Abstract
The forkhead box (Fox) family of transcription factors share homology in the winged helix/forkhead DNA-binding domain and play important roles in regulating cellular proliferation, differentiation, longevity, and cellular transformation. Forkhead box M1B (FoxM1B) is a ubiquitously expressed member of the Fox transcription factor family whose expression is restricted to proliferating cells and that mediates hepatocyte entry into DNA synthesis and mitosis during liver regeneration. Recent cDNA microarray studies indicated that age-related defects in cellular proliferation are associated with diminished expression of the FoxM1B transcription factor. Here, we show that increased levels of FoxM1B in regenerating liver of old transgenic mice restore the sharp peaks in hepatocyte DNA replication and mitosis that are the hallmarks of young regenerating mouse liver. Restoration of the young regenerating liver phenotype is associated with increased expression of numerous cell cycle regulatory genes that include cyclin D1, cyclin A2, cyclin F, cyclin B1, cyclin B2, Cdc25B, and p55cdc. Cotransfection assays in the human hepatoma HepG2 cell line demonstrated that FoxM1B protein stimulated expression of both the cyclin B1 and cyclin D1 promoters, suggesting that these cyclin genes are a direct FoxM1B transcriptional target. These results suggest that FoxM1B controls the transcriptional network of genes that are essential for cell division and exit from mitosis. Our results indicate that reduced expression of the FoxM1B transcription factor contributes to the decline in cellular proliferation observed in the aging process.
The mammalian liver is one of the few adult organs capable of completely regenerating itself in response to injury through the release of growth factors that stimulate reentry of terminally differentiated hepatocytes into the cell cycle (1–3). Liver regeneration induced by a two-thirds partial hepatectomy (PHx) results in synchronous induction of sharp peaks in hepatocyte DNA replication (S phase) and mitosis (M phase), which requires participation of the IL-6-signaling pathway (3–5). The forkhead box (Fox) family of transcription factors (6) shares homology in the winged-helix DNA-binding domain (7), and its members play important roles in regulating transcription of genes involved in cellular proliferation, differentiation, metabolic homeostasis, longevity, and cellular transformation (8–18). The mammalian (human) Fox family member FoxM1B (previously known as HFH-11B or trident) is a ubiquitously expressed transcription factor restricted to proliferating cells of the mouse embryo (including liver) and is essential for embryonic development (19), but its expression diminishes during postnatal cellular differentiation (20). In regenerating liver, FoxM1B expression is reactivated before DNA replication (S phase) and sustained throughout the period of hepatocyte proliferation (20). Liver regeneration studies with transgenic mice, in which the transthyretin (TTR) promoter functioned to prematurely express FoxM1B (TTR-FoxM1B), revealed accelerated hepatocyte entry into S phase and mitosis, which was associated with altered expression of cell cycle-promoting genes (21).
Analysis of cDNA microarrays shows that diminished proliferation of fibroblasts from elderly patients is caused by defects in the mitotic machinery, which ultimately result in chromosome instability and mutations, leading to a variety of diseases found in the elderly (22). Age-related defects in cellular proliferation are associated with diminished expression of the FoxM1B (HFH-11) transcription factor and numerous cell cycle regulatory genes (22). Because many of these diminished cell cycle progression genes are regulated by the FoxM1B transcription factor (21, 23), we proposed the hypothesis that reduced FoxM1B expression contributes to the proliferation defects observed in aging. In this study, we show that elevated levels of FoxM1B in regenerating liver of old transgenic mice increase the percentage of proliferative hepatocytes to levels similar to those observed in young regenerating mouse liver. Elevated FoxM1B levels in regenerating liver of old transgenic (tg) mice are associated with increased expression of cell cycle regulatory genes required for hepatocyte progression through DNA replication and mitosis. Our results indicate that reduced expression of the FoxM1B transcription factor contributes to the decline in cellular proliferation observed in the aging process.
Materials and Methods
Partial Hepatectomy Surgery, Immunohistochemical Staining, and Western Blot Analysis.
