA mutants grown on LB-ampicillin-chloramphenicol plate medium were inoculated into a flask containing 20 ml LB-ampicillin-chloramphenicol (50 μg ml−1 of each antibiotic) broth. The culture was shaken (250 r.p.m.) at 37 °C overnight. Then the culture was added to 400 ml LB broth containing no selection antibiotics and was shaken at 250 r.p.m. at 37 °C for 2 h. IPTG (1 mM) was added and the culture was incubated for another 2.5 h. Cells were harvested by centrifugation (5000 g, 10 min), washed with 20 ml buffer containing 25 mM Tris/HCl (pH 7.5) and resuspended in 30 ml of the same buffer. The cells were then disrupted by freezing and thawing followed by 5 min sonication at 4 °C with a Vibracell VCX 400 model sonifier (Sonics & Materials). Cell debris was removed by centrifugation at 12 000 g for 10 min. Proteins were purified at 4 °C by using an AKTApurifier 10 system with UNICORN 4.10 software (Amersham Biosciences) essentially as described by Chen et al. (2004). BSA (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa) (Amersham Biosciences) were used as the standards for molecular mass determination of the purified proteins on a HiLoad 16/60 Superdex 75 pre-grade gel filtration column (1.6×60 cm, Amersham Biosciences) (Chen et al., 2004).
SDS-PAGE analysis
SDS-PAGE was carried out in Laemmli’s buffer (Laemmli, 1970) with 12.5 % polyacrylamide. Perfect protein markers (Novagen) were used. Electrophoresis was performed in a Hoefer SE 260 Mighty Small II Mini Vertical unit (Amersham Biosciences). Gels were stained for proteins with Coomassie brilliant blue R-250 (Bio-Rad).
Enzyme assay of the mutant proteins
Azoreductase activity was determined by measuring the decrease in A430 of MR with a Hewlett Packard 8453 UV–visible spectrophotometer at room temperature (23 °C). A typical reaction mixture (2.0 ml) contained 25 mM potassium phosphate buffer (pH 7.1), 25 μM MR and 0.1 mM NADH. The reaction was initiated by addition of the enzyme. Initial velocity was determined by monitoring the change in the amount of substrate in the first 2 min in a glass cuvette of 1.0 cm light path. The 23.4 mM−1 molar absorption coefficient for MR at 430 nm (ε= cm−1) was used. Proteins were quantified using the bicinchoninic acid assay (Pierce) with BSA as the standard (Chen et al., 2004).
Kinetic constants of the mutant proteins
Initial velocities of the enzymic reaction were determined by varying the concentrations of one substrate, MR (from 5 to 40 μM) or NADH (from 50 to 400 μM), while the concentration of the other substrate was kept constant (NADH 400 μM or MR 40 μM). Km and Vmax values were obtained from Lineweaver–Burk plots. Each data point was analysed three times.
Computational analysis
Homology modelling
The homology models for mutants were generated using SWISS-MODEL (http://swissmodel.expasy.org/workspace/index.php) with the wild-type protein structure (2HPV) as a template. The structures were minimized by the GROMOS 43B1 force field; algorithm for minimization, steepest descent; maximum force to stop (kJ mol−1), 1.0; number of steps, 200 (Schwede et al., 2003). Basic structure validation was checked using PROCHECK (European Bioinformatics Institute, Cambridge, UK).
In silico analysis
To align the AzoA (2HPV) sequence with related proteins of known 3D structure in the PDB (Protein Data Bank), SAS (http://www.ebi.ac.uk/thornton-srv/databases/sas/) was used (Milburn et al., 1998). The protein–FMN interactions were verified and plotted using the LPC (Ligand Protein Contacts) online server (http://bip.weizmann.ac.il/oca-bin/lpccsu) (Sobolev et al., 1999) and LIGPLOT v.4.4.2 (Wallace et al., 1995), respectively. SuperPose (version 1.0) was used for structural superposition and RMSD (root-mean-square deviation) calculation of the wild-type AzoA and the modelled mutant proteins (Maiti et al., 2004). Two scoring functions, DrugScoreCSD (http://pc1664.pharmazie.uni-marburg.de/drugscore/) and the stand-alone X-Score (Velec et al., 2005), were adopted to score FMN binding affinity in the protein–ligand complexes. PyMOL (0.99RC6) (http://www.pymol.org) was used to visualize the per-atom score contributions and all other structure figures. LPC and HBPLUS (McDonald & Thornton, 1994) were used to make careful predictions of hydrogen bonds.
