Abstract
Azo dyes are a predominant class of colourants used in tattooing, cosmetics, foods and consumer products. A gene encoding NADPH-flavin azoreductase (Azo1) from the skin bacterium Staphylococcus aureus ATCC 25923 was identified and overexpressed in Escherichia coli. RT-PCR results demonstrated that the azo1 gene was constitutively expressed at the mRNA level in S. aureus. Azo1 was found to be a tetramer with a native molecular mass of 85 kDa containing four non-covalently bound FMN. Azo1 requires NADPH, but not NADH, as an electron donor for its activity. The enzyme was resolved to dimeric apoprotein by removing the flavin prosthetic groups using hydrophobic-interaction chromatography. The dimeric apoprotein was reconstituted on-column and in free stage with FMN, resulting in the formation of a fully functional native-like tetrameric enzyme. The enzyme cleaved the model azo dye 2-[4-(dimethylamino)phenylazo]benzoic acid (Methyl Red) into N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid. The apparent Km values for NADPH and Methyl Red substrates were 0·;074 and 0·057 mM, respectively. The apparent Vmax was 0·4 µM min−1 (mg protein)−1. Azo1 was also able to metabolize Orange II, Amaranth, Ponceau BS and Ponceau S azo dyes. Azo1 represents the first azoreductase to be identified and characterized from human skin microflora.
INTRODUCTION
Azo dyes are characterized by one or more R1-N=N-R2 bonds and are enzymically metabolized to aromatic amines by reductive mechanisms in prokaryotic and eukaryotic organisms (Nakayama et al., 1983). There are thousands of synthetic azo dyes used in the textile, pharmaceutical, cosmetics and food industries, of which more than 500 contain potentially carcinogenic aromatic amines in their chemical formulation (Platzek et al., 1999). When these compounds enter the human body through ingestion of food, dermal contact and inhalation, they are metabolized via azoreductases to aromatic amines by the intestinal microflora and in the mammalian liver (Levine, 1991; Chung et al., 1992; Platzek et al., 1999). Synthetic azo compounds have been identified in tattoo colourants, including a derivative of 3,3-dichlorobenzidine, azo derivatives of 2-amino-4-nitrotoluene and phthalocyanine (Von Lehmann & Pierchalla, 1988; Baumler et al., 2000). Aromatic amines, such as benzidine, induce urinary bladder cancer in humans and tumours in some experimental animals (Combes & Haveland-Smith, 1982; Chung, 1983). Human skin contains a characteristic complement of microbial species, which do no harm by their presence under usual circumstances. The microflora of the human skin is variable with regard to bacterial strains and species. The quantity varies in the range of 6 × 102 to 2 × 106 bacterial cells cm−2, which depends on age and colonization site (Kloos & Schleifer, 1975). Staphylococcus aureus is widespread in nature and commonly found on human skin, skin glands and mucous membranes (Nagase et al., 2002). The skin microbiota plays a very important role in preventing many pathogens from colonizing the skin and causing disease, in a similar manner to the ‘barrier effect’ produced by intestinal microbes.
Dermal exposure of consumers to azo dyes by injection of tattoo ink and the topical application of colourants as well as wearing coloured textiles should be evaluated for safety, since, potentially, aromatic amines metabolized from aromatic amine-based dyes by the microflora are easily absorbed by the skin (Levine, 1991; Chung et al., 1992; Platzek et al., 1999). There is little information about the ability of the skin microflora to metabolize azo dyes. Platzek et al. (1999) found that skin bacteria were able to cleave azo dyes under in vitro conditions. However, to our knowledge, azoreductases or their corresponding genes have not been identified or described in the skin microflora. It has been suggested that S. aureus may serve as a model strain for measuring the ability of human skin flora to reduce azo dyes (Platzek et al., 1999). To better understand the metabolism of tattoo pigments, topically applied cosmetics and textile colourants by skin microflora and the potential toxicity of the reaction products to the human body, it is essential to discover the relevant genes and characterize their products within skin microflora.
In this study, we describe for the first time the cloning and identification of the gene, azo1, encoding a novel aerobic tetrameric FMN-dependent azoreductase, from S. aureus. Characterization of the purified Azo1 overexpressed in Escherichia coli demonstrated that this azoreductase was able to decolorize a wide range of structurally complex sulfonated azo dyes.
METHODS
Bacterial strains, plasmids and growth conditions
S. aureus ATCC 25923 was grown in brain heart infusion (BHI) broth or on BHI agar plates and used for inoculum and genomic DNA preparation. E. coli TOP10F′ (Invitrogen), NovaBlue (DE3) (Novagen), and BL21-Gold(DE3)pLysS (Stratagene) were used for recombinant DNA studies. E. coli strains were cultured at 37 °C in Luria–Bertani (LB) medium with appropriate antibiotics (50 µg ml−1). The plasmids pCR2.1-TOPO (Invitrogen) and pET-11d (Stratagene) were used for cloning and expression, respectively.
Twenty millilitres of S. aureus from BHI broth was inoculated into 200 ml BHI medium supplemented with individual azo dye at a final concentration of 0·2 mM and incubated at 37 °C for various times in air without shaking. The supernatants were used to assay azo dye reduction by measuring residual absorption at the appropriate wavelength for each azo dye, as described in the enzyme assay description below.
Cloning of S. aureus azo1 gene and expression of Azo1 in E. coli
Genomic DNA of S. aureus was isolated essentially as described by Wang et al. (2002). Plasmids from E. coli Top10F′ and NovaBlue (DE3) were isolated using a Qiaprep Spin Miniprep kit (Qiagen). A DNA fragment containing the putative S. aureus azo1 was obtained by PCR with the genomic DNA of S. aureus used as template. The forward primer included a NcoI site before the start codon: 5′-catgccatggctatgaaaggattaattattattggc-3′ (Saa-forward). The reverse primer included a BamHI site downstream of the azo1 stop codon, beginning at 660 bp: 5′-gcggatccgcttactattggtgcattag-3′ (Saa-reverse). PCR was performed in a Mastercycler gradient (Eppendorf). The amplification conditions were one cycle at 95 °C for 3 min, 30 cycles with each cycle including 30 s of melting at 95 °C, 30 s of annealing at 50 °C and 60 s of extension at 72 °C, and one final extension cycle at 72 °C for 5 min. The PCR products were examined by 1% agarose gel electrophoresis.