Generation of TTR-FoxM1B transgenic CD-1 mice, which used the −3 kb TTR promoter to constitutively express the FoxM1B transgene in hepatocytes, was described previously (21). Twelve-month-old wild-type (wt), two-month-old wt, and 12-month-old TTR-FoxM1B tg CD-1 mice were subjected to PHx to induce liver regeneration as described previously (21). Briefly, a midventral laparotomy was performed on each mouse under anesthesia, two-thirds of the liver was resected surgically (removal of left lateral, left median, and right median liver lobes), and the surgical incision was sutured closed. An i.p. injection of PBS containing 10 mg/ml BrdUrd (Sigma; 50 μg/g body weight) was administered 2 h before harvesting the remnant regenerating liver. Three mice at each time point were killed by using CO2 asphyxiation at the following intervals after PHx: 24, 32, 36, 40, 44, and 48 h. The regenerating livers were harvested and divided into three portions: One to isolate total RNA (21), one to isolate total protein extract (24), and one used for paraffin embedding. Determination of the number of hepatocytes undergoing DNA synthesis was performed by mAb detection of BrdUrd incorporation (Roche Molecular Biochemicals) of regenerating liver (5-μm paraffin sections) by using microwave retrieval to enhance antigenic activity as described (21). In each regenerating liver, we counted the number of BrdUrd-positive nuclei per 1,000 hepatocytes, and the mean number of BrdUrd-positive cells and SD were calculated for each time point by using three regenerating liver samples as described (21).
For Western blot analysis, 50 μg of total liver protein (24) was separated by SDS/PAGE and transferred to Protran membrane (Schleicher & Schuell). The primary FoxM1B (HFH-11) antibody was amplified by biotin-conjugated anti-rabbit IgG (Bio-Rad), and signals were developed with enhanced chemiluminescence (ECL; Amersham Pharmacia). Three regenerating liver sections at 48 h after PHx were stained with hematoxylin and eosin and examined for mitotic figures (mitosis). Hepatocyte mitosis is expressed as the mean number of mitotic figures found per 1,000 hepatocytes ± SD as described (21).
RNase Protection and HepG2 Cell Cotransfection Assays.
RNase protection assays were performed by hybridizing 20–40 μg of total regenerating liver RNA with [32P]UTP-labeled antisense RNA probes (human FoxM1B transgene, mouse FoxM1B, and mouse cyclophilin) followed by RNase 1 digestion, electrophoresis, and autoradiography as described (21, 23, 24). Expression of the cyclin genes was determined by RNase protection, with RNA isolated from regenerating liver, and RNA protection probes were purchased from PharMingen and used as recommended by the manufacturer. RNase protection assays to examine expression of Cdc25B and p55Cdc genes during liver regeneration also were included (probes were derived from Atlas array and purchased from CLONTECH). The BIOMAX 1D program (Eastman Kodak) was used for quantification of scanned x-ray films. RNase protection assay gels also were exposed to phosphorimaging screens for 1 or 2 days and scanned with a Storm 860 phosphorimager (Amersham Pharmacia). The imagequant program (Amersham Pharmacia) was used for quantification of phosphorimager scans. Expression levels were normalized to either cyclophilin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. Mean mRNA levels ± SD were determined from three distinct regenerating livers.
Human hepatoma HepG2 cells were cultured in Ham's F-12 supplemented with 7% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, MEM amino acids (all from GIBCO/BRL), and 0.5 units/ml human insulin (Humulin; Eli Lilly). HepG2 cells were grown in six-well culture plates (35-mm wells), and DNA constructs were transfected into cells with Fugene6 (Roche Molecular Biochemicals). HepG2 cells were transfected with 200 ng of either cytomegalovirus (CMV)-FoxM1B cDNA (20) or CMV only (control) expression vectors, and 1,500 ng of the following cyclin promoters linked to luciferase reporter constructs: −1745 cyclin D1 (25), −1050 cyclin B1, and −229 cyclin B1 (26). Protein extracts were prepared from transfected HepG2 cells from 24 to 36 h after DNA transfection and used to measure luciferase enzyme activity. Luciferase activity was detected by using the Dual-Luciferase Assay System (Promega), which uses the CMV-Renilla luciferase plasmid (20 ng) to normalize for fluctuations in transfection efficiency. Results are presented as mean ± SD of seven separate experiments, with the control (CMV only) set at 1.0.