RESULTS
Identification of residues critical for FMN binding in AzoA
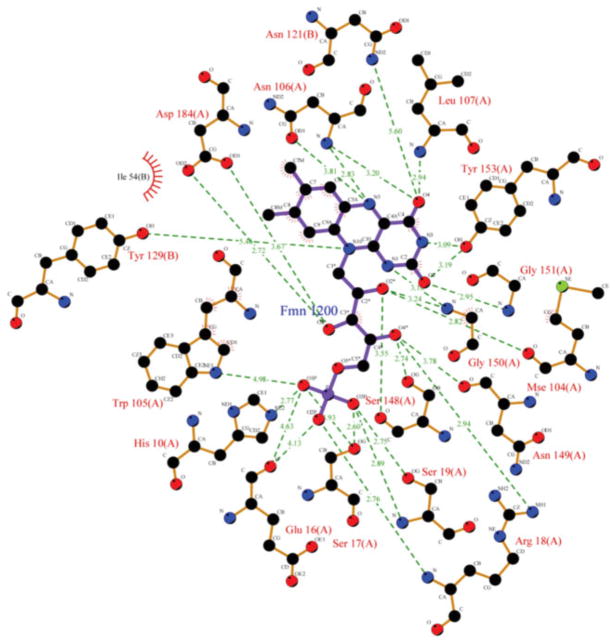
As generally observed in flavoenzymes, the FMN in AzoA is bound by a network of hydrogen bonds and hydrophobic interactions (Fig. 1). To determine which amino acid residue or which type of contact contributes most to stabilizing FMN in the structure, interatomic ligand–protein contacts were analysed in detail using the LPC online server (http://bip.weizmann.ac.il/oca-bin/lpccsu) with the wild-type AzoA (2HPV). All of the protein residues in contact with FMN are listed in Table 2 along with the types of contact. As a whole, the bound FMN is stabilized by 22 residues (17 in chain A and 5 in chain B), which are involved in hydrogen bonds and hydrophobic interactions (Table 2, Fig. 1). Among these 22 residues, eight, Trp-105, Asn-106, Leu-107, Gly-150, Gly-151, Tyr-153, Asn-121(B) and Tyr-129(B), are involved in the binding of the FMN isoalloxazine ring. The 2,4-pyrimi-dinedione moiety of the isoalloxazine ring forms hydrogen bonds with the side-chain atoms of these eight residues, while the dimethylbenzene portion of the isoalloxazine ring forms hydrophobic contacts with the enzyme. The ribityl chain of FMN is hydrogen bonded to 10 residues: to the main chain atoms of the eight residues His-10, Glu-16, Ser-17, Arg-18, Ser-19, Ser-148, Asn-149 and Asp-184, and to the side-chain atoms of Glu-16 and Mse-104. The phosphate moiety is also hydrogen bonded to His-10, Glu-16, Ser-17, Arg-18, Ser-19 and Trp-105.
Fig. 1.
Schematic diagram showing the interactions between the FMN and amino acid residues in AzoA (2HPV). Hydrogen bonds are indicated with green broken lines (red residue numbers) and van der Vaals’ interactions by red hatching (black residue numbers). Each atom is coloured according to atom type.
Table 2.
Residues in contact with the ligand FMN1200 in the wild-type (2HPV)
| Residue* | Dist. (Å)† | Surf. (Å2)‡ | Specific contacts§ | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| HB | Arom | Phob | DC | ||||
| 10A | His* | 2.8 | 26.0 | + | − | − | − |
| 16A | Glu | 4.1 | 4.7 | + | − | − | − |
| 17A | Ser* | 2.6 | 27.7 | + | − | − | − |
| 18A | Arg* | 2.8 | 54.5 | + | − | − | + |
| 19A | Ser* | 2.7 | 26.0 | + | − | − | − |
| 103A | Pro* | 3.7 | 12.5 | − | − | − | + |
| 104A | Met | 2.8 | 39.1 | + | − | + | + |
| 105A | Trp* | 3.4 | 41.6 | + | + | + | + |
| 106A | Asn* | 2.8 | 36.7 | + | − | − | + |
| 107A | Leu* | 2.9 | 36.3 | + | − | + | + |
| 148A | Ser* | 2.7 | 42.8 | + | − | − | − |
| 149A | Asn* | 3.7 | 6.6 | + | − | − | − |
| 150A | Gly* | 3.1 | 26.3 | + | − | − | − |
| 151A | Gly* | 3.0 | 25.8 | + | − | − | − |
| 153A | Tyr* | 3.1 | 31.0 | + | − | − | − |
| 184A | Asp* | 2.7 | 35.2 | + | − | − | − |
| 185A | His* | 5.7 | 0.7 | − | − | − | − |
| 54B | Ile* | 3.5 | 31.9 | − | − | + | − |
| 59B | Leu* | 4.6 | 14.4 | − | − | + | − |
| 62B | Trp* | 5.0 | 11.4 | − | − | + | − |
| 121B | Asn* | 5.6 | 1.6 | + | − | − | + |
| 129B | Tyr* | 5.5 | 11.0 | + | − | − | − |
Indicates the residues contacting the ligand by their side-chain (including CA atoms).
