Abstract
We analyzed whole-genome sequences of 8 enterovirus A71 isolates (EV-A71). We confirm the circulation of genogroup C and the new genogroup E in West Africa. Our analysis demonstrates wide geographic circulation and describes genetic exchanges between EV-A71 and autochthonous EV-A that might contribute to the emergence of pathogenic lineages.
Keywords: enterovirus, acute flaccid paralysis, Africa, recombination, phylogenetic analysis, enterovirus A71, whole-genome analysis, viruses
Enterovirus A71 (EV-A71; species Enterovirus A, genus Enterovirus, family Picornaviridae) is a common etiologic agent of hand, foot and mouth disease in young children. In addition, EV-A71 has been associated with severe and sometimes fatal neurologic diseases, including aseptic meningitis, encephalitis, and poliomyelitis-like acute flaccid paralysis (AFP) (1,2).
EV-A71 is classified info 7 genogroups (A–G). Genogroup A includes the prototype strain BrCr that was isolated in the United States in 1969 (1,2). Most EV-A71 isolates belong to genogroups B or C, which are each further divided into subgenogroups (1,2). Subgenogroups B4, B5, and C4 are mainly restricted to countries in Asia, whereas C1 and C2 circulate primarily in Europe and the Asia-Pacific region (1). Genogroup D and the newly proposed genogroup G appear to be indigenous to India, whereas genogroups E and F were recently discovered in Africa and Madagascar, respectively (3).
Although EV-A71 has been reported in many parts of the world, its epidemiology remains largely unexplored in Africa. An EV-A71 outbreak was documented in 2000 in Kenya, where HIV-infected orphans were infected by EV-A71 genogroup C (4). Several AFP cases have been associated with EV-A71 infection during 2000–2013 throughout Africa: in Democratic Republic of the Congo (5) (2000, n = 1); Nigeria (6) (2004, n = 1, genogroup E); Central African Republic (7) (2003, n = 1, genogroup E); Cameroon (8) (2008, n = 2, genogroup E); Niger (9) (2013, n = 1, genogroup E); and Senegal, Mauritania, and Guinea (9) (2013–2014, n = 3, subgenogroup C2). Four additional EV-A71 strains were obtained from captive gorillas in Cameroon during 2006–2008 (n = 2, genogroup E) (10) and from healthy children in Nigeria in 2014 (n = 2, genogroup E) (11). Molecular identification of all these isolates was based only on the analysis of sequences of the viral protein (VP) 1 capsid protein region.
Recombination events may be associated with the emergence and global expansion of new groups of EV-A71 that have induced large outbreaks of hand, foot and mouth disease with high rates of illness and death (12). For EV-A71, genetic exchanges have been described both within a given genogroup and with other types of enterovirus A (EV-A), usually in nonstructural genome regions P2 and P3 (1,12,13). However, before 2017, no complete genome sequence of EV-A71 detected in Africa has been reported, diminishing the power of such analysis. We examined the complete genome of most EV-A71 isolates reported to date in Africa to characterize the evolutionary mechanisms of genetic variability.
The Study
We sequenced the full genome of 8 EV-A71 isolates obtained from patients with AFP (Table): isolates 14-157, 14-250, 13-365, 13-194, and 15-355 from West Africa and isolates 08-041, 08-146, and 03-008 from Central Africa. We isolated and typed these isolates as previously described (7–10) and obtained nearly complete genomic sequences using degenerated primers (13) and additional primers designed for gene-walking (available on request) or unbiased sequencing methods (14). We determined the 5′-terminal sequences by means of a RACE kit (Roche, Munich, Germany). We deposited viral genomes in GenBank (accession numbers in Table) and submitted sequence alignments under BioProject PRJNA422891. We aligned sequences using ClustalW software (http://www.clustal.org).
Table. Description of enterovirus isolates from patients with acute flaccid paralysis in Africa that were sequenced for characterization of enterovirus A71.
