Abstract
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate the vast majority of fast synaptic transmission in the nervous system. When the iGluR ion channel is in the open or conducting conformation, it is non-selective for monovalent cations, driving membrane excitation. Often the channel is also permeable to Ca2+. This process of Ca2+ permeation and its physiological and pathological consequences depend strongly on the specific iGluR subtype as well as the specific subunits in the oligomeric complex. Recent evidence has highlighted additional levels of diversity to this process including a dependence on specific auxiliary subunits in non-NMDARs and post-translational modifications in NMDARs. Various de novo missense mutations associated with neurological disease in NMDAR subunits have been identified in regions critical to Ca2+ influx. These features highlight the dynamics of Ca2+ influx mediated by iGluRs and its critical role in synaptic physiology and pathology.
Keywords: AMPA receptors, kainate receptors, NMDA receptors, Ca2+ permeability, auxiliary subunits, post-translational modification
1. Introduction
We have learned in school, or should have learned, that a fundamental determinant of ion channel function is the ions it allows to pass when in the open or conducting state [1]. Consider ionotropic glutamate receptors (iGluRs), the ligand-gated ion channels that mediate fast excitatory synaptic transmission in the brain. Their ion channel is cation non-selective, being about equally permeable to both K+ and Na+ ions [2]. At rest, activation or opening of iGluRs is excitatory because of the strong driving force for Na+ influx. The iGluR ion channel evolutionarily originated from a two transmembrane K+ channel [3]. If iGluRs retained their ancient permeability properties of being selectively permeable to K+ rather than being cation non-selective, the contribution of iGluRs to membrane physiology would be the exact opposite: K+-selective iGluRs would be inhibitory rather than excitatory.
Being non-selective to monovalent cations mediates the role of iGluRs in brain excitation. iGluRs also often affect cell-to-cell signaling independent of direct membrane excitation. Some iGluR subtypes can function as metabotropic receptors, acting independent of ion flux [4]. In many instances, the iGluR ion channel is permeable not only to monovalent cations but also Ca2+. Although the charge associated with Ca2+ may contribute to excitation, Ca2+ predominantly acts as a biochemical signaling molecule. Indeed, this Ca2+ signaling often dominates how we think of iGluR function, contributing to their role in synaptic dynamics, neuronal development, and cellular pathology [5,6]. After reviewing some basic features of Ca2+ permeation, I will focus on recent efforts that have further highlighted the regulation and dynamic role of Ca2+ permeation in iGluRs.
2. The permeation pathway in iGluRs
There are three iGluR subtypes: AMPA (AMPAR), kainate (KAR), and NMDA (NMDARs) receptors, which are composed of various subunits (Figure 1). These receptors share a similar overall structure [7–9], including the topology of the pore-forming or ion permeation pathway.
Figure 1. Structure of the permeation pathway and relative Ca2+ permeability in iGluRs.
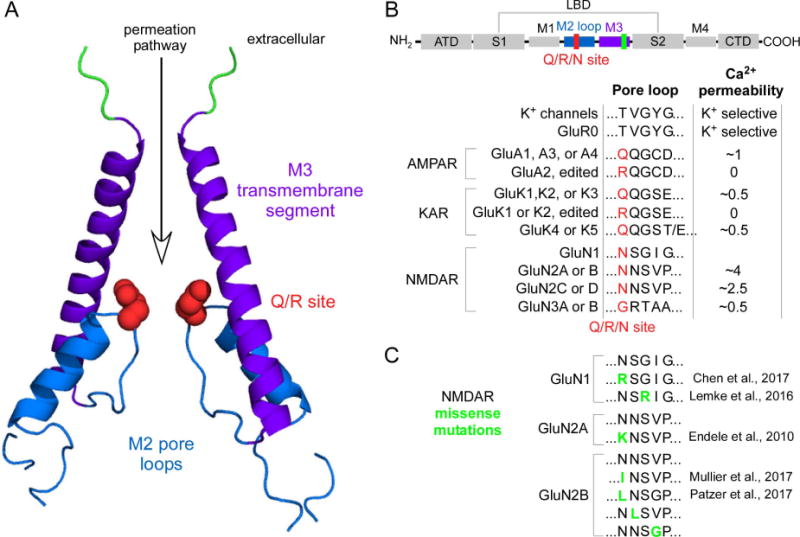
(A) Open state structure of an AMPAR (GluA2) (PDB, 5WEO) [15]. The permeation pathway in iGluRs is mainly defined by the M3 transmembrane segment and the M2 pore loop. At the tip of M2, and contributing to the channel’s narrow constriction, resides the Q/R/N site (red)(Q/R site, non-NMDARs; N site, NMDARs), a key determinant of ion permeation in iGluRs. In NMDARs, an additional site in the GluN1 M3 segment, the DRPEER motif, also contributes to Ca2+ influx [12]. Homologous residues in the AMPAR structure are highlighted in green.