The PCR products were directly cloned into the pCR2.1-TOPO vector and sequenced. Both strands of the DNA inserts were sequenced by walking from the ends of the inserts by using plasmid- and insert-specific primers. For overexpression of Azo1 in E. coli, the PCR products were cleaved with NcoI and BamHI (New England BioLabs). The digested DNA was purified from the agarose gel and ligated into pET-11d with a rapid DNA ligation kit (Roche). E. coli NovaBlue (DE3) was transformed with the resulting plasmids. The plasmids (pAZO1) were subsequently isolated and introduced into E. coli BL21-Gold(DE3)pLysS by transformation. DNA sequence analysis, translation and alignment with related genes and proteins were carried out using the Lasergene program (Version 5, dnastar). The GenBank program blast was utilized to find similar genes or proteins.
Enzyme assays
Azoreductase activity was assayed by measuring the decrease in optical density at suitable wavelengths with a Hewlett Packard 8453 UV–visible spectrophotometer at 23 °C. The reaction mixture (2 ml) contained 25 mM potassium phosphate buffer (pH 7·1), 25 µM azo dye, 0·1 mM NADPH and a suitable amount of enzyme. The reaction was initiated by addition of the enzyme. Initial velocity was determined by monitoring the change in the amount of substrate in the first 2 min in a glass cuvette of 1 cm light path. The following molar absorption coefficients were used: 23 360 M−1 cm−1 (Methyl Red at 430 nm), 18 200 M−1 cm−1 (Orange II at 482 nm), 22 600 M−1 cm−1 (Amaranth at 520 nm), 20 700 M−1 cm−1 (Orange G at 477 nm), 22 900 M−1 cm−1 (Ponceau BS at 502 nm) (Nakanishi et al., 2001; Blumel et al., 2002) and 33 470 M−1 cm−1 (Ponceau S at 520 nm). One unit (U) of enzyme activity was defined as the amount of enzyme required to degrade one µmol azo dye per minute. Proteins were quantified using the bicinchoninic acid assay (Pierce) with BSA as standard.
RNA isolation and RT-PCR
RNA was isolated from S. aureus grown overnight in BHI, BHI+Methyl Red (0·2 mM), or BHI+ Orange II (0·2 mM), essentially using the RNAzol B procedure (Chapes et al., 1994). The RT-PCR reaction mixture (25 µl) consisted of 50 pmol each primer (Sas1-forward, 5′-ggcagtgcacaagtgaattc-3′ and Sas1-reverse, 5′-ccatgatagtttggcgttcc-3′), 4 µg RNA, 0·1 mM (each) deoxynucleoside triphosphate, 1 U Taq DNA polymerase and 10 U AMV reverse transcriptase in PCR buffer with MgCl2. cDNA synthesis by reverse transcription was accomplished at 37 °C for 1 h. The PCR was performed under the following conditions: initial denaturation (95 °C, 2 min), 40 cycles of denaturation (94 °C, 1 min), annealing (55 °C, 1·5 min) and extension (72 °C, 1 min), followed by a 10 min final extension (72 °C). The reaction products were analysed by 2% agarose gel electrophoresis.
Enzyme purification
Induction of target protein and preparation of cell extracts were performed using a similar procedure to that described previously (Chen et al., 2004). Protein purification was performed at 4 °C by using an AKTApurifier 10 system with unicorn 4.10 software (Amersham Biosciences). Sixty millilitres of the supernatant (549 mg protein) was applied to a HiPrep 16/10 Q XL anion-exchange column (Amersham Biosciences) equilibrated with 25 mM Tris/HCl (pH 7·5) buffer. The peak fractions of azoreductase activity eluted at around 0·3 M NaCl (21 ml) were pooled. One volume of 10 mM potassium phosphate (pH 7·1) containing 2·0 M (NH4)2SO4 was mixed with one volume of the pooled fractions. Forty millilitres of the sample was applied to a HiPrep 16/10 Phenyl FF hydrophobic interaction column (Amersham Biosciences) equilibrated with the same buffer plus 1·0 M (NH4)2SO4. A linear gradient of 400 ml (NH4)2SO4 sulfate from 1·0 to 0 M in the same buffer was applied, followed by further elution with 100 ml buffer. Fractions with azoreductase activity (30 ml) were pooled and concentrated to a volume of about 8 ml by using an Amicon Ultra-15 (Millipore). The sample was dialysed and buffered with 25 mM Tris/HCl (pH 8·0). The sample was then applied in 2 ml portions to a Mono Q HR 5/5 column (Amersham Biosciences) equilibrated with the same buffer. Fractions of 0·5 ml were collected and those containing major enzyme activity were pooled (1·0 ml, 0·35 M NaCl). Final purification was achieved by gel filtration on a HiLoad 26/60 Superdex 200 pre grade column (Amersham Biosciences) equilibrated and eluted with 25 mM potassium phosphate (pH 7·1) plus 0·1 M NaCl (Chen et al., 2004).
Identification of the prosthetic group
One hundred microlitres of the purified enzyme (3 mg ml−1) in 10 mM potassium phosphate buffer (pH 7·0) was heated at 100 °C for 10 min in the dark. After cooling on ice, the denatured protein was removed by centrifugation at 14 500 g for 10 min in a microfuge at 4 °C. The resulting yellow supernatant (5 µl) was analysed by TLC employing a silica gel 60 F-254 plate (2 mm thickness; Merck) and chloroform, glacial acetic acid and water (6 : 7 : 1 by vol.) as the solvent phase (Lake & Goodwin, 1976). FAD and FMN (1 µl, 0·2 mM each) were used as standards, and the migration of the compounds was monitored by the characteristic fluorescence in UV light (312 nm).
SDS-PAGE and N-terminal amino acid sequencing of the protein
SDS-PAGE (12·5%, w/v) was conducted under normal conditions. Protein markers (Novagen) were used as standards. Gels were stained for the presence of proteins using Coomassie brilliant blue R-250 (Bio-Rad). The proteins were transferred onto a PVDF membrane in a Hoefer TE77 Semi-Dry Transfer Unit (Amersham Biosciences). The transferred proteins were visualized by Coomassie staining and excised with a razor blade. N-terminal amino acid sequencing was performed by an Applied Biosystems model 477A gas-phase sequencer equipped with an automatic on-line phenylthiohydantoin analyser.
Preparation of apoenzyme and reconstruction of flavoprotein
Eight millilitres of 25 mM potassium phosphate (pH 7·1) containing 1 M (NH4)2SO4, 1 M KBr and 1 mM EDTA was mixed with 2 ml purified enzyme (5 mg). The sample was applied to the HiPrep 16/10 Phenyl FF column equilibrated with the same buffer. The column was washed with 40 ml starting buffer. Flavin was eluted with 60 ml of the same buffer adjusted to pH 4·0 with 1 M phosphoric acid. After flavin elution, the column was immediately washed with 40 ml starting buffer, omitting 1 M KBr. Four hundred millilitres of a linear gradient of (NH4)2SO4 from 1 to 0 M in the same buffer was applied. This was followed by further elution with 100 ml starting buffer. The fractions (30 ml) containing apoenzyme were concentrated (Van Berkel et al., 1988).
