Abstract
Transposable elements have been used widely in the past 20 years for gene transfer and insertional mutagenesis in Drosophila. Transposon-based technology for gene manipulation and genomic analysis currently is being adopted for vertebrates. We tested the ability of Minos, a DNA transposon from Drosophila hydei, to transpose in mouse tissues. Two transgenic mouse lines were crossed, one expressing Minos transposase in lymphocytes under the control of the CD2 promoter/locus control region and another carrying a nonautonomous Minos transposon. Only mice containing both transgenes show excision of the transposon and transposition into new chromosomal sites in thymus and spleen cells. In addition, expression of Minos transposase in embryonic fibroblast cell lines derived from a transposon-carrying transgenic mouse resulted in excision of the transposon. These results are a first step toward a reversible insertional mutagenesis system in the mouse, opening the way to develop powerful technologies for functional genomic analysis in mammals.
Keywords: mouse tissues, tissue-specific transposase expression, tissue-specific transposon mobilization
The availability of genetic methodologies for functional genomic analysis, such as insertional mutagenesis and gene/enhancer trapping, is crucial for the study of gene function and genome organization of complex eukaryotes. Of the three “classical” model animals, the fly, the worm, and the mouse, efficient transposon-based insertion methodologies have been developed for Drosophila melanogaster and for Caenorhabditis elegans. The introduction of P element-mediated transgenesis and insertional mutagenesis in Drosophila by Rubin and Spradling 20 years ago (1) transformed Drosophila genetics and formed the paradigm for developing equivalent methodologies in other eukaryotes. However, the P element has a very restricted host range, and, therefore, other elements have been used in the past decade as vectors for gene transfer and/or mutagenesis in a variety of complex eukaryotes, including nematodes, plants, fish, and a bird (2–5).
Transposons belonging to the Tc1/mariner superfamily have been shown to be functional in different backgrounds, including mammalian cells. The Tc1 transposon from C. elegans is active in a human cell line (6) whereas Sleeping Beauty (SB), derived from salmonids, was shown to be active in a human cell line (7) and in mouse embryonic stem cells (8). More recently, SB has been reported to transpose from plasmids into the chromosomes of liver cells of live mice (9).
Minos is a mobile element of the Tc1/mariner superfamily that was isolated from Drosophila hydei and shown to be active in several dipteran and lepidopteran insect species (10–14). In addition to insects, Minos is also highly active in human cells in culture, where it can be used for genomewide, transposon-mediated mutagenesis (TRAMM; ref. 15). The development of an efficient insertional mutagenesis system in the mouse would provide an important tool for mouse functional genetic analysis, comparable to the P element in the fruit fly. We demonstrate here that Minos transposase produced from a transgene catalyzes transposition of a Minos transposon from one chromosomal site into new chromosomal sites in transgenic mice. This is the first step toward the development of a transposon-based mutagenesis system in mammals.
Materials and Methods
Plasmid Constructions.
The helper plasmid CD2/ILMi was constructed by subcloning the transposase cDNA (15) as an XbaI-blunt fragment into the vector SVA(−). The SVA(−) vector is a derivative of the VA vector (16), with extended multiple cloning sites. Transposon MiCMVGFP was constructed as follows: The plasmid pMiLRTetR (17) was cut with BamHI and religated to remove the tetracycline resistance gene between the Minos ends, resulting in plasmid pMiLRΔBamHI. An Asp718/SacI fragment from pMILRΔBamHI, containing the Minos inverted repeats and original flanking sequences from D. hydei, was cloned into plasmid pPolyIII-I-lox [created by insertion of the loxP oligo (ATAACTTCGTATAGCATACATTATACGAAGTTAT) into the Asp718 site of the vector pPolyIII-I (accession no. M18131)], resulting in plasmid ppolyMiLRΔBamHI. The final construct (pMiCMVGFP, Fig. 1) used for the generation of transgenic mice was created by inserting into the SpeI site of ppolyMiLRΔBamHI the 2.2-kb SpeI fragment from plasmid pBluescriptGFP, which contains a “humanized” green fluorescent protein (GFP) gene (from CLONTECH plasmid pHGFP-S65T) driven by the cytomegalovirus (CMV) promoter and followed by the simian virus 40 intervening sequence and polyadenylation signal. Plasmid pJGD/ILMi was constructed as follows: a 1-kb EcoRV/NotI fragment containing the Minos transposase cDNA was cloned into EcoRV/NotI of plasmid pJG-3 [the puromycin-resistant variant of pJG-1 (18)]. Plasmid pJGD/ILMi carries a CMV promoter upstream of the transposase cDNA, an intron with splice site and poly(A) from the human β-globin gene and the puromycin resistance gene driven by PGK promoter and followed by the poly(A) signal from the bovine GH gene and was used as the transposase source in transfections of embryonic fibroblasts.