(B) Residues at and around the Q/R/N site in various iGluR subtypes and subunits and associated Ca2+ permeability. Top cartoon, domains in iGluR subunits: the extracellular amino-terminal (ATD) and ligand-binding (LBD) domains; the transmembrane domain (TMD), transmembrane segments M1, M3, and M4 and an M2 pore loop; and the intracellular C-terminal (CTD) domain. Single letter amino acid code is shown for the N site (in red) and four adjacent positions located to the C-terminal side.
Ca2+ permeabilities (PCa/Pmonovalent) are approximate and shown just for illustration but are primarily based on fractional Ca2+ currents (see Table 18 in Traynelis et al. (2010), where specific values and original references can be found. Additional data was taken from [41]). GluR0 is a prokaryotic iGluR that is K+ selective [3].
iGluRs are tetrameric complexes composed of the same (monoheteromeric) or two (diheteromeric) or 3 (triheteromeric) different subunits. AMPAR (GluA1-A4) and KAR (GluK1-GluK3) subunits can form monoheteromers but are typically diheteromeric assemblies in native tissue. KAR GluK4 and GluK5 form di- or tri-heteromeric complexes with the other KAR subunits. NMDAR are obligate di- or tri-heteromers, being composed of GluN1 and some combination of GluN2 (GluN2A-2D) or GluN3 (GluN3A-3B).
(C) de novo missense mutations (residues in green) near the tip of the M2 loop. See original publications for clinical phenotype.
In iGluRs, the permeation pathway is largely formed by the M3 transmembrane segment and the intracellular M2 pore loop (Figure 1A). This arrangement is like that of a K+-selective channel, but is inverted in the membrane with the pore loop on the intracellular rather than the extracellular side of the membrane. Within the M2 loop resides a key site for ion permeation, the Q/R/N site (Figures 1A & 1B)(the Q/R site for non-NMDAR subunits and the N site for NMDAR subunits)[10]. The Q/R/N site is located near the tip of the M2 loop and contributes to the narrow constriction in the pore where waters of hydration of permeant ions are stripped for recognition [1]. In non-NMDARs, the Q/R site is edited, at the mRNA level, to encode the positively charged arginine (R) in GluA2 and GluK1 and GluK2, which renders the channel Ca2+ impermeable (Figure 1B)[10]. Because of the dominance of the Q/R site in terms of pore properties (single channel current, channel block and Ca2+ permeation), AMPARs are distinguished into GluA2-containing (Ca2+-impermeable AMPARs) and GluA2-lacking (GluA1, GluA3, GluA4, or Ca2+-permeable AMPARs) AMPARs. Although KARs (GluK1-K2) are also edited, this editing is less complete [11]. For KARs, the role of Ca2+ influx, or the lack of it for edited subunits, in synaptic development and physiology remains poorly defined.
Di-heteromeric assemblies of NMDARs, containing GluN1 and some combination of GluN2 subunits, have a higher selectivity for Ca2+ than non-NMDARs (Figure 1B). This difference is due only in part to the Q/R/N site [2]. One determinant of this higher selectivity is the DRPEER motif in the GluN1 M3 segment [9,12]. Although elements that contribute to ion permeation in NMDARs are largely known, the mechanism of this process remains unclear: it follows Goldman-Hodgkin-Katz (GHK) assumptions at physiological Ca2+ [13], but deviates at lower and higher concentrations [2]. Structures of iGluRs especially at higher resolutions and in the open state and molecular dynamics simulations will provide new insights into the mechanistic basis of Ca2+ permeation in iGluRs.