The protein was reconstructed directly on the column. After the release of the original flavin from the column, the pH of the column was equilibrated back to 7·1, as described above. Forty millilitres of the starting buffer containing 1 mM FMN, FAD, or riboflavin was passed through the column at a flow rate of 0·5 ml min−1. The column was placed at 4 °C overnight and then washed with 40 ml starting buffer, omitting 1 MKBr, to remove the unbound excess flavin. The elution of the protein was done as above (Van Berkel et al., 1988).
Identification of Methyl Red and its metabolites
Two assay mixtures containing 0·4 mM NADPH and 50 µM Methyl Red in 20 ml of 25 mM sodium phosphate buffer (pH 7·1) with or without the purified enzyme were prepared. Methyl Red, N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid (Sigma) solutions (50 mg l−1), using acetonitrile as solvent, were prepared. The assay mixture containing the enzyme was then incubated at room temperature until no decrease in optical density at 430 nm was noted. The two assay mixtures were extracted three times with equal volumes of ethyl acetate, subsequent to adjusting the pH to 3 with 1 M HCl. The extracts were evaporated in a rotary evaporator at 40 °C and the remaining trace solvent was removed by evaporation at room temperature overnight. Each residue was dissolved in 1 ml acetonitrile and filtered through a 0·2 µm pore-size syringe filter. Forty microlitres of the samples was analysed with a Hewlett Packard HPLC 1050 equipped with a model variable-wavelength detector (detection wavelength was 250 nm) and a reverse-phase Inertsil 5µ ODS-2 column (4·6 × 250 mm, MetaChem Technologies). The mobile phase was composed of 25 mM phosphate buffer (pH 3·0) and acetonitrile (4 : 6, v/v) with a flow rate of 0·5 ml min−1 (Moutaouakkil et al., 2003; Nakanishi et al., 2001; Wong & Yuen, 1998).
Enzyme characterization
Each data point was analysed three times with a standard deviation of less than 10 %. The effects of pH on the activity and the optimum pH of S. aureus Sa-Azo1 were determined at room temperature with the following buffers: 0·1 M sodium acetate (pH 4·2–5·4), sodium phosphate (pH 5·8–7·8) and HEPES/NaOH (pH 8·2 and 8·6). The effects of temperature on Azo1 were determined by assaying the enzyme at temperatures ranging from 10 to 65 °C. Thermostability was measured by incubating the enzyme samples in 25 mM potassium phosphate buffer, pH 7·1, for 1 h at temperatures from 30 to 70 °C. The initial velocities of the enzymic reaction were obtained by varying the concentrations of one substrate, Methyl Red (from 0·005 to 0·04 mM) or NADPH (from 0·05 to 0·4 mM), while the concentration of the other substrate was kept constant (NADPH, 0·4 mM or Methyl Red, 0·04 mM). Apparent Km and Vmax values were obtained from Lineweaver–Burk plots.
RESULTS
Degradation of azo dyes by S. aureus
When S. aureus was grown in the presence of Methyl Red, it rapidly reduced the azo dye. Only 20% Methyl Red remained after 1 h incubation, and it was completely degraded in 3 h. Orange II was reduced more slowly than Methyl Red. About 10% of the added Orange II dye was metabolized after 3 h incubation and 56%of the dye colour degraded in 20 h. The total cell numbers in Methyl Red, Orange II and without dye cultures were similar, which indicated that the azo dyes were not toxic and did not inhibit the growth of the bacterium.
Identification of the S. aureus azo1 gene
Although the ability of the skin microflora to metabolize azo dyes was demonstrated, the corresponding genes or their corresponding gene products have not been characterized (Platzek et al., 1999). Therefore we searched the S. aureus Mu50 genome sequence (Kuroda et al., 2001) in GenBank by a blast search using the entire protein sequence of 178 amino acids deduced from a main azoreductase gene of Bacillus sp. OY1–2 as the query (Suzuki et al., 2001). The search identified a single gene encoding a 188 amino acid residue (Fig. 1), which displayed moderate primary structure identity (32 %) and was similar in size to the Bacillus azoreductase. There were five place deletion and/or insertion differences between these two protein sequences. This putative azoreductase gene was amplified from the purified genomic DNA of S. aureus ATCC 25923 by PCR using a pair of primers (Saa-forward and Saa-reverse), yielding a DNA band of about 700 bp on agarose gel. The amplified DNA fragment contained a putative ORF as well as a short sequence downstream of the stop codon. The fragment was directly ligated into a TA cloning vector, pCR2.1-TOPO. Sequencing of the insert revealed that it contained a 567 bp DNA fragment with a complete ORF encoding a protein (Azo1). There are only three base pair differences between sa-azo1 from S. aureus ATCC 25923 and the corresponding gene from S. aureus Mu50. However, both protein sequences translated from the genes were identical.
Fig. 1.
Comparison of the deduced amino acid sequences of S. aureus Azo1 (top) and Bacillus sp. OY1–2 azoreductase. Amino acid residues with different degrees of conservation (+) are indicated.
Expression of the azoreductase in E. coli
For expression of the azoreductase gene, the PCR amplified fragment was inserted via its unique NcoI and BamHI restriction sites into corresponding sites of pET-11d. A 6 bp DNA fragment had to be added to the beginning of the ORF vector to create a NcoI restriction site in the construction of the expression cassette. The functional enzyme was expressed in E. coli using a phage T7-promoter system. The cell extract supernatant from the induced culture displayed a bright yellow colour and an elevated level of azoreductase activity was detected as compared with that of the non-induced culture, indicating induction and expression in the host. This expression system was particularly useful, since the cells grew and synthesized the flavoprotein continuously without disruption to cellular growth.
Analysis of azo1 expression by S. aureus
To determine whether the azo1 gene is expressed by S. aureus ATCC 25923, transcription of azo1 was examined by RT-PCR. A 233 bp transcript of azoA was detected in all three RNA samples (not shown). Control reactions, in which reverse transcription from DNase- and RNase-treated RNA samples yielded no products, confirmed the absence of contaminating DNA. These results demonstrated that the azo1 gene was constitutively expressed in S. aureus.