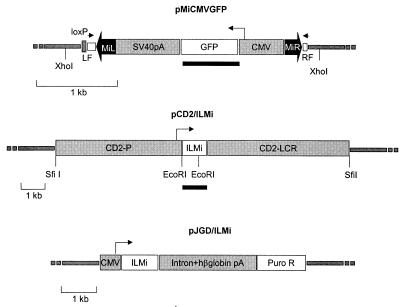
Figure 1.
Minos-derived vectors. Minos inverted terminal repeats are shown as thick, solid arrows. Open blocks outside these arrows indicate the sequences flanking the original Minos element in the D. hydei genome. Arrowheads indicate the positions of primers used to detect Minos excisions. Small arrows indicate the direction of transcription of the GFP and transposase genes. Solid bars represent fragments used as probes. All other features are described in Materials and Methods.
Generation of Transgenic Mice.
The transposase-expressing TM2 mouse line was generated by injecting the 12.5-kb SfiI fragment from the CD2/ILMi plasmid into CBA × C57 Bl/10 fertilized oocytes. Transgenic founder animals were identified by Southern blotting of DNA from tail biopsies, using the 1-kb transposase cDNA fragment as a probe and crossed with F1 CBA × C57 B1/10 mice to generate lines.
The transposon-carrying MCG line was constructed by injecting the 3.2-kb XhoI fragment from the pMiCMVGFP plasmid into FVB × FVB fertilized oocytes. Transgenic founder animals were identified by Southern blotting of DNA from tail biopsies, using GFP DNA as a probe.
Cell Culture and Transfection.
Pregnant females [day 13.5, from crosses between MCG heterozygous transgenic male and wild-type (wt) females] were killed, embryos were isolated, and part of the material was used for genotyping. The remaining embryonic tissue was minced by using a pair of scissors and immersed in a thin layer of F10/DMEM culture medium supplemented with 10% FCS and antibiotics. Two spontaneously immortalized mouse embryonic fibroblast (MEF) lines with MCG/+ genotype were obtained by subculturing of primary MEFs. They were stably transfected with 20 μg of plasmid pJGD/ILMi linearized with ScaI, using Lipofectin (GIBCO/BRL). Transfectants were selected on puromycin at a concentration of 1 μg/ml.
Northern Blot Hybridization.
Total RNA (15 μg) isolated (19) from kidney, thymus, and spleen was subjected to electrophoresis in a 1.2% agarose gel containing 15% formaldehyde. Northern blot analysis was performed as described (20).
PCR Analysis.
Genomic DNA from different tissues was isolated with the DNeasy Tissue Kit (Qiagen) according to the manufacturer's instructions.
PCRs were performed by using primers 11DML (5′-AAGTGTAAGTGCTTGAAATGC-3′) and GOUM67 (5′-GCATCAAATTGAGTTTTGCTC-3′). PCR conditions were as follows: 10 mM Tris⋅HCl (pH 8.8)/50 mM KCl/1.5 mM MgCl2/0.001% gelatin and 1.2 units Taq 2000 DNA Polymerase (Stratagene), 200 ng of template DNA, and 10 pmol of each primer per 25-μl reaction. Forty-three or 60 cycles of 30 sec at 94°C, 30 sec at 59°C, and 30 sec at 72°C were performed. PCR products were cloned into the PCRII TA cloning vector (Invitrogen) and were sequenced by using the T7 primer.