One of the hallmarks of GluN2-containing NMDARs is a strong voltage-dependent block of the pore by extracellular Mg2+ [10]. One question that has perplexed biophysicists is how the pore of NMDARs can distinguish between Ca2+, which is permeable, and Mg2+, which is largely impermeable. Determinants of both processes, Ca2+ permeation and Mg2+ block, are shared in the M2 loop. Part of the distinction between Ca2+ and Mg2+ must involve differences in hydration energy [1]: Mg2+ because of its small size and condensed charge holds on to its water of hydration tightly, whereas the larger Ca2+ holds onto these waters much less tightly. Recent efforts involving molecular dynamic simulations have started to define how elements in the M2 loop mechanistically distinguish between Ca2+ and Mg2+ [14*]. In addition, high resolution structures of the M2 loop in the open state, which are now available for AMPARs [15**], will further allow additional mechanistic insights into how the narrow constriction in NMDAR distinguishes between these two critical physiological ions.
An additional element that remains uncertain is how extracellular permeant ions, Na+ and Ca2+ in particular, access the central permeation pathway defined by the M3 transmembrane segment (Figure 1A). iGluRs are highly modular proteins (upper panel, Figure 1B). The ligand-binding domain (LBD) is positioned on the extracellular end of the ion channel and is connected to the ion channel-forming transmembrane domain (TMD) by a set of short polypeptide linkers, the LBD-TMD linkers. These linkers are highly dynamic during pore opening and contain numerous charged side chains [16]. They presumably represent pathways or portals for permeant ions to access the central permeation pathway. Nevertheless, how permeant ions cross these portals and their impact on selectivity and single channel conductance levels are unknown. This question is of interest since these linkers are promising sites for modulation of receptor function in the clinic [17].
The NMDAR GluN3 subunits, GluN3A & GluN3B, reduce Ca2+ influx when part of a di-heteromeric (GluN1 & GluN3) or triheteromeric (GluN1/GluN2/GluN3) complex [18,19]. This difference presumably reflects the composition of the residues occupying the N site (Figure 1B), though this needs to be directly tested. To do so will require approaches where the subunit composition of NMDARs is well-defined [20].
3. Subunit composition: New subunits in the mix
A major determinant of Ca2+ permeability in iGluRs is subunit composition (Figure 1B). The AMPAR subunits GluA1-A4 and KAR subunits GluK1–5 are pore-forming. Native AMPARs and KARs are most likely associated with an assortment of auxiliary subunits, non-pore forming subunits that modulate the cell biology and/or functional properties of the pore-forming subunits [21,22]. Stargazin (γ−2) was the first characterized auxiliary subunit, but these subunits are now expansive, encompassing transmembrane AMPA receptor regulatory proteins (TARPs)(γ-2, γ-3, γ-4, γ-5, γ-7, γ-8), cornichons (CNIH-1, -2, -3), GSG1L, CKAMP-44 (CKAMP-44, -39, -52, -59) and Neto (Neto1 & Neto2). These auxiliary subunits show cell-type and developmental specific patterns of expression and add many layers of diversity to non-NMDARs, altering receptor trafficking and localization, gating, and properties of the permeation pathway including polyamine block, single channel conductance, and, notable here, Ca2+ permeation.
Of the tested auxiliary subunits, stargazin (γ−2 and γ-4) and cornichon (CNIH-2) increase Ca2+ permeation in AMPARs by about two-fold [23,24]. In contrast, CNIH-1 has no effect and GSG1L decreases permeability by about 50% [24,25]. These properties parallel those for the general effect of these auxiliary subunits on receptor function: γ-2 and CNIH-2 tend to increase receptor function including decreasing the extent of desensitization and polyamine block and increasing single channel conductance, whereas GSG1L has the opposite effects. This raises two interesting points. The first is physiological: why does nature incorporate auxiliary subunits that either increase (γ-2, CNIH-2) or decrease (GSGL1) all receptor functions? Ca2+ influx through Ca2+-permeable AMPARs has a strong effect on synaptic physiology, but the parallel effects on receptor function will increase or decrease both the electrical as well as the biochemical function of Ca2+-permeable AMPARs.
The second question is a structure-function one: how do these various auxiliary subunits impact the permeation pathway? High-resolution cryo-EM structures of auxiliary subunits in complex with AMPARs highlight the close contact between membrane spanning domains [26–29] and give guidance but do not reveal mechanistic features. The effect of stargazin on receptor function depends on the residue occupying the Q/R site [30], suggesting that auxiliary subunits might alter the local structure of the narrow constriction, though they do not change pore size [31]. On the other hand, the intracellular C-terminus of CNIH-2 affects pore properties [24], possibly by directly interacting with intracellular determinants of the pore. Nevertheless, how auxiliary subunits mechanistically alter pore properties, especially Ca2+ permeation, remains unclear.