Purification of the recombinant S. aureus Azo1 expressed in E. coli
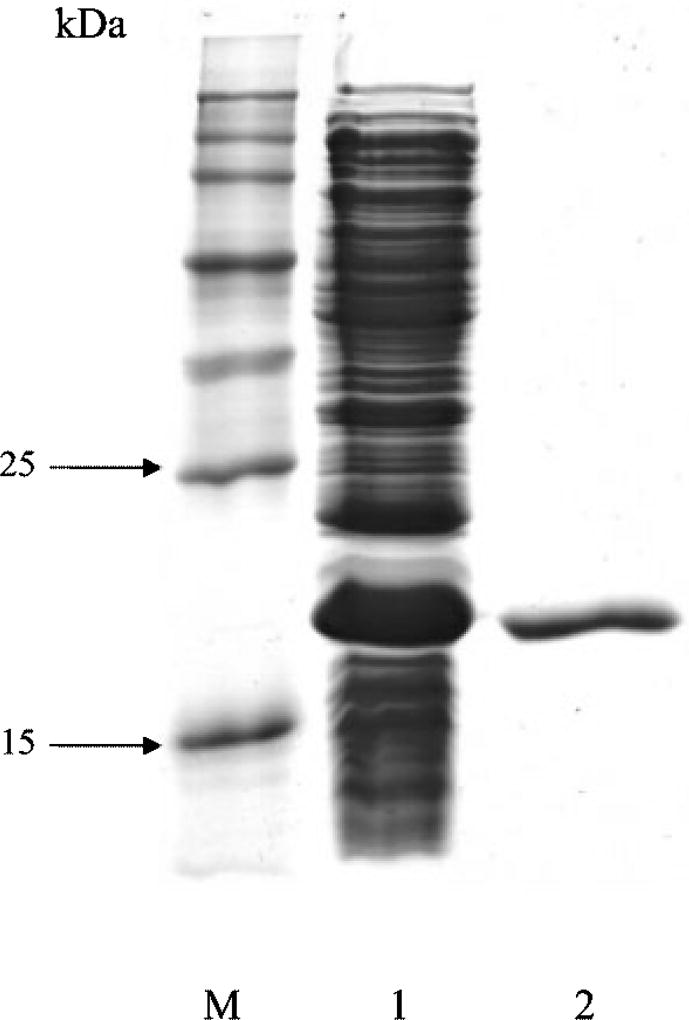
The purification procedure is summarized in Table 1. The enzyme was purified at about 9·3-fold with a yield of 20 %, which allows the proportion of the enzyme to be estimated at 10·8%of the protein present in the crude cell extract. The specific activity of the final purified S. aureus Azo1 was 0·14 U (mg protein)−1 using Methyl Red as substrate. The molecular mass of the purified azoreductase was found to be 20 kDa as shown by SDS-PAGE (Fig. 2). However, when determined by gel filtration chromatography on a HiLoad Superdex 200 column, it had an approximate size of 85 kDa. The purified protein on SDS-PAGE was transferred onto a PVDF membrane and subjected to N-terminal sequence. The first 10 amino acid residues (AMKGLIIIGS) were determined and found to be identical to the 10 residues shown for the polypeptide sequence, beginning with the second position in the N-terminal sequence, translated from the modified azoreductase ORF of S. aureus.
Table 1.
Purification of recombinant Azo1 of S. aureus expressed in E. coli Activities were determined with Methyl Red as substrate.
| Step | Total protein (mg) |
Total activity (U) |
Specific activity [U (mg protein)−1] |
Yield (%) |
|---|---|---|---|---|
| Crude cell extract | 549 | 8·32 | 0·015 | 100 |
| HiPrep Q XL | 167 | 5·68 | 0·034 | 68 |
| HiPrep Phenyl FF | 76 | 4·33 | 0·057 | 52 |
| Mono Q | 34 | 2·75 | 0·081 | 33 |
| HiLoad Superdex 200 | 12 | 1·68 | 0·14 | 20 |
Fig. 2.
SDS-PAGE of the purified Azo1 of S. aureus expressed in E. coli. Lanes: M, protein molecular mass standards; 1, crude cell extract (60 µg); 2, purified Azo1 (2 µg).
HPLC analysis of Methyl Red metabolites
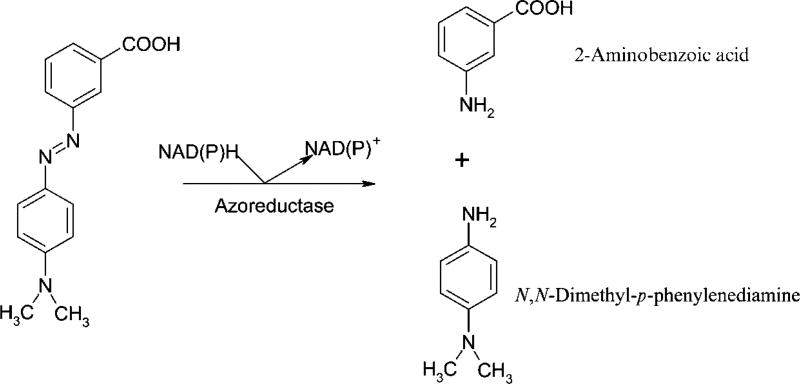
HPLC analyses of the extracts of the assay mixtures without the enzyme gave one major compound at a retention time of 24·5 min and the UV–visible spectrum of the compound showed a λmax at 500 nm and a minor peak at 287 nm, which matches those of the Methyl Red standard. The HPLC chromatogram of the extract of the reaction mixture incubated in the presence of the purified recombinant Azo1 showed the disappearance of Methyl Red and the appearance of two new major compounds with retention times of 5·2 and 7·1 min, respectively (not shown). The absorption spectrum of the 5·2 min compound showed a λmax at 246 nm and a minor peak at 292 nm, which matched the values of authentic N,N-dimethyl-p-phenylenediamine. On the other hand, the absorption spectrum of the 7·1 min compound showed λmax at 218 nm and two minor peaks at 246 and 334 nm, which matched the values of authentic 2-aminobenzoic acid. These two compounds are known metabolites in the breakdown of Methyl Red (Wong & Yuen, 1998; Moutaouakkil et al., 2003; Yan et al., 2004) (Fig. 3).
Fig. 3.
Reductive degradation of Methyl Red by Azo1 of S. aureus expressed in E. coli. Methyl Red was treated with the purified enzyme in the presence of NADPH. Products were analysed by HPLC and identified as N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid.