DNA Fluorescent in Situ Hybridization (FISH) Analysis.
Cells from minced thymus or spleen were cultured for 48 h in RPMI 1640 medium (GIBCO/BRL) supplemented with 9% FCS (GIBCO/BRL), 13.6% Hybridoma medium (GIBCO/BRL), 3.4 μg/ml lithium chloride (Merck), 7.2 μg/ml Con A (Sigma), 22.7 units/ml Heparin (Leo, Helsingborg, Sweden), 50 μM 2-mercaptoethanol, 25.4 μg/ml lipopolysaccharide (Sigma), 10 ng/ml IL-6 (PEPROTECH EC LTD). Chromosome preparations and FISH were carried out as described (21). The 737-bp SacI/NotI GFP fragment from the pMiCMVGFP construct was used as probe. The probe was labeled with Biotin (Boehringer Mannheim) and detected immunochemically with FITC. A telomeric probe for chromosome 14 (22) was labeled with digoxigenin (Boehringer Mannheim) and detected immunochemically with Texas Red.
Results and Discussion
Two transgenic mouse lines were generated to determine whether Minos can transpose in mouse tissues: one containing a Minos transposon and another containing the Minos transposase gene expressed in a tissue-specific manner. The transposon-carrying line (line MCG) contains a tandem array of a fragment consisting of a Minos transposon (MiCMVGFP, Fig. 1) containing the GFP gene under the control of the CMV promoter. The transposon was engineered such that almost all sequence internal to the inverted repeats has been replaced by the CMV/GFP cassette. Not containing the transposase-encoding gene, this transposon is nonautonomous, and can be mobilized only when a source of transposase is present. The transposase-expressing line (line TM2) contains a tandem array of a construct comprising the Minos transposase cDNA under the control of the human CD2 locus, consisting of the CD2 promoter and locus control region elements (pCD2/ILMi, Fig. 1). In transgenic mice, the human CD2 locus is transcribed at high levels in virtually all thymocytes as well as peripheral T cells (16).
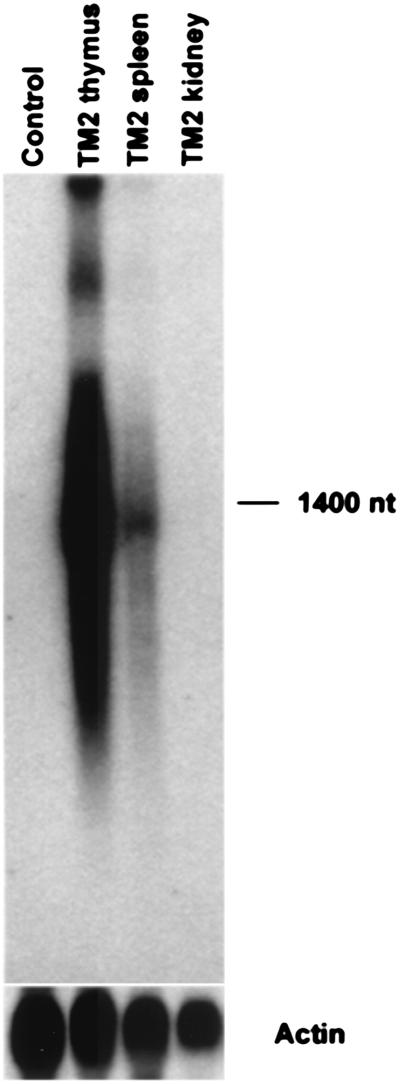
Heterozygous TM2/+ mice were tested for tissue-specific production of Minos transposase RNA by Northern blot analysis. As expected, Minos transposase mRNA was detected in thymus and spleen, the two organs with large numbers of T cells, but undetectable in other organs such as kidney (Fig. 2).
Figure 2.
Tissue-specific expression of Minos transposase in transgenic mice. Northern blot analysis of thymus, spleen, and kidney RNA isolated from TM2/+ mice (40-h exposure). Control RNA is from thymus of a nontransgenic mouse. The lower image shows the signal obtained on rehybridization of the same filter with a mouse actin probe (3-h exposure).