Dist. indicates the closest distance between atoms of FMN and the residue.
Surf. indicates the contact surface area between FMN and the residue.
HB, hydrophilic–hydrophilic contact (hydrogen bond); Arom, aromatic–aromatic contact; Phob, hydrophobic–hydrophobic contact; DC, hydrophobic–hydrophilic contact (destabilizing contact).
Based on the contact information between FMN and AzoA (2HPV) (Fig. 1), Trp-105 is thought to contribute most to stabilizing FMN in the structure. Interestingly, as shown in Table 2, Trp-105 is involved in both stabilizing and destabilizing contacts: it is involved in hydrophilic–hydrophilic (hydrogen bond), aromatic–aromatic and hydrophobic–hydrophobic contacts, which contribute to FMN stabilization, and also in hydrophobic–hydrophilic interaction, which contributes to FMN destabilization. Trp105-NE1 forms a hydrogen bond with FMN-O1P (Surf. 1.9 Å2) and also contacts FMN-C5* (Dist. 5.3 Å and Surf. 1.8 Å2), contributing to stabilization of FMN binding. However, it also causes FMN destabilization in the structure by contacts with the atoms C7M (Dist. 4.3 Å and Surf. 0.2 Å2) and C8M (Dist. 3.7 Å and Surf. 10.5 Å2) of FMN (data not shown). The CD1 of Trp-105, which is the nearest (Dist. 3.4 Å) to atoms of FMN and the residue, forms hydrophobic interactions with a set of atoms of FMN, C7 (Dist. 3.4 Å and Surf. 1.3 Å2), C7M (Dist. 3.8 Å and Surf. 6.7 Å 2), C8 (Dist. 3.4 Å and Surf. 2.2 Å 2), C9 (Dist. 3.6 Å and Surf. 2.9 Å 2) and C2* (Dist. 4.9 Å and Surf. 0.4 Å 2) (Fig. 1). The atoms CA, CG, CB, CD2 and CE2 of Trp-105 also support FMN binding in the structure. All these interactions affect all parts, including the isoalloxazine ring, the ribityl chain and the phosphate moiety, of the bound FMN.
To apply structural information derived from the related proteins of known 3D structure in the PDB to the site-directed mutagenesis of Trp-105, a FASTA alignment from a search of the sequence of AzoA (2HPV) was generated by SAS analysis (Fig. 2). The retrieved and aligned sequences from the PDB can be clustered by the ligand, FMN or FAD, into two groups. The proteins 2d5i (PDB) and 1t5b (PDB) each have a FMN ligand, and show a favourable sequence identity of 37.4 % of the entire protein sequence and appreciable alignment of secondary structure with AzoA. Note that those proteins with a FAD ligand, despite a relatively low sequence identity (<27.1 %) to AzoA, have appreciable overall alignments, based on the secondary structure of the flavin binding region. Human quinone oxidoreductase 2 (NQO2) has been known to reduce MR, indicating that it can also act as a four-electron transfer oxidoreductase like AzoA. NQO2 is expressed selectively in organs such as liver and kidney. It may function physiologically as one of several vitamin K reductases (Wu et al., 1997). Moreover, although there is no significant conservation of residues in the flavin binding region, particularly in positions 103–107 of AzoA, the corresponding residues are in contact with the flavin in the structure of the proteins. In AzoA, Trp-105 is on the turning point of loop ℓ9, made up of residues 104–110, which connects β-sheet β3 and α-sheet α3 (Fig. 2). The equivalents of Trp-105 of AzoA in the related proteins of known 3D structure are either tyrosine or glutamine. Tyrosine is the most conserved among the FMN binding proteins in Fig. 2.