The recombinant S. aureus Azo1 contains FMN as a prosthetic group
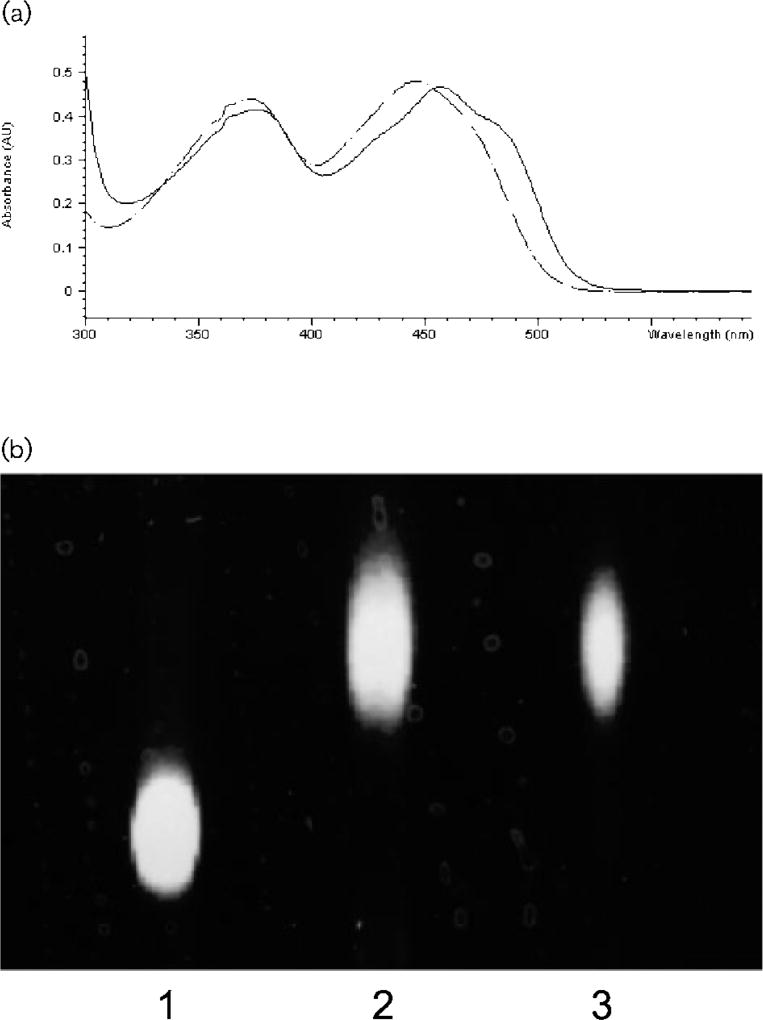
The solution of the purified Azo1 was initially yellow. This colouration was tightly bound to the protein through the various purification procedures and resistant to dialysis after several chromatography steps. The absorption spectrum of the purified enzyme had peaks at 376 and 457 nm and shoulders at 430 and 488 nm (Fig. 4a, solid line). When the enzyme was denatured in boiling water, the yellow compound was released, indicating a non-covalent association with the protein. The absorption spectrum of the supernatant of the boiling solution showed peaks at 373 and 446 nm, suggesting the presence of a flavin compound (Fig. 4a, dashed line). TLC of the flavin of the supernatant of the boiling sample gave a single fluorescent spot identical to that of the FMN standard (Fig. 4b). Thus, the prosthetic group of Azo1 is presumably FMN. Based on the extinction coefficient of 12 200 M−1 cm−1 for FMN at 446 nm (Aliverti et al., 1999), on the absorbance reading of the boiling sample (0·86 mg protein ml−1; A446 = 0·48), and the molecular mass of Azo1 (85 kDa), 38·4 µmol flavin was calculated to be associated with 10·1 µmol purified enzyme (3·8 mol of flavin bound per mol of the protein). Thus, the estimated ratio of flavin in Azo1 would be 4 mol FMN (mol protein)−1.
Fig. 4.
Detection of FMN in Azo1 of S. aureus expressed in E. coli. (a) Comparison between spectra of Azo1 of S. aureus and free FMN. Solid and dotted lines show spectra of Azo1 and free FMN, respectively. (b) TLC of the flavins. Lanes: 1, FAD; 2, FMN; 3, Azo1 of S. aureus.
Flavin removal and reconstitution with various flavins
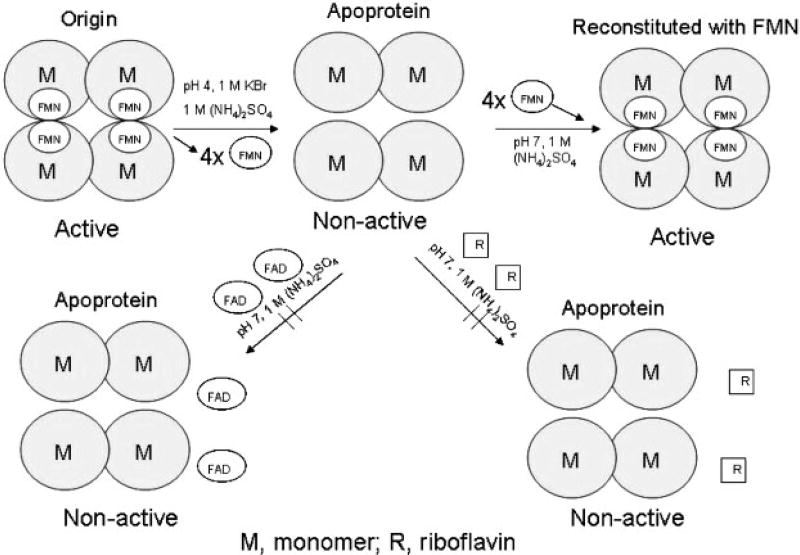
The purified tetrameric recombinant S. aureus Azo1 was bound to a hydrophobic interaction HiPrep 16/10 Phenyl FF column and the flavin of the enzyme was removed by washing the column with low-pH buffer, which was described in more detail in Methods. The bright yellow colour of the flavin can easily be seen visually and by UV detection. The size of the purified apoenzyme was determined on the gel filtration column to be approximately 42 kDa, indicating that the apoprotein was in a dimeric form (Fig. 5). The yield of the apoprotein was about 90% of the enzyme loaded onto the column.
Fig. 5.
Possible mechanisms of the dissociation and reassociation of Azo1 of S. aureus expressed in E. coli.
Azo1 could be fully reconstituted with FMN when the apoprotein was still bound to the column. Passing an excess of FMN over the column yielded a distinct fluorescent yellow band. The eluted reconstituted protein retained full azoreductase activity (Table 2) and had a size of about 85 kDa, as determined by gel filtration chromatography. Also, Azo1 could be reconstituted with FMN in solution. FAD and riboflavin were not able to bind to the protein and no reconstitution of the protein was observed when excess of the flavin was passed over the column. TLC analysis indicated that the partial reconstitution of the enzyme with an excess of FAD on the column was due to trace contamination with FMN in the commercial FAD preparation.
Table 2.
