Key Points
Question
Do racial/ethnic disparities reduce the usefulness of genetic testing for cardiomyopathy and other diseases?
Findings
In this cross-sectional study, molecular diagnostic testing data from 5729 probands over a 15-year period were analyzed. A statistically significant reduction in the detection rate of pathogenic and likely pathogenic variants for cardiomyopathy was seen in patients from underrepresented minority racial/ethnic groups compared with white patient populations.
Meaning
Disparities in access to genetic testing have already reduced the rate of detection of disease via genetic testing in underrepresented minorities; improvements in access to testing will be needed to overcome these disparities.
This cross-sectional study examines whether cardiomyopathy detection rates via genetic tests differ between white people, Asian people, and racial/ethnic minorities who are underrepresented in genetic research and clinical data.
Abstract
Importance
Individuals of all races/ethnicities have a fundamental right to access health care and benefit from advances in science and medicine, including genetic testing.
Objective
To determine whether detection rates for cardiomyopathy genetic testing differed between white people, Asian people, and underrepresented minorities (individuals of black, Hispanic, Native American, Alaskan Native, or Pacific Islander descent).
Design, Setting, and Participants
We conducted a cross-sectional analysis of the genetic panel test results of 5729 probands who had a suspected diagnosis or family history of cardiomyopathy and who had been referred for testing between October 2003 and December 2017. Testing was performed at the Laboratory for Molecular Medicine at Partners Personalized Medicine in Cambridge, Massachusetts. Results were stratified into 3 categories of self-reported race/ethnicity: white, Asian, and underrepresented minorities.
Main Outcomes and Measures
The primary outcome was whether a pathogenic or likely pathogenic variant was identified that explained the features or family history of cardiomyopathy. A secondary outcome was the number of test results that were inconclusive because of the presence of 1 or more variants of uncertain significance in the absence of an explanation for cardiomyopathy features or family history.
Results
A total of 5729 probands were studied (of whom 3523 [61.5%] were male). Of these, 4539 (79.2%) were white, 348 (6.1%) were Asian individuals, and 842 (14.7%) were underrepresented minorities. Positive detection occurred in 1314 white individuals (29.0%) compared with 155 underrepresented minorities (18.4%; χ21 = 39.8; P < .001) and 87 Asian individuals (25.0%; χ21 = 2.5; P = .12). Inconclusive results were found in 1115 white individuals (24.6%) compared with 335 underrepresented minorities (39.8%; χ21 = 83.6; P < .001) and 136 Asian individuals (39.2%; χ21 = 35.8; P < .001).
Conclusions and Relevance
These results show a significantly higher positive detection rate and a significantly lower rate of inconclusive results in white individuals in comparison with underrepresented minorities. This suggests greater clinical usefulness of genetic testing for cardiomyopathy in white persons in comparison with people of other racial/ethnic groups. This clear disparity warrants further study to understand the gaps in usefulness, which may derive from a lack of clinical testing and research in underrepresented minority populations, in the hopes of improving genetic testing outcomes for cardiomyopathy in nonwhite groups.
Introduction
Over the past 10 years, genetic testing has become commonplace in the diagnosis of monogenic diseases for many patients. However, the application of genetic testing has not been equal in all sectors of the population. Racial/ethnic disparities in research study enrollment and the delivery of health care, favoring white individuals and racial/ethnic minorities of higher socioeconomic status, have led to differences in the development and application of the evidence base that underlies the usefulness of genetic testing.
The Laboratory for Molecular Medicine at the Partners Healthcare Personalized Medicine launched the first clinical genetic test for hypertrophic cardiomyopathy in 2003 and expanded testing over the subsequent 15 years to encompass 62 genes that have been implicated in cardiomyopathy. Here we present data from 15 years of genetic testing for cardiomyopathy. With a prevalence of 1 in every 500 individuals, cardiomyopathy is one of the most common monogenic cardiac diseases in the US population. We document the association between racial/ethnic disparities and genetic testing intended to inform the care of patients.