Results and Discussion
Increased FoxM1B Levels Restore Peaks in DNA Replication and Mitosis During Liver Regeneration of Old tg Mice.
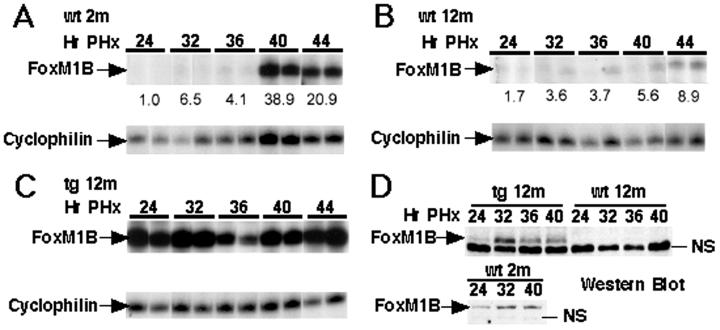
To examine the role of FoxM1B levels in proliferation during aging, we performed liver regeneration studies with old-aged wt and TTR-FoxM1B tg CD-1 mice, the latter of which constitutively express the FoxM1B mRNA in hepatocytes (21). Twelve-month-old (old-aged) wt and tg mice were subjected to PHx operations, and their regenerating livers were harvested at different intervals between 24 and 48 h after surgery (21). Hepatocyte DNA synthesis was monitored by immunohistochemical staining of BrdUrd incorporation into DNA as described (21). As a comparison, we included regenerating livers of 2-month-old (young) CD-1 mice, which exhibited a sharp S phase peak at 40 h after PHx (Fig. 1A, blue line) and corresponding induction of FoxM1B mRNA and protein levels (Fig. 2 A and D). A significant reduction in the 40-h S phase peak was observed in regenerating livers from 12-month-old wt mice (Fig. 1A, red line), and this was associated with diminished induction of FoxM1B mRNA and protein levels compared with young regenerating liver (Fig. 2 A, B, and D). In contrast, regenerating livers of 12-month-old tg mice exhibited a sharp peak in hepatocyte DNA replication at 40 h after PHx (Fig. 1A, green line). This increase in BrdUrd incorporation (Fig. 1 C and D) was associated with elevated FoxM1B mRNA and protein levels that are comparable to regenerating liver of young mice (Fig. 2 B–D). These results suggest that restoring FoxM1B levels in the livers of old mice alleviate the decline in hepatocyte entry into S phase associated with aging in regenerating liver. Although previous studies demonstrated that premature expression of FoxM1B in regenerating livers of young tg mice elicited earlier hepatocyte entry into S phase, this earlier hepatocyte DNA replication was not observed in regenerating livers of 12-month-old tg mice (21). One potential explanation for this discrepancy is that we observed delayed nuclear localization of FoxM1B protein in regenerating liver of old tg mice (data not shown), and this may contribute to preventing earlier onset of hepatocyte DNA replication.
Figure 1.