| Strain (reference) | Country of isolation | Patient age at diagnosis, y | Year | Virus | Genogroup or subgenogroup | Genbank accession no. |
|---|---|---|---|---|---|---|
| 14-157 (9) | Senegal | 3 | 2014 | EV-A71 | C2 | MG672480 |
| 14-250 (9) | Mauritania | 1.6 | 2014 | EV-A71 | C2 | MG672481 |
| 13-365 (9) | Guinea | 1.7 | 2013 | EV-A71 | C2 | MG672479 |
| 15-355 (this study) | Senegal | 2.4 | 2015 | EV-A71 | C2 | MG013988 |
| 13-194 (9) | Niger | 1.3 | 2013 | EV-A71 | E | MG672478 |
| 03-008 (7) | Central African Republic | 1.9 | 2003 | EV-A71 | E | LT719068 |
| 08-146 (8) | Cameroon | 2.6 | 2008 | EV-A71 | E | LT719066 |
| 08-041(8) | Cameroon | 1.7 | 2008 | EV-A71 | C1 | LT719067 |
| 14-254 (15) |
Senegal |
3 |
2014 |
CV-A14 |
NA |
MG672482 |
| *NA, not available. | ||||||
To investigate the genetic relationship between Africa and global EV-A71 isolates, we constructed subgenomic phylogenetic trees based on the P1, P2, and P3 regions of the genome (Figure 1). We identified viral isolates showing related sequences in 1 of these 3 regions by BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) and included them in the corresponding datasets used for analyses. We completed these datasets with a representative global set of EV-A71 sequences available in GenBank and belonging to the different EV-A71 genogroups (Technical Appendix). As expected, in the structural P1 region, the 8 isolates we studied clustered within their respective genogroups (C1, C2, and E), previously established by VP1-based typing (Figure 1, panel A). In particular, the isolates of genogroup E consistently clustered together (bootstrap value 100%), confirming their belonging to the EV-A71 type and their divergence from the other isolates belonging to the common genogroups A, B, and C. Analysis of the nonstructural P2 and P3 genome regions were in agreement with these data. However, the genetic heterogeneity, <12%, observed among the complete genome of genogroup E sequences highly suggested that they have circulated and diverged for years in a large geographic area in Africa. The unique Africa EV-A71-C1 strain clustered with other C1 strains originating worldwide, regardless of which genome region we analyzed. In contrast, the nonstructural sequences of Africa EV-A71 isolates of subgenogroup C2 did not cluster with their non-Africa C2 counterparts or with any of the existing EV-A71 genogroups. The incongruent phylogenetic relationships of Africa C2 strains in the different regions of the genome suggested that recombination events have occurred during evolution.
Figure 1.
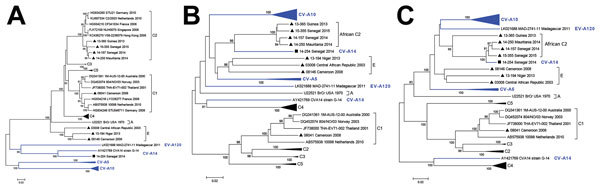
Phylogenetic relationships of the Africa enterovirus EV-A71 study strains based on A) P1 and B) P3 coding regions. An expanded version based on P1, P2, and P3 coding regions is online (LINK). Apart from the studied sequences, subgenomic datasets included their best nucleotide sequence matches identified by NCBI BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) as well as representative sequences of different EV-A71 genogroups and subgenogroups originating worldwide. Trees were constructed from the nucleotide sequence alignment using MEGA 5.0. software (http://megasoftware.net/) with the neighbor-joining method. Distances were computed using the Kimura 2-parameter model. The robustness of the nodes was tested by 1,000 bootstrap replications. Bootstrap support values >75 are shown in nodes and indicate a strong support for the tree topology. For clarity, CV-A10, CV-A5, and EV-A71 subgenogroups C3, C4, and C5 have been collapsed. Study strains are indicated by laboratory code, country of origin, and year of isolation; previously published strains are indicated by GenBank accession number, isolate code, country of origin, and year of isolation. Black triangles indicate EV-A71 strains from this study; black square indicates the CV-A14 strain from this study. Strains gathered in brackets belong to EV-A71 genogroups or subgenogroups; strains marked in blue color belong to other species of EV-A. Scale bars indicate nucleotide substitutions per site. CV, coxsackievirus; EV, enterovirus.