Effects of enzyme stages and cofactors on the azoreductase activity of S. aureus Azo1
| Enzyme stages | Cofactors added |
Specific activity [U (mg protein)−1] |
|---|---|---|
| Original enzyme* | NADPH | 0·15 |
| Original enzyme | NADH | ND† |
| Original enzyme | NADPH+FMN | 0·14 |
| Reconstructed with FMN‡ | NADPH | 0·12 |
| Apoprotein | NADPH+FMN | 0·13 |
| Apoprotein | NADPH+FAD | ND |
| Apoprotein | NADPH+riboflavin | ND |
Original enzyme, the purified intact recombinant S. aureus Azo1.
ND, not detectable.
Reconstructed with FMN, the apoprotein reconstructed with FMN on the column.
Properties of the recombinant S. aureus Azo1
Analysis of the purified Azo1 indicated that the enzyme used NADPH, but not NADH, for Methyl Red reduction (Table 2). Azo1 required its original flavin or free FMN for its activity. FAD and riboflavin had no ability to mediate oxidation of the NADPH. Azoreductase activity was not enhanced by adding free FMN to the original purified Azo1, indicating that the flavin was tightly bound to the protein.
The activity of Azo1 was determined from pH 4·2–8·6 with Methyl Red as substrate. A typical bell-shaped pH profile was obtained with an optimum between pH 6·0 and 6·6. The effect of temperature on azoreductase activity was determined from 10 to 65 °C. Maximum activity was observed between 35 and 40 °C. Thermal stability was investigated by incubating the enzyme at different temperatures for 1 h. The enzyme was stable at temperatures up to 55 °C. Inactivation occurred at 60 °C, with 67% of the enzyme activity remaining after 1 h pre-incubation and a half-life of 1·6 h. Enzymic reactions were performed by varying the concentrations of one substrate (Methyl Red or NADPH) and fixing the other substrate concentration. Apparent Km and Vmax values were obtained from Lineweaver–Burk plots. Km values of 0·074 and 0·057 mM and Vmax values of 0·39 and 0·41 U (mg protein)−1 for NADPH and Methyl Red, respectively, were obtained. In addition, a number of synthetic model azo compounds were used to determine the substrate specificity of the enzyme. The enzyme displayed the highest activity with Methyl Red [0·14 U (mg protein)−1]. The enzyme also degraded the sulfonated azo dyes Orange II [0·11 U (mg protein)−1], Amaranth [0·055 U (mg protein)−1], Ponceau BS [0·13 U (mg protein)−1] and Ponceau S [0·13 U (mg protein)−1]. No enzymic activity was observed when Orange G was used as substrate.
DISCUSSION
Decolorization of azo colourants by the skin microflora has been much less studied than similar processes mediated by intestinal bacterial species. Only one investigation has demonstrated the ability of bacteria on the human skin to metabolize azo dyes to aromatic amines which are known to easily penetrate the skin (Platzek et al., 1999).
Our studies demonstrated that S. aureus was able to grow and reduce azo dyes, indicating that azoreductase was functionally expressed in the bacterium. Unlike adapted soil bacteria isolated from azo dye-contaminated wastewater (Suzuki et al., 2001), as a skin bacterium, S. aureus presumably does not produce highly active azoreductase. It is an extremely difficult and very time-consuming process to purify sufficient amounts of protein from the bacterium for biological characterization of the enzyme. A novel approach was adapted by using homology comparison, direct cloning and overexpression of the genes encoding hypothetical proteins. We were able to identify and express a functional azoreductase gene from S. aureus in E. coli. The N-terminal methionine of the recombinant S. aureus Azo1 was cleaved by methionine aminopeptidase in the E. coli cells, as is common for heterologous proteins expressed in the E. coli host (Ben-Bassat et al., 1987). The N-terminal sequence of the purified enzyme started from the second position of the deduced protein sequence, indicating that the purified recombinant Azo1 was an intact protein when expressed in E. coli and could be similar to the corresponding native enzyme produced in S. aureus. Gel filtration chromatography revealed that AzoA exists as a tetramer (Figs 2 and 5) which was different from the dimeric azoreductases from E. coli (AcpD) (Nakanishi et al., 2001) and Enterococcus faecalis (Chen et al., 2004). Some other flavin-binding proteins, however, do exist as tetramers. This is the case for Sulfolobus shibatae type 2 isopentenyl diphosphate isomerase (Yamashita et al., 2004) and Bacillus subtilis glycine oxidase (Mortl et al., 2004). The prosthetic FMNs in the tetrameric enzyme may hold the subunits together (Van Berkel et al., 1988). During hydrophobic interaction chromatography in the presence of high concentrations of KBr and ammonium sulfate, the yellow flavin compound was eluted with decreasing pH, indicating a non-covalent association with the protein (Hefti et al., 2003). Azoreductases from Bacillus sp. OY1–2 (Suzuki et al., 2001), Xenophilus azovorans (Blumel et al., 2002) and Pigmentiphaga kullae (Blumel & Stolz, 2003) exist as monomers and enzyme-bound FMN or FAD have not been reported.