Increased FoxM1B expression in regenerating liver of old tg mice stimulates hepatocyte entry into DNA synthesis and mitosis. (A) Graphic representation of hepatocyte BrdUrd incorporation during mouse liver regeneration. Graphically presented is the BrdUrd incorporation (DNA replication) at the indicated hours after PHx with 12-month-old wt (red), 12-month-old TTR-HFH-11B tg (green), or 2-month-old wt (blue) CD-1 mice. Two hours before the regenerating livers were harvested, the mice received an i.p. injection of BrdUrd and immunohistochemical staining was used to measure the BrdUrd-incorporation rate in regenerating liver sections as described previously (21). The mean of the number of BrdUrd-positive nuclei per 1,000 hepatocytes and SD were calculated for each time point as described in Materials and Methods. Shown is the BrdUrd immunostaining of liver sections at the S phase peak (40 h after PHx) from either 12-month-old wt mice (B) or 12-month-old tg mice (C), demonstrating that restoring FoxM1B levels elicited increased BrdUrd incorporation in regenerating liver of old tg mice. (D) Graphic representation of increased hepatocyte mitosis in regenerating livers of old tg mice at the peak of mitosis (48 h after PHx). In three regenerating livers at 48 h after PHx, hepatocyte mitosis is expressed as the mean ± SD of the number of mitotic figures found per 1,000 hepatocytes (21).
Figure 2.
FoxM1B mRNA and protein levels in regenerating livers of old wt and tg mice. Time course of FoxM1B mRNA levels in regenerating liver of 2-month-old (young) wt (A), 12-month-old wt (B), and 12-month-old tg (C) CD-1 mice. Total liver RNA was prepared at the indicated hours after PHx operation (numbers along the top), and RNase protection assays were used to analyze for FoxM1B expression as described previously (21, 23). Expression of FoxM1B mRNA was normalized to cyclophilin levels, and shown below each image is the fold induction compared with young wt 24-h time point. (D) Western blot analysis using affinity-purified FoxM1B antibody (20, 21) was performed with total liver protein extracts (24) isolated from regenerating livers (at the indicated hours after PHx) of 12-month-old (12 m) wt, 12-month-old tg, or 2-month-old (2 m) wt mice. Indicated on the Western blot is the FoxM1B protein band, which migrates more slowly on the gel than a nonspecific (NS) band, which varies in intensity.
Progression into mitosis also was diminished significantly in regenerating hepatocytes of old wt mice as evidenced by only a few mitotic figures at 48 h after PHx (Fig. 1D). Increased hepatic levels of FoxM1B restored tg hepatocyte progression into mitosis at a rate similar to that found in young regenerating liver (Fig. 1D). Although we have used liver regeneration as a model system to examine cellular replication, it is important to note that expression of FoxM1B is induced during the proliferation of many cell types, such as fibroblasts (20, 22, 27). Our data therefore imply that restoring FoxM1B expression in a variety of distinct cell types will ameliorate proliferation defects observed during the aging process.