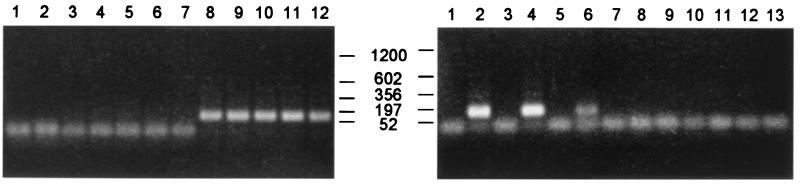
The first indication for active transposition by Minos transposase in mouse tissues was obtained by a sensitive PCR assay for transposon excision, using primers that hybridize to the nonmobile D. hydei sequences that flank the Minos transposon in our constructs (17) (see Fig. 1). In Drosophila cells, transposase-mediated excision of Minos is followed by repair of the chromatid, which usually leaves a characteristic 6-bp footprint (23). With the specific pair of primers used in our assay, this creates a diagnostic 167-bp PCR fragment (24). As shown in Fig. 3, the diagnostic band is present in tissues of double-transgenic (MCG/+ TM2/+) mice expressing the transposase, but not of MCG/+ mice not expressing transposase. The identity of the fragment was confirmed by Southern blot analysis by using a labeled DNA probe specific for the amplified sequence (data not shown). Excision is detectable mainly in thymus and spleen of the double transgenics; lower levels of excision are detectable in liver (Fig. 3). Very low levels of excision also can be detected in kidney, brain, and skeletal muscle after 15 additional cycles of amplification (data not shown). Low levels of expression of the human CD2 locus in liver and lung of transgenic mice has been documented (25). We therefore attribute the excision detected in tissues other than thymus and spleen to the presence of small numbers of T cells or to the expression of transposase in non-T cells of these tissues as a result of position effects.
Figure 3.
Transposase-dependent, tissue-specific excision of a Minos transposon in mice. Oligonucleotide primers flanking the transposon were used for PCR, and the products were analyzed by agarose gel electrophoresis. (Left) Transposase-dependent excision in the thymus. Lanes: 1, nontransgenic DNA; 2, TM2/+; 3–7, MCG/+; 8–12, MCG/+ TM2/+. (Right) Excision in various tissues of transposase-expressing mice. Lanes: 1, 3, 5, 7, 9, and 11, from MCG/+ mice; 2, 4, 6, 8, 10, and 12, from MCG/+ TM2/+ mice; 1 and 2, thymus; 3 and 4, spleen; 5 and 6, liver; 7 and 8, kidney; 9 and 10, brain; 11 and 12, muscle; 13, no DNA added. Amplification of the entire transposon is not detectable under the PCR conditions used. The lower band present in most lanes probably represents primer or primer extensions during the PCR.
In a separate experiment, the same PCR assay was used to detect Minos excision in cultured embryonic fibroblasts carrying the MCG transgene. Cells were transfected with a plasmid carrying the Minos transposase cDNA under CMV control (pJGD/ILMi, Fig. 1) and analyzed by the PCR excision assay. Excision products were detectable in transfected but not in nontransfected cells (data not shown). This result suggests that the transposon transgene is accessible to the Minos transposase not only in T cells.
To determine the nature of the excision events, PCR products from thymus and spleen of MCG/+ TM2/+ mice and from pJGD/ILMi-transfected embryonic fibroblasts were cloned and sequenced. The sequence left behind after Minos excision in Drosophila consists of the TA dinucleotide duplication that is created on Minos insertion, flanking the terminal 4 nt of the transposon (i.e., either a AcgagT or a ActcgT insertion in the TA target site). In the mouse excisions analyzed, the size and sequence of the footprints varied considerably (Fig. 4). Only 2 of the 32 footprints had the typical 6-bp sequence; the others contained extra nucleotides in addition to complete or partial versions of the typical footprint. Four events had 1–2 nt of the flanking D. hydei chromosomal sequence deleted. The differences in footprint structures observed between Drosophila and mouse may reflect the involvement of host factors in Minos excision and/or chromatid repair after excision.