Fig. 2.
Sequence alignment of the portion of AzoA (2HPV) involved in FMN binding with that of the related proteins of known 3D structure in the PDB. The equivalents of Trp-105 are indicated in the black box. *, Conserved amino acid residues.
Production and purification of the mutant proteins
SDS-PAGE indicated that AzoA wild-type and all AzoA mutants were induced and overexpressed in E. coli, with a strong band of 23 kDa on SDS-PAGE (Fig. 3a). The cell extract supernatants from the IPTG-induced cultures of wild-type, and mutants W105Y, W105Q, W105F and W105H, displayed a bright yellow colour indicating the presence of the flavin proteins. Elevated levels of azo-reductase activity were detected in the samples as compared with that of the non-induced wild-type culture, indicating that the flavin proteins with azoreductase activity were expressed in the host cells. However, no bright yellow coloration or elevated levels of enzyme activity were observed in the cell extract supernatants from the induced cultures of W105A and W105G. The recombinant proteins of wild-type and mutants with the bright yellow colour were purified to homogeneity by consecutive chromatography using anion exchangers HiPrep Q XL and Mono Q followed by a gel filtration step on HiLoad Superdex 75 (Fig. 3b) (Chen et al., 2004). On the basis of spectroscopy and protein molecular mass, the molar ratios between FMN and purified proteins for the wild-type and mutants W105Y, W105Q, W105F and W105H are similar (~2), suggesting that 2 FMN bind to 1 mutant AzoA (homodimer) (Table 3). However, no FMN was detected in the purified proteins of mutants W105A and W105G (Table 3).
Fig. 3.
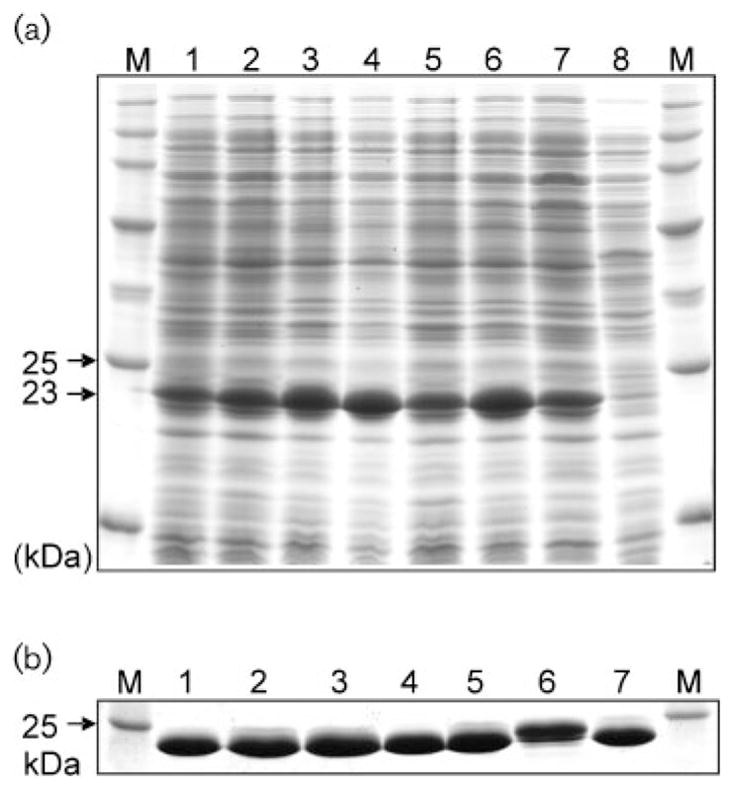
SDS-PAGE of protein profiles of cell extracts (a) and purified protein preparations (b) from wild-type and mutant AzoA of Ent. faecalis expressed in E. coli. Lanes: M, protein molecular mass standards; 1, wild-type; 2, W105Y; 3, W105Q; 4, W105F; 5, W105H; 6, W105A; 7, W105G; and 8, wild-type. Extracts in lanes 1–7 were induced with IPTG; extract in lane 8 was not induced with IPTG.
Table 3.