Increased FoxM1B Levels in Regenerating Liver of Old Transgenic Mice Alter Expression of Genes That Stimulate S Phase and M Phase Progression.
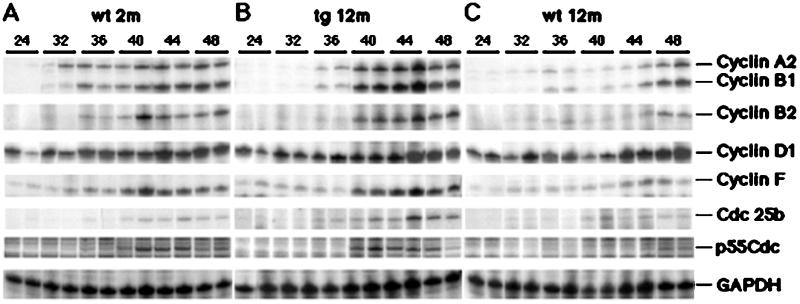
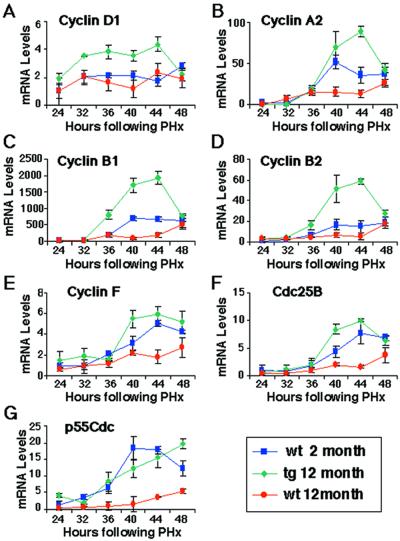
To identify cell cycle regulatory genes whose expression is increased in regenerating liver of old tg mice, RNase protection assays were performed in triplicate with potential FoxM1B target gene probes and RNA isolated from regenerating liver (Fig. 3; representative duplicate lanes are shown). Increased FoxM1B expression in old-aged regenerating tg liver was associated with elevated expression of the S phase-promoting cyclin D1 gene compared with either 12- or 2-month-old wt mice (Figs. 3 and 4A). Cyclin D1/cyclin-dependent kinase (cdk) 4/6 complexes are critical for S phase entry through phosphorylation of retinoblastoma (RB) protein and activation of the E2F transcription factor, which, in turn, activates its proliferation-specific target genes (reviewed in ref. 28). Elevated FoxM1B levels also restored increased expression of cyclin A2 in regenerating liver of old tg mice (Figs. 3 and 4B). The formation of the cyclin A2/cdk2 complex has been implicated in maintaining phosphorylation of RB during S phase, suggesting that this complex is required for progression through DNA replication (28). Furthermore, the cyclin A2/cdk2 complex is essential for progression into mitosis because it phosphorylates the cdh1 subunit of the ubiquitin-ligase anaphase-promoting complex (APC), which prevents APC-mediated degradation of cyclin B at the end of S phase and allows its accumulation to promote entry into mitosis (28). Taken together, these data indicate that restoring FoxM1B expression in regenerating liver of old mice stimulates induction of S phase-promoting cyclin D1 and cyclin A2, which serve to facilitate hepatocyte entry and progression through S phase.
Figure 3.
Increased FoxM1B expression in regenerating liver of old tg mice stimulates expression of cell cycle-promotion genes. RNase protection assay demonstrates increased expression of cell cycle-promotion genes in regenerating liver of old tg mice. Total RNA was prepared from regenerating livers from 2-month-old wt mice (A), 12-month-old tg mice (B), and 12-month-old wt mice (C) at indicated hours after PHx (numbers along the top). The liver RNA samples were used for RNase protection assays (in triplicate) to determine expression of the indicated cyclin genes as well as the Cdc25B and p55Cdc genes as described in Materials and Methods. Note that representative duplicate samples are shown.
Figure 4.
Graphic presentation of normalized mean mRNA levels (±SD) of cell cycle regulatory genes. Shown are mean mRNA levels of cyclin D1 (A), cyclin A2 (B), cyclin B1 (C), cyclin B2 (D), cyclin F (E), Cdc25B (F), and p55cdc (G) at various intervals in regenerating liver (in triplicate). The regenerating liver was obtained from 2-month-old wt mice (blue), 12-month-old tg mice (green), or 12-month-old wt mice (red). Expression levels of cell cycle regulatory genes were normalized to the GAPDH mRNA levels. The expression level of the 2-month-old regenerating mouse liver at 24 h after PHx was set at 1.0.