The most prominent auxiliary subunits in KARs are Netos (Neto1 & Neto2). Although Netos can alter pore block, they have no apparent effect on Ca2+ permeability [32]. Interestingly, Netos apparently reduce polyamine block by enhancing its permeation apparently by changing the structure of the pore loop [33]. What is needed to fully clarify this mechanism of action are high resolution structures of KAR-Neto complexes.
4. Dynamic regulation of Ca2+ permeation in NMDARs at synapses
Given the importance of Ca2+ influx to synaptic physiology and dynamics, nature finds way to increase functional diversity. Indeed, the magnitude of Ca2+ permeation in NMDARs can be regulated in a use-dependent manner at synapses [34,35]. This dynamic action is associated with the GluN2B subunit. At least one of the pathways for it occurs via protein kinase A modulation [34] of Ser1166 in the GluN2B C-terminal domain (CTD) [36*]. This dynamic action has clear biological effects impacting synaptic plasticity, learning and memory, and emotional responses to stress [34–36*].
How phosphorylation of the intracellular CTD modulates pore properties of the receptor is completely unknown. Functional studies of the CTD have been hindered by a lack of structural information, as the CTD has been excluded from all available iGluR structures [37]. Further complicating a structural understanding is that the GluN2B CTD contains significant regions of intrinsic disorder [38,39**]. The CTD in NMDARs interacts with lipids [40*], which might in some way disrupt the overall structure of the transmembrane domain, where interactions among pore forming elements are important to permeation properties [41]. Alternatively, the CTD is directly connected to the M4 transmembrane segment, which has strong effects on receptor function [42]. Conformational changes in the CTD, induced by post-translation modifications, might alter the orientation of M4 in the membrane, which in turn alters pore properties. In any case, how the conformation of the CTD, driven by post-translational modifications, affects receptor function including pore properties and receptor gating is an area of intense investigation [43]. To define how the CTD impacts receptor function will require integration of various approaches including electrophysiological and optical approaches as well as single molecule techniques that allow details of the conformation of the CTD under various conditions to be ascertained.
5. De novo and inherited missense mutations in NMDAR in neurological disorders
The role of iGluRs in neurological diseases has a long history. Recently, an ever expanding list of de novo and inherited missense mutations in NMDAR and AMPAR subunits have been identified in patients with neurological disorders including epileptic encephalopathy, intellectual disability, autism, schizophrenia, and motor dysfunction [44–46]. Functionally, most of these mutations have not been characterized in detail, though when characterized, they often have clear effects on receptor function [47–49].
Missense mutations have been identified in the M2 pore loop (Figure 1C) and, when tested, affect Mg2+ block [50–54]. In the one instance where tested, which occurred for the positively charged lysine (K) at the N site in GluN2A (Figure 1C), Ca2+ permeation was strongly affected [50]. Given the central role of M2 in Ca2+ permeation, other M2 loop missense mutations presumably also alter Ca2+ though this remains untested. Understanding how these missense mutations might alter Ca2+ influx has strong implications for their clinical pathology as well as the mechanism of the permeation process itself. As such, it remains an area of intense interest.
6. Concluding remarks
While the Ca2+ permeability of iGluRs and its biological importance was discovered some time ago, it remains an important topic of study. Many mechanistic questions remain such as how the conformational status of the CTD impacts pore properties. Further, Ca2+ permeation is not a static property solely dictated by the subunit composition of traditional subunits but depends also on auxiliary subunits, and we do not understand how these changes in Ca2+ permeation induced by different auxiliary subunits impact synaptic dynamics including plasticity. Finally, how missense mutations in the M2 loop affect Ca2+ permeation and clinical pathologies has not been well established.
Acknowledgments
This work was supported by a NIH RO1 grant from NINDS (NS088479) (LPW). I thank Drs. Stuart Cull-Candy and Geoffrey Swanson, and Johansen Amin and Kelvin Chan for helpful discussions and/or comments on the manuscript.