Methods
We conducted a cross-sectional analysis of 7409 probands referred for genetic testing. For analyses, we grouped black, Hispanic, Native American, Alaska Native, Hawaiian, and other South Pacific Islander individuals in a single category termed underrepresented minorities (URM) and did not consider those of mixed, unspecified, or other races/ethnicities. We analyzed probands for the detection rate for cardiomyopathy, which we defined as the percentage of probands with a positive report because of 1 or more pathogenic or likely pathogenic variants identified to be causative for the proband’s clinical presentation or family history of cardiomyopathy. This study was approved by Partners Healthcare institutional review board and conducted under a protocol for which consent procedures were waived.
For comparison, we analyzed the detection rates of other diseases, including hearing loss and RASopathies (ie, Noonan syndrome and related disorders). Additionally, we analyzed the rate of inconclusive findings, which resulted largely from the presence of 1 or more variants of uncertain significance in the absence of a pathogenic or likely pathogenic variant. Categorical variables were assessed using a χ2 analysis or Fisher exact test. We used SAS version 9.4 (SAS Institute) and regarded a 2-tailed P value of less than .05 as significant.
Results
Of the 7409 probands identified, 5729 were described as white, Asian, or URM. Of these, 4539 were white (of whom 2753, or 61.1%, were male), 348 were Asian (of whom 210, or 61.0%, were male), 565 were black, 237 were Hispanic, and 40 were Native American, Alaskan, Hawaiian, and other South Pacific Islander. Collectively, the URM group included 472 males (56.6%). Approximately 5 of every 6 patients tested by the Laboratory for Molecular Medicine were from the United States or Canada, and the remaining portion were from other countries. An additional 204 persons of mixed racial/ethnic background and 1476 individuals of unspecified or other races/ethnicities (combined, 22.6% of the 7409 probands) were not included in the analysis.
The Table provides characteristics of the cohort stratified by race/ethnicity. These analyses demonstrate a statistically significant reduction in detection rate for URM people (155/842; 18.4%), compared with the detection rate for white individuals (1314/4539; 29.0%; χ21 = 39.8; P < .001). Asian individuals also had a reduced detection rate compared with white individuals (87/348; 25.0%), but these findings were not statistically significant (χ21 = 2.5; P = .12).
Table. Genetic Testing for Cardiomyopathy, Hearing Loss, and RASopathies.
| Characteristic or Outcome | Racial/Ethnic Patient Subgroups | ||||
|---|---|---|---|---|---|
| White | Underrepresented Minorities | P Valuea | Asian | P Valuea | |
| Cardiomyopathy | |||||
| Patient characteristic | |||||
| No. | 4539 | 842 | NA | 348 | NA |
| Age, y, median (range) | 41.0 (0.5-93.0) | 20.0 (0.5-89.0) | NA | 41.7 (0.5-84.0) | NA |
| Male, No. (%) | 2753 (60.7) | 472 (56.1) | .01 | 210 (60.3) | .91 |
| Test results, No. (%)b | |||||
| Positive | 1314 (29.0) | 155 (18.4) | <.001 | 87 (25.0) | .12 |
| Inconclusive | 1115 (24.6) | 335 (39.8) | <.001 | 136 (39.1) | <.001 |
| Negative | 1987 (43.8) | 330 (39.2) | NA | 118 (33.9) | NA |
| Hearing Loss | |||||
| Patient characteristics | |||||
| No. | 1579 | 465 | NA | 164 | NA |
| Age, y, median (range) | 6 (0.5-76.0) | 6 (0.5-73.0) | NA | 6 (0.5-64.0) | NA |
| Male, No. (%) | 778 (49.3) | 242 (52.0) | .29 | 80 (48.8) | .90 |
| Test results, No. (%)b | |||||
| Positive | 360 (22.8) | 72 (15.5) | <.001 | 62 (37.8) | <.001 |
| Inconclusive | 500 (31.7) | 169 (36.3) | .06 | 55 (33.5) | .62 |
| Negative | 694 (44.0) | 222 (47.7) | NA | 42 (25.6) | NA |
| RASopathies | |||||
| Patient characteristics | |||||
| No. | 1887 | 382 | NA | 105 | NA |
| Age, y, median (range) | 5 (0.5-68.0) | 2 (0.5-40.0) | NA | 6 (0.5-73.1) | NA |
| Male, No. (%) | 1015 (53.8) | 210 (55.0) | .67 | 44 (41.9) | .02 |
| Test results, No. (%)b | |||||
| Positive | 502 (26.6) | 128 (33.5) | .01 | 40 (38.1) | .01 |
| Inconclusive | 81 (4.3) | 22 (5.8) | .21 | 4 (3.8) | .81 |
| Negative | 1285 (68.1) | 229 (59.9) | NA | 58 (55.2) | NA |
Abbreviation: NA, not applicable.