Proliferating fibroblasts from elderly patients also display a block in the G2/M phase transition of the cell cycle, which results from diminished expression of genes required for progression into mitosis (22). We also found that regenerating liver of 12-month-old wt mice displayed reduced hepatocyte entry into mitosis (Fig. 1D), suggesting that they exhibited diminished expression of genes required to progress into mitosis. We next used RNase protection assays to demonstrate that increased FoxM1B expression in regenerating liver of old tg mice displayed increased expression of M phase-promoting genes (Figs. 3 and 4). At the peak of hepatocyte DNA replication, regenerating liver from old tg mice displayed significant induction of cyclin B1 and cyclin B2 compared with either 2- or 12-month-old regenerating liver (Figs. 3 and 4 C and D). The cyclin B proteins associate with cdc-2 (cdk1) to mediate cell cycle progression from the G2 phase into mitosis (reviewed in ref. 29). Concomitant with induction of cyclin B levels, significant increases in cyclin F levels were evident in regenerating liver of 2-month-old wt and 12-month-old tg mice (Figs. 3 and 4E). Cyclin F is critical for M phase progression because it associates with the cyclin B complexes to facilitate their nuclear translocation (30). Elevated FoxM1B levels in 2-month-old wt and 12-month-old tg mice also elicited a more pronounced increase of Cdc25B mRNA levels between 40- to 48-h time points after the PHx operation (Figs. 3 and 4F). Cdc25b regulates M phase progression by activating the mitotic kinase cdk1/cyclin B by means of dephosphorylation (31–33), and, therefore, its induced expression is essential to promote entry into mitosis. Finally, regenerating liver of old tg mice displayed induced expression of p55Cdc comparable to young mice (Figs. 3 and 4G). The p55Cdc protein regulates ubiquitin-ligase APC-mediated degradation of the cyclin proteins, whose elimination is required for the completion of mitosis (29). Consistent with the role of FoxM1B in regulating the G2/M phase transition, Foxm1b/Hfh11/Trident-deficient (−/−) embryos display an abnormal polyploid phenotype resulting from endoreduplication in embryonic hepatocytes and cardiomyocytes (day 13 post coitum), suggesting that FoxM1B functions to regulate cell cycle progression into mitosis (19). It is tempting to speculate that this aberrant endoreduplication in Foxm1b −/− embryos is a result of reduced expression of cyclin B, Cdc25B, and p55Cdc genes, which are necessary for progression and completion of mitosis. Taken together, our liver-regeneration studies indicate that increased FoxM1B expression in old tg mice increases expression of M phase-promoting cyclin B1, cyclin B2, cyclin F, Cdc25B, and p55Cdc genes, which are required for M phase progression and exit from mitosis.
Cotransfection Assays Demonstrate That FoxM1B Activates Cyclin D1 and Cyclin B1 Promoter Expression.
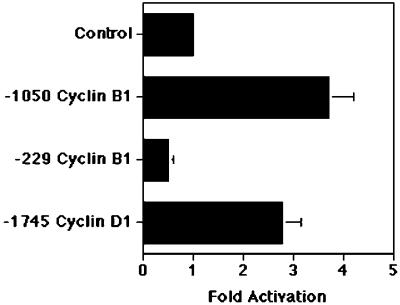
Because the human cyclin D1 promoter (−1095 to −1083) and the human cyclin B1 promoter (−806 to −817) contained potential binding sites for the FoxM1B transcription factor (20, 34), we wanted to examine whether FoxM1B could stimulate expression of these cyclin promoters. We used human hepatoma HepG2 cells for cotransfection assays with the CMV FoxM1B cDNA expression vector and the cyclin promoters linked to luciferase reporter constructs: −1745 cyclin D1 (25), −1050 cyclin B1, and −229 cyclin B1 (26) as described in Materials and Methods. Following normalization with the CMV-Renilla luciferase control, the mean fold induction of cyclin promoters is presented graphically in Fig. 5, where the CMV promoter vector cotransfection (control) was set at 1.0. These cotransfection assays showed that the FoxM1B expression vector provided a 3.7-fold induction in cyclin B1 promoter activity and that retention of the FoxM1B-binding sequence in the cyclin B1 promoter is required for transcriptional activation (Fig. 5). Furthermore, FoxM1B provided a 2.8-fold induction in cyclin D1 promoter expression in HepG2 cell cotransfection assays (Fig. 5). These studies demonstrated that FoxM1B is able to transcriptionally activate the cyclin B1 and cyclin D1 promoters, suggesting that FoxM1B functions to directly regulate transcription of these cyclin genes.
Figure 5.