Abbreviations
- AMPAR
AMPA receptors
- KAR
kainate receptors
- NMDAR
NMDA receptors
- ATD
amino-terminal domain
- LBD
ligand-binding domain
- TMD
transmembrane domain
- CTD
C-terminal domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
One star, of special interest
Two stars, of outstanding interest
- 1.Hille B. Ion channels of excitable membranes. Sinauer Associates, Inc; MA: 2001. [Google Scholar]
- 2.Jatzke C, Watanabe J, Wollmuth LP. Voltage and concentration dependence of Ca2+ permeability in recombinant glutamate receptor subtypes. J Physiol. 2002:538, 25–39. doi: 10.1113/jphysiol.2001.012897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999:402, 817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- 4.Valbuena S, Lerma J. Non-canonical Signaling, the Hidden Life of Ligand-Gated Ion Channels. Neuron. 2016;92(2):316–329. doi: 10.1016/j.neuron.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14(6):383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty A, Murphy S, Coleman N. The Role of NMDA Receptors in Neural Stem Cell Proliferation and Differentiation. Stem Cells Dev. 2017;26(11):798–807. doi: 10.1089/scd.2016.0325. [DOI] [PubMed] [Google Scholar]
- 7.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511(7508):191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344(6187):992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 2011;34(3):154–163. doi: 10.1016/j.tins.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe J, Beck C, Kuner T, Premkumar L, Wollmuth LP. DRPEER: A motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci. 2002:22, 10209–10216. doi: 10.1523/JNEUROSCI.22-23-10209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneggenburger R. Simultaneous measurement of Ca2+ influx and reversal potentials in recombinant N-methyl-D-aspartate receptor channels. Biophys J. 1996:70, 2165–2174. doi: 10.1016/S0006-3495(96)79782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Mesbahi-Vasey S, Veras L, Yonkunas M, Johnson JW, Kurnikova MG. All atom NMDA receptor transmembrane domain model development and simulations in lipid bilayers and water. PLoS One. 2017;12(6):e0177686. doi: 10.1371/journal.pone.0177686. One of the first efforts to define the molecular interaction of Mg2+ with residues at the tip of the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 2017 doi: 10.1038/nature23479. The first open state structure of the ion channel of any iGluR. Will stimulate new studies of permeation in iGluRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30(35):11792–11804. doi: 10.1523/JNEUROSCI.5382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang TM, Brown BM, Deng L, Sellers BD, Lupardus PJ, Wallweber HJA, Gustafson A, Wong E, Volgraf M, Schwarz JB, Hackos DH, et al. A novel NMDA receptor positive allosteric modulator that acts via the transmembrane domain. Neuropharmacology. 2017:121, 204–218. doi: 10.1016/j.neuropharm.2017.04.041. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci. 2001:21, 1228–1237. doi: 10.1523/JNEUROSCI.21-04-01228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Otano I, Larsen RS, Wesseling JF. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci. 2016;17(10):623–635. doi: 10.1038/nrn.2016.92. [DOI] [PubMed] [Google Scholar]
- 20.Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81(5):1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70(2):178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greger IH, Watson JF, Cull-Candy SG. Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins. Neuron. 2017;94(4):713–730. doi: 10.1016/j.neuron.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Kott S, Sager C, Tapken D, Werner M, Hollmann M. Comparative analysis of the pharmacology of GluR1 in complex with transmembrane AMPA receptor regulatory proteins gamma2, gamma3, gamma4, and gamma8. Neuroscience. 2009;158(1):78–88. doi: 10.1016/j.neuroscience.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 24.Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M, Cull-Candy SG. Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci. 2012;32(29):9796–9804. doi: 10.1523/JNEUROSCI.0345-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGee TP, Bats C, Farrant M, Cull-Candy SG. Auxiliary Subunit GSG1L Acts to Suppress Calcium-Permeable AMPA Receptor Function. J Neurosci. 2015;35(49):16171–16179. doi: 10.1523/JNEUROSCI.2152-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 2016;353(6294):83–86. doi: 10.1126/science.aaf8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2017;94(3):569–580 e565. doi: 10.1016/j.neuron.