Figure 4.
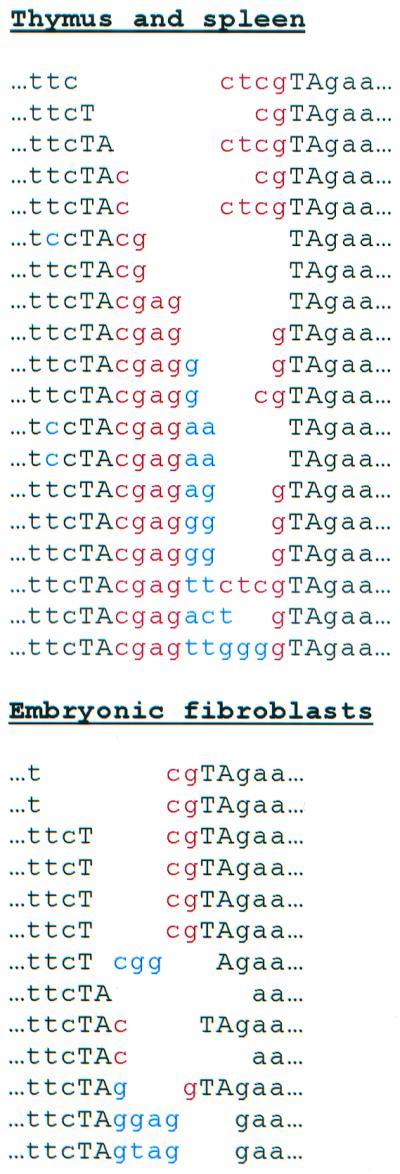
Footprints left behind at chromosomal sites after Minos excision. DNA was extracted from thymus and spleen of a double-transgenic mouse (Upper) or from an embryonic fibroblast cell line from a MCG/+ mouse after transfection with a transposase-expressing plasmid (Lower) and used as template for PCR with the flanking primers. PCR-amplified bands were cloned, and 32 clones (19 from thymus and spleen and 13 from fibroblast cells) were sequenced. TA is the target-site duplication. Nucleotides in red correspond to the ends of the transposon terminal repeats; nucleotides in blue are of unknown origin. The flanking nucleotides and TA repeats are aligned.
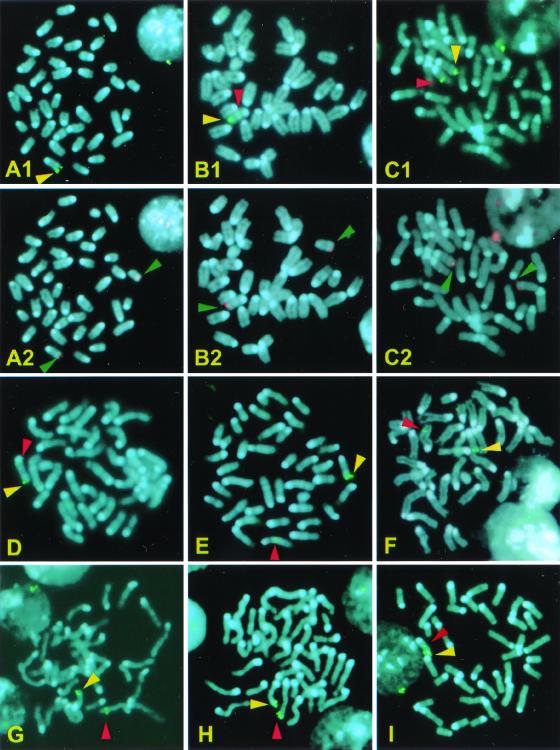
Detection of transposase-dependent excision in thymus and spleen suggested that transposition also may take place in these tissues. The detection of transposition events is not straightforward because every transposition event is unique, and, as a result, the tissue in which transposition has occurred will be a mosaic of cells with unique transpositions. Indeed, Southern blot analysis failed to show transposition events in the thymus of double transgenics (data not shown), indicating that, if such mosaics exist, they consist of small numbers of clonally related cells. Therefore, we used FISH in metaphase nuclei from the thymus and spleen to detect individual transposition events. A GFP fragment was used as a probe to detect relocalization of transposons into new chromosomal positions. The initial position of the array of transposons was at the tip of chromosome 14, at a position indistinguishable from the telomere, as shown by colocalization in metaphase and interphase chromosomes with a probe specific for telomeric sequences of chromosome 14 (Fig. 5 A1 and A2). A total of 3,114 metaphases from five MCG/+ TM2/+ mice were analyzed; 1,688 were from spleen and 1,426 were from thymus. Nineteen of these metaphases (11 from spleen and 8 from thymus) showed transposition. In addition to the signal at the tip of chromosome 14, pairs of dots were present in these metaphases on chromosomes other than 14 or on a new position on chromosome 14. Representative metaphases are shown in Fig. 5 B1– I. Morphological analysis of the chromosomes carrying new insertions showed that all events except one are independent from each other, i.e., they represent different transpositions. Analysis of the positive metaphases with a probe specific for the telomere of chromosome 14 indicated that transpositions do not involve translocation of telomeric material (Fig. 5 B1, B2, C1, and C2). As controls, 2,440 metaphases from thymus and spleens of five MCG/+ mice were screened; no transpositions were detectable in those samples.
Figure 5.