In silico analysis of the specific contacts of residue 105 with ligand FMN in the wild-type and the mutants
| Protein | Residue (105) | Dist. (Å)* | Surf. (Å2)† | Specific contacts‡ | FMN§ | FMN binding affinity | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| HB | Arom | Phob | DC | DrugScore|| | X-Score¶ | |||||
| Wild-type | Trp | 3.4 | 41.6 | + | + | + | + | ++ | −201753 (5.30) | 6.52 |
| W105Y | Tyr | 3.4 | 47.9 | + | + | + | − | ++ | −210603 (5.32) | 6.16 |
| W105Q | Gln | 3.6 | 30.7 | − | − | + | + | ++ | −189194 (5.28) | 5.93 |
| W105F | Phe | 3.5 | 43.6 | − | + | + | − | ++ | −207471 (5.31) | 6.14 |
| W105H | His | 1.8 | 62.6 | − | + | + | + | ++ | −200886 (5.30) | 5.90 |
| W105A | Ala | 3.6 | 19.1 | − | − | + | − | − | −176275 (5.24) | 5.82 |
| W105G | Gly | 3.6 | 15.0 | − | − | − | − | − | −171712 (5.23) | 5.69 |
Dist. indicates the closest distance between atoms of the ligand and the residue.
Surf. indicates the contact surface area between the ligand and the residue.
HB, hydrophilic–hydrophilic contact (hydrogen bond); Arom, aromatic–aromatic contact; Phob, hydrophobic–hydrophobic contact; DC, hydrophobic–hydrophilic contact (destabilizing contact).
++, contains 2 FMN per dimer.
The more negative the DrugScore value, the greater the FMN binding affinity. The scores in parentheses are negative logarithm units (−log).
A higher value of X-Score is more favourable.
Kinetic characterization and FMN binding of the mutant proteins
Table 4 compares kinetic constants of the wild-type and mutants with MR as the substrate. Among the mutants W105Y, W105Q, W105F and W105H, Km values were similar to the wild-type values of 158 and 25.4 μM for NADH and MR, with ranges from 24.7 to 28.2 μM for MR and from 166 to 175 μM for NADH, respectively, which indicates that these mutations in Trp-105 do not affect binding constants for either MR or NADH. The Vmax value of mutant W105Y was comparable to that of the wild-type. In the mutants W105Q, W105F and W105H, in which the number of FMN binding per AzoA was not changed (Table 3), Vmax values were significantly decreased. However, the mutants W105A and W105G, which completely lacked FMN, showed no enzyme activity (Table 4). These data indicated that Trp-105 is involved in FMN stability and sequential electron transfer from NADH to MR via FMN mediation rather than NADH- and MR-binding capabilities.
Table 4.
Kinetic parameters of wild-type and the mutants
| Enzyme | Km (MR) (μM) | Km (NADH) (μM) | Vmax (MR) [U (mg protein)−1] |
|---|---|---|---|
| Wild-type | 25.4±2.1 | 158±13 | 88.6±6.5 |
| W105Y | 27.6±1.5 | 166±17 | 68.7±6.7 |
| W105Q | 28.2±3.2 | 169±10 | 1.3±0.1 |
| W105F | 24.7±1.7 | 175±14 | 2.9±0.2 |
| W105H | 25.1±2.0 | 171±14 | 10.8±0.9 |
| W105A | – | – | – |
| W105G | – | – | – |
Since Trp-105 of wild-type AzoA is involved in both stabilizing and destabilizing FMN in the active site, mutational analysis on the residue can provide valuable information on the role of the specific amino acid side-chain in FMN binding and in enzyme activity. Table 3 shows the effects of residue Trp-105 substitutions on FMN binding affinity. Analysis of the mutants indicates that Trp-105 of AzoA is mainly responsible for FMN binding affinity. The effect of Trp-105 mutations on the affinity of the complex depends on the type of residue that is introduced. The W105A and W105G mutants exhibited the greatest changes in FMN binding. The W105G mutant, in which the side-chain was removed, completely lost the ability to bind FMN (Table 3). Even the introduction of a methyl side-chain produced a dramatic decrease in FMN binding affinity, resulting in loss of affinity for the flavin. The substitution of Ala for Trp-105 removes the interactions established by the Trp side-chain atoms, causing weaker FMN binding. Furthermore, an Ala in this position decreases the contact surface area between FMN and the residue, causing more accessibility to the solvent. However, introduction of the bulky side-chains did not affect FMN binding. In the mutants W105Y, W105Q, W105F and W105H, the FMN is bound with approximately equal affinity to the wild-type (Table 3, Fig. 4).