Results are based on the χ2 statistic. Statistical analysis compared underrepresented minority and Asian groups individually with white individuals.
Positive reflects the identification of pathogenic or likely pathogenic variant(s) that explain the cause of disease; inconclusive reflects the identification of a variant of uncertain significance in the absence of an explanation of disease; negative reflects the absence of pathogenic, likely pathogenic, or uncertain significance variants.
We compared this reduction with other diseases, including hearing loss and RASopathies (ie, Noonan syndrome and related disorders). Similar differences were seen in hearing loss, although in this case, Asian individuals (62/164; 37.8%) had a higher detection rate than white individuals (360/1579; 22.8%; χ21 = 18.2; P < .001). For RASopathies, detection rates were higher in both the URM group (128/382; 33.5%) and the Asian group (40/105; 38.1%) than in the white group (502/1887; 26.6%; χ21 = 7.6 and P = .01 for comparisons of the URM and white groups; χ21 = 6.6 and P = .01 for comparisons of the Asian and white groups). These results are shown in the Table.
Additionally, the rate of inconclusive results was higher for the URM group in comparison with the white group for cardiomyopathy (URM: 335/842; 39.8%; vs white: 1115/4539; 24.6%), hearing loss (URM: 169/465; 36.3%; vs white: 500/1579; 31.7%), and RASopathies (URM: 22/382; 5.8%; vs white: 81/1887; 4.3%) testing. However, the difference was only statistically significant for cardiomyopathy (χ21 = 83.6; P<.001). Inconclusive results for cardiomyopathy testing in Asian individuals was also higher than in white individuals (Asian: 136/348; 39.2%; vs white: 1115/4539; 24.6%; χ21 = 35.8; P < .001). Results are shown in the Table.
Discussion
We posit the following explanation for the reduction in positive test rates in URM individuals for cardiomyopathy and hearing loss but not RASopathies. Given dominant inheritance and reduced reproductive fitness of affected individuals, most children born with a RASopathy disorder have unaffected parents, and therefore most causative variants occur de novo. This type of variant typically does not require a prior evidence base to implicate the variant as likely pathogenic in a diagnostic setting (the tested patient is affected with a disease). As such, one would expect that prior efforts in research and clinical testing would not affect detection rates. This is consistent with the observation that the positive RASopathy detection rate in white people is not higher than the rate in URM individuals.
In contrast, for hearing loss and cardiomyopathy, reproductive fitness is not substantially reduced by the disorder, which leads to much rarer observations of de novo occurrence that can be used as evidence for pathogenicity. Furthermore, a large percentage of variants that are pathogenic for hearing loss and cardiomyopathy are missense variants, which are difficult to distinguish from benign variants that are not disease-causing and which require multiple case observations and/or functional studies to implicate the variants in disease. Thus, these disorders are likely to suffer more significantly from a poor evidence base as observed in the URM populations tested for cardiomyopathy and hearing loss. Interestingly, RASopathy detection rates in the URM and Asian groups were actually statistically significantly higher than in the white group, which may be because the small number of patients of these races/ethnicities sent for testing have a more convincing phenotype and are therefore more likely to have positive test results.
Also, we found that the detection rate in Asian individuals was higher than white individuals for hearing loss, yet similar to URM individuals for cardiomyopathy. We believe this difference is because the Asian population has been well-studied for the genetic basis of hearing loss, with several founder mutations identified. (GJB2 p.Val37Ile is the most common cause of genetic hearing loss in the Asian population.) In contrast, to our knowledge, Asian individuals have not been as well studied for cardiomyopathy, especially given that basic testing requires a large expensive sequencing panel; this is unlike hearing loss, for which many positive individuals can be identified through simple genetic tests.