FoxM1B directly activates transcription of the cyclin D1 and cyclin B1 promoter in cotransfection assays. HepG2 cells were cotransfected with either the CMV-FoxM1B cDNA expression vector or CMV empty vector and the cyclin promoters linked to luciferase reporter constructs: −1745 cyclinD1 (25), −1050 cyclin B1, and −229 cyclin B1 (26). Cell protein extracts were prepared and assayed for luciferase enzyme activity as described in Materials and Methods. The CMV-Renilla luciferase vector was included as a normalization control for transfection efficiency. Normalized fold induction of cyclin B1 and cyclin D1 promoter expression in response to FoxM1B cDNA cotransfection is presented graphically, with CMV-empty vector control set at 1.0. Seven distinct transfection experiments were performed and used to determine mean fold induction ± SD.
Elevated FoxM1B Levels Increased Expression of Cell Cycle Regulatory Genes Associated with Age-Related Proliferation Defects.
Analysis of cDNA microarrays shows that diminished proliferation exhibited by fibroblasts from either elderly patients or genetically aged patients with Hutchinson—Gilford progeria is associated with reduced expression of cell cycle genes as well as a decline in FoxM1B levels (22). These studies indicate that an underlying mechanism of the aging process involves defective induction of cell cycle-promotion genes and dysfunction of the mitotic machinery, which causes accumulation of proliferating cells blocked in the G2/M phase transition of the cell cycle. These proliferation defects ultimately result in chromosome instability and mutations, leading to a variety of diseases found in the elderly population. Our current study demonstrates that diminished expression of FoxM1B mRNA and protein is also found in regenerating livers of 12-month-old mice. Although the mechanism for this decline in FoxM1B expression remains uncharacterized, it is possible that age-related decreases in hepatic expression of insulin-like growth factor 1 (IGF-1) contribute to the decline in FoxM1B levels in regenerating liver of old mice (35). Nevertheless, our liver-regeneration studies determine that restoring hepatic expression of FoxM1B alone in old tg mice is sufficient to restore hepatocyte progression into DNA synthesis and mitosis to levels similar to those found in young regenerating mouse liver. Furthermore, stimulation of hepatocyte proliferation in response to elevated FoxM1B levels was associated with increased expression of numerous cell cycle-promoting genes. Restoring FoxM1B levels increased expression of S phase-promoting cyclin D1 and cyclin A2 and stimulated expression of M phase-promoting cyclin F, cyclin B1, cyclin B2, and Cdc25B as well as p55Cdc, which is required for exit from mitosis. Because all young, proliferating cells display elevated FoxM1B levels (20, 27, 34), our data imply that increased FoxM1B expression alone in numerous distinct cell types of the elderly is sufficient to potentiate transcription of the cell cycle-promotion genes and alleviate age-related proliferation defects. Furthermore, cotransfection assays demonstrate that FoxM1B activates transcription of cyclin B1 and cyclin D1 promoters, suggesting that these cyclin genes are a direct target of the FoxM1B transcription factor. Taken together, our studies demonstrate that FoxM1B controls the transcriptional network of genes that regulate cell cycle progression.
Although hepatocyte expression of the c-myc transcription factor in transgenic mice stimulates hepatocyte replication during liver regeneration, its constitutive expression causes aberrant hepatocyte proliferation without liver injury because c-myc localizes to the nucleus in the absence of proliferative signals (36). Unlike the c-myc transcription factor, overexpression of the FoxM1B (HFH-11B) transcription factor alone is insufficient to induce differentiated cells to enter the cell cycle because its nuclear localization requires proliferation-specific signals, and, therefore, FoxM1B transgene protein remains in the cytoplasm of quiescent hepatocytes (21). This feature makes the FoxM1B transcription factor a likely candidate for therapeutic intervention to ameliorate defective proliferation observed in the elderly population because its aberrant expression in quiescent cells will not induce unwanted cellular proliferation. Future transgenic mouse studies in which FoxM1B is expressed ubiquitously would allow determination of whether sustaining FoxM1B expression during the aging process leads to an increase in longevity.