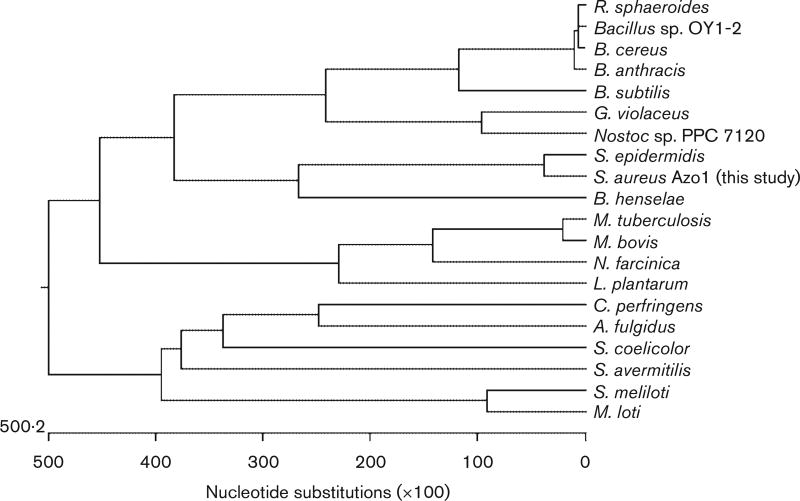
Azo1 was further annotated and searched for homologues from other organisms in the DNA and protein databases. We found that Azo1 has the highest homology (80 %) to a hypothetical protein from another more closely related skin bacterium Staphylococcus epidermidis. However, it has moderate homology (range 30–35 %) with other azoreductases and hypothetical proteins from other bacteria. Based on the multiple alignments of Azo1, protein sequences of the related azoreductases, and related hypothetical protein sequences, an unrooted phylogenetic tree was generated. Fig. 6 shows that Azo1 was clustered with hypothetical proteins from the skin bacteria S. epidermidis and Bartonella henselae. Azo1 has 32% homology to a hypothetical FMN-dependent azobenzene reductase (Yhda protein) from B. subtilis, whose crystal structure has been newly discovered (PDB coordinate: 1NNI). More recently, the three-dimensional structure of a homodimer NAD(P)H-dependent FMN reductase of a Saccharomyces cerevisiae YLR011w gene product, which has minor azoreductase activity to cleave Ethyl Red, has shown structural homology to B. subtilis Yhda protein (Liger et al., 2004). Both B. subtilis Yhda and Sac. cerevisiae YLR011W have conserved amino acid residues at several crucial positions that contribute to FMN binding. These same residues are conserved in Azo1 and other real and hypothetical azoreductases from various bacteria (Figs 1 and 6). It was suggested that the azoreductase from Bacillus sp. strain OY1–2 contains an NAD(P)H-binding motif (GXGXXG) at positions 106–111 within the protein (Fig. 1). This motif was not conserved in the protein sequences of Azo1 or other members of this azoreductase family (Figs 1 and 6). The vast majority of flavodoxin domains whose structure is known have a six-residue flavodoxin key fingerprint motif (T/S-X-T-G-X-T) in common (Liger et al., 2004). However, the hypothetical FMN phosphate-binding loop sequences (T/S-X-T-G-X-T) were not found in the B. subtilis Yhda protein, the Sac. cerevisiae YLR011 wp, or Azo1. Instead of this, several amino acid residues in the sequences (P-X-Y-H/N-2X-P/S-G/A-X-L-K-N-A/S-D-L/I-D) form another loop between the β3 and α3 helix in the Sac. cerevisiae YLR011 wp that interacts with both parts of FMN, and these residues are conserved in this enzyme family. This indicates that this conserved sequence may serve as a new signature sequence for these novel flavin enzymes.
Fig. 6.
Unrooted phylogenetic tree of S. aureus Azo1. Based on tblastn results, protein sequences showing similarities to Azo1 were aligned using the Jutun Hein method. The unrooted tree was generated using MegAlign of the dnastar software package. GenBank accession numbers are as follows: Archaeoglobus ulgidus, AE000972; Bacillus anthracis, AE017225; Bacillus cereus, AE017005; Bartonella henselae, BX897699; Bacillus subtilis, AB071366; Bacillus sp. OY1–2, AB032601; Clostridium perfringens, BA000016; Gloeobacter violaceus, BA000045; Lactobacillus plantarum, AL935261; Mycobacterium bovis, BX248344; Mesorhizobium loti, BA000012; Mycobacterium tuberculosis, AE000516; Nocardia farcinica, AP006618; Nostoc sp. PPC 7120, BA000019; Rhodobacter sphaeroides, AY150311; Staphylococcus aureus Azo1 (this study), AY545994; Streptomyces avermitilis, AP005049; Streptomyces coelicolor, AL939107; Staphylococcus epidermidis, AE016745; Sinorhizobium meliloti, AL595985.
HPLC analysis indicated that the decolorization of Methyl Red by Azo1 is the result of cleavage of the molecule to N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid. These compounds were reported as metabolites of Methyl Red degradation by Klebsiella pneumoniae and Acetobacter liquefaciens (Wong & Yuen, 1998), and by azoreductases from E. coli (Nakanishi et al., 2001), Enterobacter agglomerans (Moutaouakkil et al., 2003) and Rhodobacter sphaeroides (Yan et al., 2004). These results proved that the purified Azo1 catalysed the reductive cleavage of the azo bond of Methyl Red in the presence of NADPH serving as an electron donor (Fig. 3). Among several cloned and expressed azoreductases, the enzymes from E. coli (Nakanishi et al., 2001), Ent. faecalis (Chen et al., 2004), X. azovorans KF46F (Blumel et al., 2002) and P. kullae K24 (Blumel & Stolz, 2003) use NADH as electronic donor for their enzymic activities. On the other hand, azoreductases from Bacillus sp. OY1–2 (Suzuki et al., 2001) and S. aureus are NADPH-dependent enzymes (Table 2). Recently, an azoreductase from R. sphaeroides AS1.1737 with NADH as a donor was expressed and characterized (Yan et al., 2004). Since the azoreductase from R. sphaeroides AS1.1737 had similar sizes and homologies to azoreductases from both Bacillus sp. OY1–2 (Suzuki et al., 2001) and S. aureus, it was placed in the same enzyme family (Fig. 6); however, the specificity of its electron donor should be re-examined.
Two further azoreductases belonging to the same family, from E. coli (Nakanishi et al., 2001) and Ent. faecalis (Chen et al., 2004), are FMN-dependent enzymes. No information is available regarding other azoreductases from Bacillus sp. OY1–2 (Suzuki et al., 2001) and R. sphaeroides AS1.1737 (Yan et al., 2004). In this study, we discovered that Azo1 contained a tightly bound flavin and that its activity was strictly dependent on that flavin, indicating that it is a flavin-dependent azoreductase. Dissociation and reconstitution of the enzyme demonstrated that the flavin prosthetic group was required for its function. Since azoreductases from Bacillus sp. OY1–2 (Suzuki et al., 2001) and R. sphaeroides AS1.1737 (Yan et al., 2004) fall into the same enzyme family as Azo1, presumptively they are also flavin-dependent enzymes. Besides these flavin-dependent azoreductases, two monomeric flavin-free azoreductases from X. azovorans and P. kullae K24 have also been described (Zimmermann et al., 1982, 1984; Blumel et al., 2002; Blumel & Stolz, 2003), indicating that there are at least two different types of azoreductase in bacteria.
Acknowledgments
We thank Anna J. Williams for assistance with HPLC analyses. We also thank Drs Christopher A. Elkins and Karen Beenken for their critical review of the manuscript.
Abbreviation
- Azo1
NADPH-flavin azoreductase
Footnotes
The GenBank/EMBL/DDBJ accession number for the nucleotide sequence of azo1 of S. aureus ATCC 25923 reported in this paper is AY545994.