A second challenge encountered in genetic testing is the receipt of an inconclusive test result because of the identification of 1 or more variants of uncertain significance (VUSs) in the absence of an explanation for disease. Receiving a VUS can lead to confusion for patients and physicians and can sometimes lead to improper use of the results for clinical decision making. We examined rates of inconclusive test results in cardiomyopathy, hearing loss, and RASopathies. We found that cardiomyopathy had a statistically significant increased rate of inconclusive test results in both the URM subgroup and the Asian subgroup compared with the white subgroup (Figure). Cardiomyopathy and hearing loss genes encode many large structural proteins that are highly susceptible to benign variation, whereas the RASopathies are largely caused by variants in the small, highly conserved signaling proteins of the Ras-MAP kinase pathway, which are largely devoid of benign variants. The main reason for the inability to interpret the variants of uncertain significance seen in URM individuals compared with white individuals is that limitations exist in the databases of normal genetic variation that aid in ruling out pathogenicity when allele frequencies are too high to be consistent with pathogenicity. The negative impact of the slower development of databases of allele frequencies in diverse populations has been previously published. (Interestingly, while the introduction of diverse population databases as provided by the 1000 Genomes Project, Exome Aggregation Consortium, and the Genome Aggregation Database have reduced the rate of VUSs per kilobase of DNA sequences, the rate per test has not dropped because the number of genes included in genetic tests continues to rise, sustaining the rate of VUS results.)
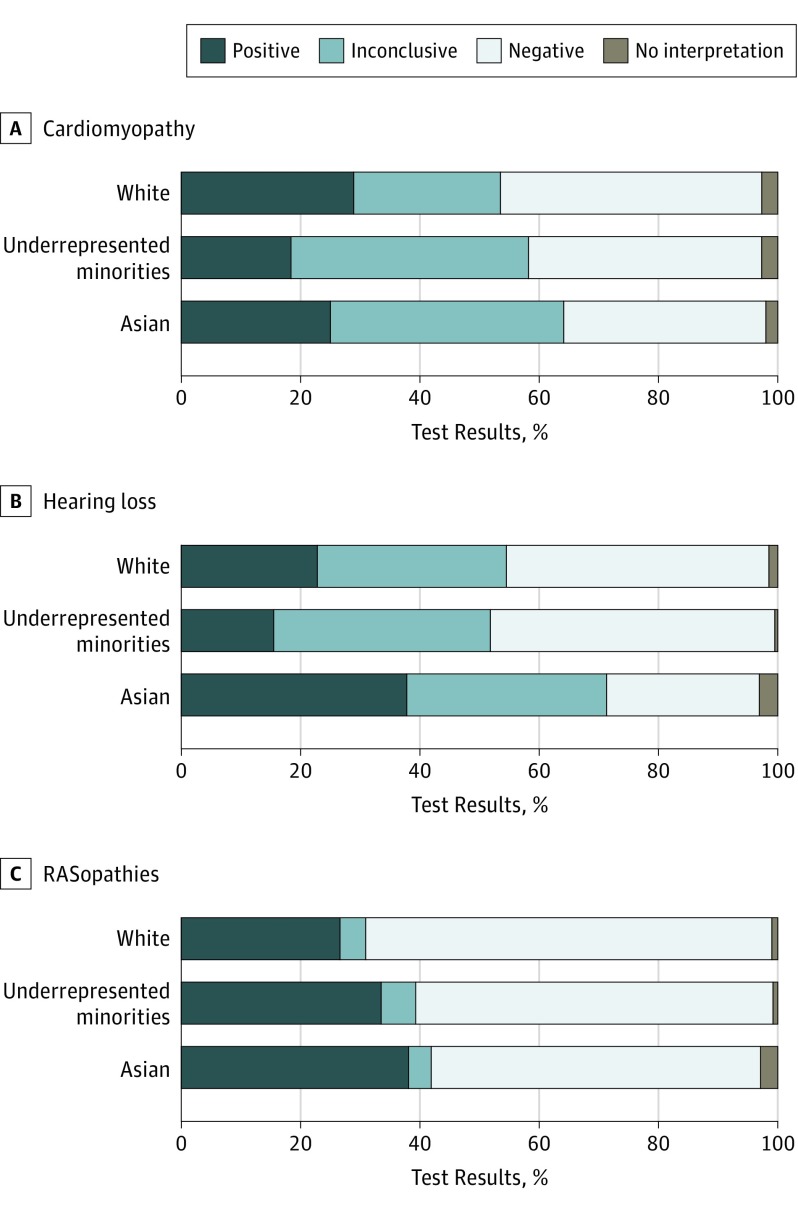
Figure. Genetic Testing Results by Racial/Ethnic Group.