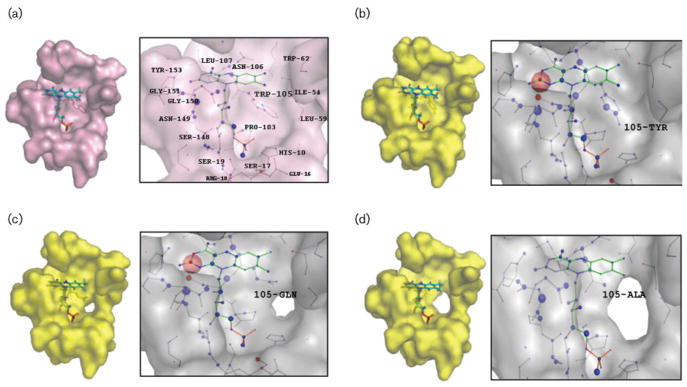
Fig. 4.
The active-site pockets and FMN environment of AzoA (2HPV) and its mutants. The panels show the FMN geometries and the per-atom score contributions of 2HPV (a), W105Y (b), W105Q (c), and W105A (d), respectively. FMN, TRP-105, TYR-105, GLN-105 and ALA-105 molecules are represented by stick models. In the visualization of the per-atom score contributions, favourably interacting atoms are surrounded by blue spheres whereas unfavourable interactions are shown in red. The sizes of the spheres correspond to the values of the contributing per-atom scores.
In silico analysis of the wild-type and mutant proteins
The modelling set of the six single-point mutants of AzoA at position 105 was prepared by homology modelling with 2HPV (AzoA) as the template. In silico analysis of the modelled mutants provides further insight into the structural consequences of these amino acid substitutions. The relationship between FMN binding affinity and enzyme activity is directly addressed through the 3D structure comparison of the mutant proteins with the wild-type AzoA. None of the substitutions introduced large changes to the mutants and the small changes that were observed were localized within the immediate vicinity of the substitution. The local RMSDs of the residues involved in the active site were more than 0.2 Å, with the exception of W105Y (0.04 Å), while the global RMSDs of the α carbons of the mutant proteins were less than 0.06 Å.
The FMN (oxidized form) binding affinity of each mutant was calculated using two different scoring functions, the knowledge-based DrugScore and the empirical-based X-score. The scores of the FMN binding affinity and the per-atom score contributions, together with the LPC analysis, can be used as a measure of the relative stability of the bound FMN in the structures. Specifically, the visualization of the per-atom score contributions gives us further insight into the overall structural impact of the mutations. The computational scores show a quantitative correlation with the experimentally determined FMN binding affinities. The analysis shows that the two substitutions with the smaller side-chains, W105A and W105G, cause the largest decrease in FMN binding affinity relative to that of the wild-type. In the case of mutant W105A, the score was decreased from 5.30 to 5.24 (wild-type vs W105A) in DrugScore. Strong unfavourable interactions are observed at position Met-104, where the N3-C2=O2 locus of FMN comes too close to the residue (Fig. 4). The distance between M104-CE and FMN-N3 is 2.11 Å (5.20 Å in the wild-type). In LPC analysis, Gly-105 cannot contribute interactions for the stabilization of FMN (Table 3). This mutation results in an inactive enzyme that was unable to bind to FMN, together with the W105A mutant.
The modelled structures also suggested that changing the side-chain size at position 105 could significantly affect the solvent exposure of the bound FMN cofactor. On LPC analysis, the exposed surface of the FMN systematically increased from 134.4 to 156 Å 2 (wild-type vs W105G) as the size of the side-chain at position 105 was decreased, with the change in exposed area being localized mainly to the dimethylbenzene portion of the isoalloxazine ring.
DISCUSSION
FMN has an important functional role in AzoA: transfer of NADH-derived electrons to azo dye substrates (Chen et al., 2004; Liu et al., 2007). AzoA shares features with the catalytic apparatus of other azoreductases, AzoR (Ito et al., 2006) from E. coli and paAzoR1 (Wang et al., 2007) from Pseudomonas aeruginosa. FMN is bound to approximately the same regions of these proteins, making the overall conformation of FMN and the orientation of isoalloxazine moiety very similar in each; the si face of the isoalloxazine ring of FMN is solvent-accessible, whereas its re face is buried in the protein. These features are in favour of the ping-pong mechanism: a NAD(P)H molecule or substrate is stacked between an aromatic amino acid and the isoalloxazine ring of flavin, which was previously postulated for the proteins. All the enzymes employ a Tyr residue for the pi-pi stacking that facilitates the transfer of a hydride from NAD(P)H to FMN, which is the first stage of the reduction reaction.