FISH analysis of Minos transpositions in thymus and spleen. Chromosomes were stained with 4′,6-diamidino-2-phenylindole. A1 and A2 are from the same MCG/+ metaphase nucleus, probed with a GFP and a telomere 14 specific probe, respectively. B–I are nuclei from thymus or spleen of MCG/+ TM2/+ mice. Nuclei in B1, C1, and D–I are probed with the GFP probe. B2 and C2 are the same nuclei as B1 and C1, respectively, probed with the telomere 14 specific probe. Yellow arrowheads indicate the original integration site of the transposon transgene, near the telomere of chromosome 14. Green arrowheads indicate the telomeres of chromosome 14. Red arrowheads indicate transposition events.
This work demonstrates that a transposase expressed from a transgene can mobilize a transposon to jump into new chromosomal sites in mammalian tissues. The observed efficiency of Minos transposition in mouse T cells was low compared with Minos transposition in human HeLa cells (15) and to SB transposition in mouse embryonic stem cells (8). Limits in detection of transposition events, lower transposition frequencies because of the particular expression cassettes and cell type chosen (T cells), or a combination of these factors may explain these differences. Detection of transposition by FISH can lead to underestimation of transposition frequencies if a high proportion of transpositions occur near the original site of the element. Such “local insertions,” within a few kilobases from the original site, are quite frequent in Drosophila with P elements (26) and in embryonic stem cells with SB (8). Low transposition frequencies also could result from both low and very high levels of transposase protein (high transposase levels can cause reexcision and loss of new transpositions), decreased accessibility of the transposon, the requirement of host-encoded factors necessary for transposition, or a combination of the above. Levels of transposase protein can be optimized by modifying codon usage and can be controlled by using an inducible promoter. Localization of the transposon in open chromatin, for example, by introducing a highly transcribed gene between the inverted repeats and/or mobilizing a transposon that is inserted in a highly transcribed gene by recombination in embryonic stem cells, is a strategy to increase transposon accessibility. Once transposition efficiency is optimized, Minos-based transposons would be powerful tools for gene manipulation and functional genomic analysis in the mouse. Combined with the ability to express transposase in mouse tissues in a regulatable manner by using available genetic engineering technologies (27), Minos would be a tool for insertional gene inactivation and gene tagging, enhancer trapping, and exon trapping. In addition, the ability to induce Minos excision by regulated transposase expression would allow genetic manipulation of targeted genes, such as regulatable reversion of gene disruptions.
Acknowledgments
We thank E. M. E. Smit for providing the chromosome 14 probe and help with the FISH analysis, K. Kourouniotis, T. de Wit, and A. Babaratsas for expert technical help, and S. Oehler for critically reading the manuscript. This work was supported by European Union Grant BIO4-CT98-0203 (to C.M., F.G., and C.S.). L.Z. was a recipient of a European Molecular Biology Organization Short Term Fellowship.