A, Percentages of people of white, Asian, and underrepresented minority (URM) backgrounds with positive, inconclusive, or negative test results for cardiomyopathy. The difference between positive results in the white and URM groups was significant (white: 1314/4539; 29.0% vs URM: 155/842; 18.4%; χ21 = 39.8; P < .001). B, Percentages of people of white, Asian, and URM backgrounds with positive, inconclusive, or negative test results for hearing loss. Significant differences were found between positive results in the white and URM groups (white: 360/1579; 22.8% vs URM: 72/465; 15.5%; χ21 = 11.5; P = .001) and the white and Asian groups (white: 360/1579; 22.8% vs Asian: 62/164; 37.8%; χ21 = 18.2; P < .001). C, Percentage of people of white, Asian, and URM backgrounds with positive, inconclusive, or negative test results for RASopathies. Significant differences were found between positive results in the white and URM groups (white: 502/1887; 26.6% vs URM: 128/382; 33.5%; χ21 = 7.6; P = .01) and the white and Asian groups (white: 502/1887; 26.6% vs Asian: 40/105; 38.1%; χ21 = 6.6; P = .01).
Limitations
There are several limitations to the study. One limitation is the difference in sample size between white, Asian, and URM probands. Because this study was conducted at a clinical molecular laboratory, the population evaluated was not randomly sampled. For this reason, the proportion of individuals from each racial/ethnic group is reflective of differences in referrals for genetic testing. Also, race/ethnicity was self-reported in this study and may not fully reflect biologic genetic ancestry. Lastly, there was a significant difference in the age of URM individuals referred for testing compared with the ages of the white and Asian subgroups, which leaves age as a potential confounder. This should be adjusted for in future studies with sufficient sample sizes.
Conclusions
To our knowledge, this represents the first empirical analysis of differences in detection rate and inconclusive result rates for cardiomyopathy between different racial/ethnic groups. Our data suggest that cardiomyopathy testing has a statistically significant lower detection rate in URM individuals, which is likely because of the reduction of primary data from URM individuals in both the research and clinical testing settings. Furthermore, the rate of inconclusive test results is also higher in URM individuals, further undermining the utility of genetic testing in these populations and creating additional disparities for these populations beyond the fundamental lack of use of genetic testing already documented for URM individuals. To counter these challenges, we encourage recruitment of members of URM groups into both research and clinical practice to enable these populations to equally contribute to and benefit from genetic testing in the care and treatment of cardiomyopathy and other genetic disorders.
References
- 1.McKenna WJ, Maron BJ, Thiene G. Classification, epidemiology, and global burden of cardiomyopathies. Circ Res. 2017;121(7):722-730. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Noguchi Y, Yashima T, Ohno K, Kitamura K. Hereditary hearing loss and deafness genes in Japan. J Med Dent Sci. 2010;57(1):1-10. [PubMed] [Google Scholar]
- 3.Ouyang XM, Yan D, Yuan HJ, et al. The genetic bases for non-syndromic hearing loss among Chinese. J Hum Genet. 2009;54(3):131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallant E, Francey L, Tsai EA, et al. Homozygosity for the V37I GJB2 mutation in fifteen probands with mild to moderate sensorineural hearing impairment: further confirmation of pathogenicity and haplotype analysis in Asian populations. Am J Med Genet A. 2013;161A(9):2148-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng Q, Huang S, Liang Y, et al. Concurrent genetic and standard screening for hearing impairment in 9317 southern Chinese newborns. Genet Test Mol Biomarkers. 2016;20(10):603-608. [DOI] [PubMed] [Google Scholar]
- 6.Chu CW, Chen YJ, Lee YH, Jaung SJ, Lee FP, Huang HM. Government-funded universal newborn hearing screening and genetic analyses of deafness predisposing genes in Taiwan. Int J Pediatr Otorhinolaryngol. 2015;79(4):584-590. [DOI] [PubMed] [Google Scholar]
- 7.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shields AE, Burke W, Levy DE. Differential use of available genetic tests among primary care physicians in the United States: results of a national survey. Genet Med. 2008;10(6):404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cragun D, Weidner A, Lewis C, et al. Racial disparities in BRCA testing and cancer risk management across a population-based sample of young breast cancer survivors. Cancer. 2017;123(13):2497-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]