To examine further recombination events, we analyzed EV-A71-C2 study strains by similarity plot against potential parental genomes (Figure 2). This analysis showed that sequences 14-157, 14-250, and 15-355 had high similarity (>95%). By contrast, 13-365 diverged from the other C2 isolates around nt 5600 in the P3 region, suggesting a recombination breakpoint. The analysis showed high sequence similarity (>97%) between the studied EV-A71-C2 isolates and other subgenogroup C2 strains over the P1 capsid region. Conversely, in the noncapsid region, sequence similarity between Africa EV-A71-C2 isolates and classical subgenogroup C2 isolates (e.g., GenBank accession no. HQ647175) was much lower (66%–77%). This finding confirmed a recombination event of the Africa EV-A71 C2 lineage with an unknown enterovirus, the most likely breakpoint being located between nt 3596 and 3740, within the 2A gene. Sequence identity of EV-A71-C2 study strains with their closest related viruses (coxsackievirus A10 [CV-A10], CV-A5, EV-A120, and EV-A71 genogroup E strains) in the 3′ half of the genome was <87.7%. Of note, we found much higher sequence identity with the full-genome sequence of CV-A14 isolate in our database, obtained in 2014 from a patient with AFP in Senegal (15). This strain features a high similarity value (>97%) with the 3′ half of the genomes of EV-A71-C2 West Africa strains (Figure 2), indicating that their P3 regions share a recent common ancestor. Because these strains belong to 2 different types, this finding strongly suggests that genetic exchanges occurred through intertypic recombination. This result cannot be a result of cross-contamination during the sequencing process because the CV-A14 and EV-A71 isolates were sequenced on 2 different platforms.
Figure 2.
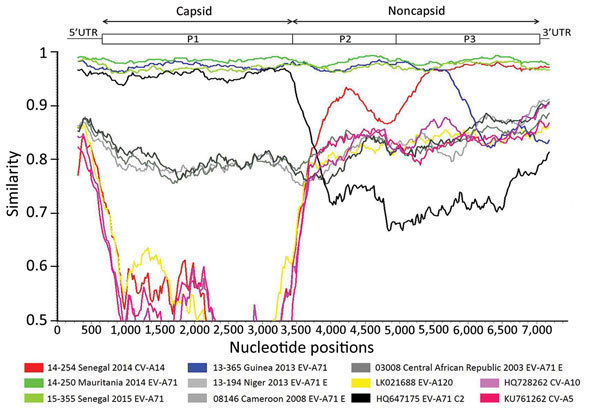
Identification of recombinant sequences in the genome of EV-A71 C2 isolates from patients with acute flaccid paralysis in Africa (14-157, 14-250, 13-365, 15-355) by similarity plot against potential parent genomes (CV-A14 strain 14-254; EV-A71 genogroup E strains 13-194, 08-146, and 03-008) and from GenBank (CV-A10, CV-A5, EV-A120). Similarity plot analysis was performed using SimPlot version 3.5.1 (http://sray.med.som.jhmi.edu/SCRoftware/simplot) on the basis of full-length genomes. For the analysis, we used a window of 600 nt moving in 20-nt steps. Approximate nt positions in the enterovirus genome are indicated. The enterovirus genetic map is shown in the top panel. We used the genome of EV-A71 study strain 14-157 as a query sequence. UTR, untranslated region.
Conclusions
Genogroup E was previously identified and characterized only on the basis of VP1 analysis (3). This study confirms the circulation in West and Central Africa of EV-A71 isolates belonging to the new genogroup E on the basis of the characterization of whole genomes. The divergence among isolates indicates that this genogroup has been extensively circulating in Africa. We also suggest that the common ancestor of EV-A71-C2 strains in West Africa has undergone recombination with >1 EV-A circulating in Africa. Genogroup E and recombinant C2 appear to be indigenous to Africa; they have not yet been detected elsewhere. Further exploration of environmental or clinical samples using deep sequencing technology would be of interest to determine the extent of EV-A71 circulation in Africa in the absence of AFP cases. Systematic surveillance based on full-genome sequencing could also serve to monitor these viruses for potential recombinations and to study their role in the emergence of new EV-A71 variants in Africa.
More information about the enterovirus isolates used for phylogenetic analysis of enterovirus A71 in Africa.
Acknowledgments
We thank Karla Prieto and Catherine Pratt, who assisted in obtaining nearly complete genomes of West Africa strains, and Joseph Chitty for analysis of the next-generation sequencing data.
The next-generation sequencing equipment at Institut Pasteur of Dakar was provided by the Defense Biological Product Assurance Office under the Targeted Acquisition of Reference Materials Augmenting Capabilities Initiative. This work was supported by the IPD, the Pasteur Institute’s Transverse Research Program PTR484, Actions Concertées Inter-Pasteuriennes A22-16, Fondation Total Grant S-CM15010-05B, Roux Howard Cantarini postdoctoral fellowship, and Grant Calmette and Yersin from the International Directorate of the Institut Pasteur.
About the Author
Biography
Dr. Fernandez-Garcia is a scientist with a PhD degree in virology and is involved in research and surveillance of enterovirus infections at Institut Pasteur of Dakar, Senegal. Her research interests include infectious disease epidemiology and public health microbiology.