Abbreviations
- SB
Sleeping Beauty
- FISH
fluorescent in situ hybridization
- GFP
green fluorescent protein
Note Added in Proof.
After this manuscript was submitted, Fisher et al. (28) reported regulated transposition of Sleeping Beauty, a transposable element from fish, in the male mouse germ line.
References
- 1.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 2.Zwaal R R, Broeks A, van Meurs J, Groenen J T, Plasterk R H. Proc Natl Acad Sci USA. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walbot V. Curr Opin Plant Biol. 2000;3:103–107. doi: 10.1016/s1369-5266(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 4.Raz E, van Luenen H G, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 5.Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H. Nat Biotechnol. 1998;16:1050–1053. doi: 10.1038/3497. [DOI] [PubMed] [Google Scholar]
- 6.Schouten G J, Van Luenen H G, Verra N C, Valerio D, Plasterk R H. Nucleic Acids Res. 1998;12:3013–3017. doi: 10.1093/nar/26.12.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivics Z, Hackett P B, Plasterk R H, Izsvak Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 8.Luo G, Ivics Z, Izsvak Z, Bradley A. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yant S R, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay M. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 10.Franz G, Savakis C. Nucleic Acids Res. 1991;19:6646. doi: 10.1093/nar/19.23.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loukeris T G, Arca B, Livadaras I, Dialektaki G, Savakis C. Proc Natl Acad Sci USA. 1995;92:9485–9489. doi: 10.1073/pnas.92.21.9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 13.Catteruccia F, Nolan T, Loukeris T G, Blass C, Savakis C, Kafatos F C, Crisanti A. Nature (London) 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu K, Kamba M, Sonobe H, Kanda T, Klinakis A G, Savakis C, Tamura T. Insect Mol Biol. 2000;9:277–281. doi: 10.1046/j.1365-2583.2000.00182.x. [DOI] [PubMed] [Google Scholar]
- 15.Klinakis A G, Zagoraiou L, Vassilatis D K, Savakis C. EMBO Rep. 2000;1:416–421. doi: 10.1093/embo-reports/kvd089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. J Immunol Methods. 2000;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 17.Klinakis A G, Loukeris T G, Pavlopoulos A, Savakis C. Insect Mol Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
- 18.Drabek D, Guy J, Craig R, Grosveld F. Gene Ther. 1997;4:93–100. doi: 10.1038/sj.gt.3300366. [DOI] [PubMed] [Google Scholar]
- 19.Chomozynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Mulder M P, Wilke M, Langeveld A, Wilming L G, Hagemeijer A, van Drunen E, Zwarthoff E C, Riegman P H, Deelen W H, van den Ouweland A M, et al. Hum Genet. 1995;96:133–141. doi: 10.1007/BF00207368. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Mohapatra G, Miller J, Hanahan D, Lander E, Gold P, Pinkel D, Gray J. Genomics. 1997;45:42–47. doi: 10.1006/geno.1997.4904. [DOI] [PubMed] [Google Scholar]
- 23.Arca B, Zabalou S, Loukeris T G, Savakis C. Genetics. 1997;145:267–279. doi: 10.1093/genetics/145.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catteruccia F, Nolan T, Blass C, Muller H M, Crisanti A, Kafatos F C, Loukeris T G. Proc Natl Acad Sci USA. 2000;97:2157–2162. doi: 10.1073/pnas.040568397. . (First Published February 11, 2000; 10.1073/pnas.040568397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang G, Wotton D, Owen M J, Sewell W A, Brown M H, Mason D Y, Crumpton M J, Kioussis D. EMBO J. 1988;6:1675–1682. doi: 10.1002/j.1460-2075.1988.tb02995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Spradling A C. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 28.Fisher S E J, Wienholds E, Plasterk R H A. Proc Natl Acad Sci USA. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. . (First Published May 29, 2001; 10.1073/pnas.121569298) [DOI] [PMC free article] [PubMed] [Google Scholar]