References
- Aliverti A, Curti B, Vanoni MA. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol Biol. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- Baumler W, Eibler ET, Hohenleutner U, Sens B, Sauer J, Landthaler M. Q-switched laser and tattoo pigment: first results of the chemical and photophysical analysis of 41 compounds. Lasers Surg Med. 2000;26:13–21. doi: 10.1002/(sici)1096-9101(2000)26:1<13::aid-lsm4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumel S, Stolz A. Cloning and characterization of the gene coding for the aerobic azoreductase from Pigmentiphaga kullae K24. Appl Microbiol Biotechnol. 2003;62:186–190. doi: 10.1007/s00253-003-1316-5. [DOI] [PubMed] [Google Scholar]
- Blumel S, Knackmuss H-J, Stolz A. Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl Environ Microbiol. 2002;68:3948–3955. doi: 10.1128/AEM.68.8.3948-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapes SK, Baharka AA, Hart ME, Smeltzer MS, Iandolo JJ. Differential RNA regulation by staphylococcal enterotoxins A and B in murine macrophages. J Leukocyte Biol. 1994;55:523–529. doi: 10.1002/jlb.55.4.523. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang RF, Cerniglia CE. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif. 2004;34:302–310. doi: 10.1016/j.pep.2003.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K-T. The significance of azo-reduction in the mutagenesis and carcinogenesis of azo dyes. Mutat Res. 1983;114:269–281. doi: 10.1016/0165-1110(83)90035-0. [DOI] [PubMed] [Google Scholar]
- Chung K-T, Stevens SE, Jr, Cerniglia CE. The production of azo dyes by the intestinal microflora. Crit Rev Microbiol. 1992;18:175–190. doi: 10.3109/10408419209114557. [DOI] [PubMed] [Google Scholar]
- Combes RD, Haveland-Smith RB. A review of the genotoxicity of food, drug and cosmetic colours and other azo, triphenylmethane and xanthene dyes. Mutat Res. 1982;98:101–248. doi: 10.1016/0165-1110(82)90015-x. [DOI] [PubMed] [Google Scholar]
- Hefti MH, Vervoort J, van Berkel WJH. Deflavination and reconstitution of flavoproteins: tacking fold and function. Eur J Biochem. 2003;270:4227–4242. doi: 10.1046/j.1432-1033.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- Kloos WE, Schleifer KH. Isolation and characterization of staphylococci from human skin: II. Description of four new species; Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int J Syst Bacteriol. 1975;25:62–79. [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, 34 other authors Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Lake BD, Goodwin HJ. Lipids. In: Smith I, Seakin JMT, editors. Chromatographic and Electrophoretic Techniques. Vol. 1. Bath, UK: Pitman Press; 1976. pp. 345–366. [Google Scholar]
- Levine WG. Metabolism of azo dyes: implication for detoxification and activation. Drug Metab Rev. 1991;23:253–309. doi: 10.3109/03602539109029761. [DOI] [PubMed] [Google Scholar]
- Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, van Tilbeurgh H. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J Biol Chem. 2004;279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- Mortl M, Diederichs K, Welte W, Molla G, Motteran L, Andriolo G, Pilone MS, Pollegioni L. Structure-function correlation in glycine oxidase from Bacillus subtilis. J Biol Chem. 2004;279:29718–29727. doi: 10.1074/jbc.M401224200. [DOI] [PubMed] [Google Scholar]
- Moutaouakkil A, Zeroual Y, Zzayri FZ, Talbi M, Lee K, Blaghen M. Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch Biochem Biophys. 2003;413:139–146. doi: 10.1016/s0003-9861(03)00096-1. [DOI] [PubMed] [Google Scholar]
- Nagase N, Sasaki A, Yamashita K, Shimizu A, Wakita Y, Kitai S, Kawano J. Isolation and species distribution of Staphyloccocci from animal and human skin. J Vet Med Sci. 2002;64:245–250. doi: 10.1292/jvms.64.245. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Yatome C, Ishida N, Kitade Y. Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J Biol Chem. 2001;276:46394–46399. doi: 10.1074/jbc.M104483200. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Kimura T, Kodama M, Nagata C. Generation of hydrogen peroxide and superoxide anion from active metabolites of naphthylamines and aminoazo dyes: its possible role in carcinogenesis. Carcinogenesis. 1983;4:765–769. doi: 10.1093/carcin/4.6.765. [DOI] [PubMed] [Google Scholar]
- Platzek T, Lang C, Grohmann G, Gi U-S, Baltes W. Formation of a carcinogenic aromatic amine from an azo dye by human skin bacteria. Hum Exp Toxicol. 1999;18:552–559. doi: 10.1191/096032799678845061. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yoda T, Ruhul A, Sugiura W. Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-1 isolated from soil. J Biol Chem. 2001;276:9059–9065. doi: 10.1074/jbc.M008083200. [DOI] [PubMed] [Google Scholar]
- Van Berkel WJ, Van den Berg WA, Muller F. Large-scale preparation and reconstitution of apo-flavoproteins with special reference to butyryl-CoA dehydrogenase from Megasphaera elsdenii. Hydrophobic-interaction chromatography. Eur J Biochem. 1988;178:197–207. doi: 10.1111/j.1432-1033.1988.tb14444.x. [DOI] [PubMed] [Google Scholar]
- Von Lehmann G, Pierchalla P. Tatowierungsfarbstoffe. Derm Beruf Umwelt. 1988;36:152–156. [PubMed] [Google Scholar]
- Wang RF, Kim S-J, Robertson LH, Cerniglia CE. Development of a membrane-array method for the detection of human intestinal bacteria in fecal samples. Mol Cell Probes. 2002;16:341–350. doi: 10.1006/mcpr.2002.0432. [DOI] [PubMed] [Google Scholar]
- Wong PK, Yuen PY. Decolourization and biodegradation of N,N′-dimethyl-p-phenylenediamine by Klebsiella pneumoniae RS-13 and Acetobacter liquefaciens S-1. J Appl Microbiol. 1998;85:79–87. doi: 10.1046/j.1365-2672.1998.00479.x. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Hemmi H, Ikeda Y, Nakayama T, Nishino T. Type 2 isopentenyl diphosphate isomerase from a thermo-acidophilic archaeon Sulfolobus shibatae. Eur J Biochem. 2004;271:1087–1093. doi: 10.1111/j.1432-1033.2004.04010.x. [DOI] [PubMed] [Google Scholar]
- Yan B, Zhou J, Wang J, Du C, Hou H, Song Z, Bao Y. Expression and characteristics of the gene encoding azoreductase from Rhodobacter sphaeroides AS1.1737. FEMS Microbiol Lett. 2004;236:129–136. doi: 10.1016/j.femsle.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Zimmermann T, Kulla HG, Leisinger T. Properties of purified Orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur J Biochem. 1982;129:197–203. doi: 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann T, Gasser F, Kulla HG, Leisinger T. Comparison of two bacterial azoreductases acquired during adaptation to growth on azo dyes. Arch Microbiol. 1984;138:37–43. doi: 10.1007/BF00425404. [DOI] [PubMed] [Google Scholar]