Although azoreductases share a set of common features with flavodoxins with respect to the flavin binding, a significant difference is in the flavin isoalloxazine ring binding site. In flavodoxin, the isoalloxazine rings are sandwiched between aromatic amino acids, tryptophan and tyrosine, flanking the re face and si face, respectively (Lostao et al., 2003; Murray & Swenson, 2003). AzoA, however, does not have the combined net effect of Trp and Tyr on FMN binding. No aromatic residues seem to be in the si face directly affecting flavin ring binding (Fig. 1). Tyr-129 is in a position at a distance of 5.34 Å from the OH of tyrosine to N5 of FMN and at a 68° angle to the si face of FMN, so that this aromatic residue cannot contribute to the flavin ring binding. Therefore, if AzoA Trp-105 is a homologue of the Trp involved in FMN stabilization of flavodoxin (Lostao et al., 1997), the position of AzoA Trp-105 is intriguing. This aromatic amino acid residue appears to interact with all moieties of FMN, with its indole functional group being involved in the interactions. This contribution to FMN binding is probably due to the combination of the residue position and its side-chain specificity in AzoA. The presence of an aromatic side-chain at the flavin binding site is a common feature of many flavin binding proteins and is considered essential for the flavin cofactor binding and hydrophobic active site environment (Ito et al., 2006; Liu et al., 2007; Wang et al., 2007). It is known to promote a stable and highly negative redox potential for the electron-transporting activity of the enzyme (Li & Meighen, 1995).
The FMN binding affinity and enzyme activity of the mutants were revealed to be correlated closely with the side-chain length of the introduced amino acids. AzoA did not appear to be tolerant of the substitution of Trp-105 by short side-chain amino acids. Complete loss of flavin of the mutants W105A and W105G was probably due to a decrease in the number of FMN–protein interactions caused by the introduced short side-chain amino acids, resulting in no enzyme activity. On the other hand, substitution of Trp-105 by the aromatic amino acids Tyr and Phe did not affect the bound FMN content when compared to the wild-type AzoA (Table 3). In the mutant W105Y, the free phenolic group of the Tyr-105 side-chain interacts with all the moieties of FMN, similar to Trp-105 of the wild-type AzoA. However, in spite of binding an approximately equal amount of FMN as compared to the wild-type, the mutants with bulky side-chains showed decreased catalytic activity. The results may be related to structural perturbations induced by the bulky side-chain substitutions. At least in part, local adjustments within the FMN binding site are likely to be responsible for the lower enzyme activities observed in the mutants. In fact, conformational changes could affect not only the binding of FMN, NADH and/or substrates like MR, but also the interactions among them. However, since structural changes were localized within the FMN binding area, it is most likely that the mutations caused changes in the position of the bound FMN, which negatively affected the interactions between FMN and NADH and/or substrate, resulting in the decrease in enzyme activity. Although none of the substitutions introduced significant changes to the mutants, relatively small changes in the protein structure within the immediate vicinity of the substitution were observed, leaving FMN possibly more accessible to solvent. The exposed surface of the FMN was gradually increased as the size of the side-chain at position 105 was decreased, with the change in exposed area being localized mainly in the dimethylbenzene portion of the isoalloxazine ring.
In conclusion, we performed this study to elucidate the structural and functional role of the residue Trp-105 in the AzoA reductase. The results demonstrated that Trp-105 plays important roles in both the FMN binding and catalytic ability of AzoA. This study provides a step forward in determining the catalytic properties of AzoA in FMN stabilization and enzyme activity. Further mutagenesis studies are needed to elucidate the role of other potential amino acid residues in the enzyme.