Acknowledgments
We thank P. Raychaudhuri, N. Hay, V. Kalinichenko, F. Rausa, M. Major, and D. Hughes for critically reviewing the manuscript and F. Rausa for help with the cotransfection experiments. We also thank John Cogswell for providing us with the −1050 and −229 human cyclin B1 promoter and Chris Albanese and Richard Pestell for providing us with the −1745 cyclin D1 human promoter luciferase construct. This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK54687 (to R.H.C.).
Abbreviations
- FoxM1B
forkhead box M1B
- PHx
partial hepatectomy
- S phase
DNA synthesis
- M phase
mitosis
- TTR
transthyretin
- tg
TTR-FoxM1B transgenic
- wt
wild type
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CMV
cytomegalovirus
- cdk
cyclin-dependent kinase
- BrdUrd
bromodeoxyuridine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fausto N, Laird A D, Webber E M. FASEB J. 1995;9:1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- 2.Taub R. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 3.Michalopoulos G K, DeFrances M C. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 4.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 5.Yamada Y, Kirillova I, Peschon J J, Fausto N. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaestner K H, Knochel W, Martinez D E. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 7.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 8.Costa R H, Kalinichenko V V, Lim L. Am J Physiol Lung Cell Mol Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- 9.Duncan S A. Dev Dyn. 2000;219:131–142. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 10.Lai E, Prezioso V R, Tao W F, Chen W S, Darnell J E., Jr Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 11.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 12.Kaestner K H. Trends Endocrinol Metab. 2000;11:281–285. doi: 10.1016/s1043-2760(00)00271-x. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann E, Knochel W. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Geerdes D W, Vogt P K. Oncogene. 2000;19:4815–4821. doi: 10.1038/sj.onc.1203834. [DOI] [PubMed] [Google Scholar]
- 15.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 16.Perl A K, Whitsett J A. Clin Genet. 1999;56:14–27. doi: 10.1034/j.1399-0004.1999.560103.x. [DOI] [PubMed] [Google Scholar]
- 17.Scheidler S, Fredericks W J, Rauscher F J, III, Barr F G, Vogt P K. Proc Natl Acad Sci USA. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaret K. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 19.Korver W, Schilham M W, Moerer P, van den Hoff M J, Dam K, Lamers W H, Medema R H, Clevers H. Curr Biol. 1998;8:1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- 20.Ye H, Kelly T F, Samadani U, Lim L, Rubio S, Overdier D G, Roebuck K A, Costa R H. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye H, Holterman A, Yoo K W, Franks R R, Costa R H. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ly D H, Lockhart D J, Lerner R A, Schultz P G. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Hung N-J, Costa R H. Hepatology. 2001;33:1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 24.Rausa F M, Tan Y, Zhou H, Yoo K, Stolz D B, Watkins S, Franks R R, Unterman T G, Costa R H. Mol Cell Biol. 2000;20:8264–8282. doi: 10.1128/mcb.20.21.8264-8282.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 26.Cogswell J P, Godlevski M M, Bonham M, Bisi J, Babiss L. Mol Cell Biol. 1995;15:2782–2790. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korver W, Roose J, Clevers H. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour J W, Dean D C. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 29.Zachariae W, Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 30.Kong M, Barnes E A, Ollendorff V, Donoghue D J. EMBO J. 2000;19:1378–1388. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson I, Hoffmann I. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 32.Sebastian B, Kakizuka A, Hunter T. Proc Natl Acad Sci USA. 1993;90:3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trembley J H, Ebbert J O, Kren B T, Steer C J. Cell Growth Differ. 1996;7:903–916. [PubMed] [Google Scholar]
- 34.Yao K M, Sha M, Lu Z, Wong G G. J Biol Chem. 1997;272:19827–19836. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Bennett S A, Ingram R L, Sonntag W E. Endocrinology. 1995;136:4551–4557. doi: 10.1210/endo.136.10.7664676. [DOI] [PubMed] [Google Scholar]
- 36.Factor V M, Jensen M R, Thorgeirsson S S. Hepatology. 1997;26:1434–1443. doi: 10.1002/hep.510260610. [DOI] [PubMed] [Google Scholar]