Footnotes
Suggested citation for this article: Fernandez-Garcia MD, Volle R, Joffret M-L, Sadeuh-Mba SA, Gouandjika-Vasilache I, Kebe O, et al. Genetic characterization of enterovirus A71 circulating in Africa. Emerg Infect Dis. 2018 Apr [date cited]. https://doi.org/10.3201/eid2404.171783
These first authors contributed equally to this article.
These senior authors contributed equally to this article.
References
- 1.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–90. 10.1016/S1473-3099(10)70194-8 [DOI] [PubMed] [Google Scholar]
- 2.Chang P-C, Chen S-C, Chen K-T. The current status of the disease caused by enterovirus 71 infections: Epidemiology, pathogenesis, molecular epidemiology, and vaccine development. Vol. 13. Int J Environ Res Public Health. 2016;13:890. 10.3390/ijerph13090890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessaud M, Razafindratsimandresy R, Nougairède A, Joffret ML, Deshpande JM, Dubot-Pérès A, et al. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS One. 2014;9:e90624. 10.1371/journal.pone.0090624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty R, Iturriza-Gómara M, Musoke R, Palakudy T, D’Agostino A, Gray J. An epidemic of enterovirus 71 infection among HIV-1-infected orphans in Nairobi. AIDS. 2004;18:1968–70. 10.1097/00002030-200409240-00018 [DOI] [PubMed] [Google Scholar]
- 5.Junttila N, Lévêque N, Kabue JP, Cartet G, Mushiya F, Muyembe-Tamfum JJ, et al. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J Med Virol. 2007;79:393–400. 10.1002/jmv.20825 [DOI] [PubMed] [Google Scholar]
- 6.Oyero OG, Adu FD. Non-polio enteroviruses serotypes circulating in Nigeria. Afr J Med Med Sci. 2010;39(Suppl):201–8. [PubMed] [Google Scholar]
- 7.Bessaud M, Pillet S, Ibrahim W, Joffret ML, Pozzetto B, Delpeyroux F, et al. Molecular characterization of human enteroviruses in the Central African Republic: uncovering wide diversity and identification of a new human enterovirus A71 genogroup. J Clin Microbiol. 2012;50:1650–8. 10.1128/JCM.06657-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadeuh-Mba SA, Bessaud M, Massenet D, Joffret ML, Endegue MC, Njouom R, et al. High frequency and diversity of species C enteroviruses in Cameroon and neighboring countries. J Clin Microbiol. 2013;51:759–70. 10.1128/JCM.02119-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Garcia MD, Kebe O, Fall AD, Dia H, Diop OM, Delpeyroux F, et al. Enterovirus A71 genogroups C and E in children with acute flaccid paralysis, West Africa. Emerg Infect Dis. 2016;22:753–5. 10.3201/eid2204.151588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadeuh-Mba SA, Bessaud M, Joffret ML, Endegue Zanga MC, Balanant J, Mpoudi Ngole E, et al. Characterization of Enteroviruses from non-human primates in cameroon revealed virus types widespread in humans along with candidate new types and species. PLoS Negl Trop Dis. 2014;8:e3052. 10.1371/journal.pntd.0003052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faleye TO, Adewumi MO, Coker BA, Nudamajo FY, Adeniji JA. Direct detection and identification of enteroviruses from faeces of healthy Nigerian children using a cell-culture independent RT-seminested PCR assay. Adv Virol. 2016;2016:1412838. [DOI] [PMC free article] [PubMed]
- 12.McWilliam Leitch EC, Cabrerizo M, Cardosa J, Harvala H, Ivanova OE, Koike S, et al. The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol. 2012;86:2676–85. 10.1128/JVI.06065-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoke-Fun C, AbuBakar S. Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol. 2006;6:74. 10.1186/1471-2180-6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kugelman JR, Wiley MR, Mate S, Ladner JT, Beitzel B, Fakoli L, et al. ; US Army Medical Research Institute of Infectious Diseases; National Institutes of Health; Integrated Research Facility–Frederick Ebola Response Team 2014–2015. National Institutes of Health; Integrated Research Facility–Frederick Ebola Response Team 2014–2015. Monitoring of Ebola virus Makona evolution through establishment of advanced genomic capability in Liberia. Emerg Infect Dis. 2015;21:1135–43. 10.3201/eid2107.150522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Garcia MD, Kebe O, Fall AD, Ndiaye K. Identification and molecular characterization of non-polio enteroviruses from children with acute flaccid paralysis in West Africa, 2013-2014. Sci Rep. 2017;7:3808. 10.1038/s41598-017-03835-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
More information about the enterovirus isolates used for phylogenetic analysis of enterovirus A71 in Africa.