Acknowledgments
We thank Drs Fatemeh Rafii and Robin Stingley for their critical review of the manuscript. This study was funded by the United States Food and Drug Administration and the National Center for Toxicological Research and supported in part by appointments (Haiyan Xu and Ohgew Kweon) to the Postgraduate Research Fellowship Program and an appointment to the summer internship program (Siwei Chen) at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Abbreviations
- MR
methyl red
- PDB
Protein Data Bank
- RMSD
root-mean-square deviation
- LPC
ligand protein contacts
References
- Chen H. Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci. 2006;7:101–111. doi: 10.2174/138920306776359786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang RF, Cerniglia CE. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif. 2004;34:302–310. doi: 10.1016/j.pep.2003.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hopper SL, Cerniglia CE. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology. 2005;151:1433–1441. doi: 10.1099/mic.0.27805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KT. The significance of azo-reduction in the mutagenesis and carcinogenesis of azo dyes. Mutat Res. 1983;114:269–281. doi: 10.1016/0165-1110(83)90035-0. [DOI] [PubMed] [Google Scholar]
- Deller S, Sollner S, Trenker-El-Toukhy R, Jelesarov I, Gubitz GM, Macheroux P. Characterization of a thermostable NADPH : FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis. Biochemistry. 2006;45:7083–7091. doi: 10.1021/bi052478r. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer SB. Inflammatory bowel disease. N Engl J Med. 1996;334:841–848. doi: 10.1056/NEJM199603283341307. [DOI] [PubMed] [Google Scholar]
- Ito K, Nakanishi M, Lee WC, Sasaki H, Zenno S, Saigo K, Kitade Y, Tanokura M. Three-dimensional structure of AzoR from Escherichia coli. An oxidoreductase conserved in micro-organisms. J Biol Chem. 2006;281:20567–20576. doi: 10.1074/jbc.M513345200. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li Z, Meighen EA. Tryptophan 250 on the alpha subunit plays an important role in flavin and aldehyde binding to bacterial luciferase. Effects of W→Y mutations on catalytic function. Biochemistry. 1995;34:15084–15090. doi: 10.1021/bi00046a014. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Chen H, Shaw N, Hopper SL, Chen L, Chen S, Cerniglia CE, Wang BC. Crystal structure of an aerobic FMN-dependent azoreductase (AzoA) from Enterococcus faecalis. Arch Biochem Biophys. 2007;463:68–77. doi: 10.1016/j.abb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Lostao A, Gomez-Moreno C, Mayhew SG, Sancho J. Differential stabilization of the three FMN redox forms by tyrosine 94 and tryptophan 57 in flavodoxin from Anabaena and its influence on the redox potentials. Biochemistry. 1997;36:14334–14344. doi: 10.1021/bi971384h. [DOI] [PubMed] [Google Scholar]
- Lostao A, Daoudi F, Irun MP, Ramon A, Fernandez-Cabrera C, Romero A, Sancho J. How FMN binds to Anabaena apoflavodoxin: a hydrophobic encounter at an open binding site. J Biol Chem. 2003;278:24053–24061. doi: 10.1074/jbc.M301049200. [DOI] [PubMed] [Google Scholar]
- Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Milburn D, Laskowski RA, Thornton JM. Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng. 1998;11:855–859. doi: 10.1093/protein/11.10.855. [DOI] [PubMed] [Google Scholar]
- Murray TA, Swenson RP. Mechanism of flavin mononucleotide cofactor binding to the Desulfovibrio vulgaris flavodoxin. 1. Kinetic evidence for cooperative effects associated with the binding of inorganic phosphate and the 5′-phosphate moiety of the cofactor. Biochemistry. 2003;42:2307–2316. doi: 10.1021/bi026967s. [DOI] [PubMed] [Google Scholar]
- Platzek T, Lang C, Grohmann G, Gi US, Baltes W. Formation of a carcinogenic aromatic amine from an azo dye by human skin bacteria in vitro. Hum Exp Toxicol. 1999;18:552–559. doi: 10.1191/096032799678845061. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol. 2001;56:69–80. doi: 10.1007/s002530100686. [DOI] [PubMed] [Google Scholar]
- Velec HF, Gohlke H, Klebe G. DrugScore (CSD)-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem. 2005;48:6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Hagemeier C, Rahman N, Lowe E, Noble M, Coughtrie M, Sim E, Westwood I. Molecular cloning, characterisation and ligand-bound structure of an azoreductase from Pseudomonas aeruginosa. J Mol Biol. 2007;373:1213–1228. doi: 10.1016/j.jmb.2007.08.048. [DOI] [PubMed] [Google Scholar]
- Wu K, Knox R, Sun XZ, Joseph P, Jaiswal AK, Zhang D, Deng PS, Chen S. Catalytic properties of NAD(P)H : quinone oxidoreductase-2 (NQO2), a dihydronicotinamide riboside dependent oxidoreductase. Arch Biochem Biophys. 1997;347:221–228. doi: 10.1006/abbi.1997.0344. [DOI] [PubMed] [Google Scholar]