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E. Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 2016;536(7614):108–111. doi: 10.1038/nature18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Zhao Y, Wang Y, Shekhar M, Tajkhorshid E, Gouaux E. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 2017;170(6):1234–1246 e1214. doi: 10.1016/j.cell.2017.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korber C, Werner M, Hoffmann J, Sager C, Tietze M, Schmid SM, Kott S, Hollmann M. Stargazin interaction with alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) receptors is critically dependent on the amino acid at the narrow constriction of the ion channel. J Biol Chem. 2007;282(26):18758–18766. doi: 10.1074/jbc.M611182200. [DOI] [PubMed] [Google Scholar]
- 31.Soto D, Coombs ID, Gratacos-Batlle E, Farrant M, Cull-Candy SG. Molecular mechanisms contributing to TARP regulation of channel conductance and polyamine block of calcium-permeable AMPA receptors. J Neurosci. 2014;34(35):11673–11683. doi: 10.1523/JNEUROSCI.0383-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher JL, Mott DD. The auxiliary subunits Neto1 and Neto2 reduce voltage-dependent inhibition of recombinant kainate receptors. J Neurosci. 2012;32(37):12928–12933. doi: 10.1523/JNEUROSCI.2211-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown PM, Aurousseau MR, Musgaard M, Biggin PC, Bowie D. Kainate receptor pore-forming and auxiliary subunits regulate channel block by a novel mechanism. J Physiol. 2016;594(7):1821–1840. doi: 10.1113/JP271690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skeberdis VA, Chevaleyre V, Lau CG, Goldberg JH, Pettit DL, Suadicani SO, Lin Y, Bennett MV, Yuste R, Castillo PE, Zukin RS. Protein kinase A regulates calcium permeability of NMDA receptors. Nat Neurosci. 2006:9, 501–510. doi: 10.1038/nn1664. Epub 2006 Mar 2012. [DOI] [PubMed] [Google Scholar]
- 35.Sobczyk A, Svoboda K. Activity-dependent plasticity of the NMDA-receptor fractional Ca2+ current. Neuron. 2007;53(1):17–24. doi: 10.1016/j.neuron.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 36*.Murphy JA, Stein IS, Lau CG, Peixoto RT, Aman TK, Kaneko N, Aromolaran K, Saulnier JL, Popescu GK, Sabatini BL, Hell JW, et al. Phosphorylation of Ser1166 on GluN2B by PKA is critical to synaptic NMDA receptor function and Ca2+ signaling in spines. J Neurosci. 2014;34(3):869–879. doi: 10.1523/JNEUROSCI.4538-13.2014. A detailed characterization of both the molecular action as well as physiological outcome of changes in Ca2+ permeation due to post-translational modifications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou HX, Wollmuth LP. Advancing NMDA Receptor Physiology by Integrating Multiple Approaches. Trends Neurosci. 2017;40(3):129–137. doi: 10.1016/j.tins.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19(4):566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Choi UB, Kazi R, Stenzoski N, Wollmuth LP, Uversky VN, Bowen ME. Modulating the intrinsic disorder in the cytoplasmic domain alters the biological activity of the N-methyl-D-aspartate-sensitive glutamate receptor. J Biol Chem. 2013;288(31):22506–22515. doi: 10.1074/jbc.M113.477810. One of the first efforts to define the structural changes induced by phosphorylation of the GluN2B C-terminal domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Wilding TJ, Lopez MN, Huettner JE. Chimeric Glutamate Receptor Subunits Reveal the Transmembrane Domain Is Sufficient for NMDA Receptor Pore Properties but Some Positive Allosteric Modulators Require Additional Domains. J Neurosci. 2016;36(34):8815–8825. doi: 10.1523/JNEUROSCI.0345-16.2016. Taking advantage of chimeras, the authors identify a key structural motif in the C-terminal domain of NMDARs that interacts with the lipid bilayer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegler Retchless B, Gao W, Johnson JW. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci. 2012;15(3):406–413. S401–402. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amin JB, Salussolia CL, Chan K, Regan MC, Dai J, Zhou HX, Furukawa H, Bowen ME, Wollmuth LP. Divergent roles of a peripheral transmembrane segment in AMPA and NMDA receptors. J Gen Physiol. 2017;149(6):661–680. doi: 10.1085/jgp.201711762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zachariassen LG, Katchan L, Jensen AG, Pickering DS, Plested AJ, Kristensen AS. Structural rearrangement of the intracellular domains during AMPA receptor activation. Proc Natl Acad Sci U S A. 2016;113(27):E3950–3959. doi: 10.1073/pnas.1601747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol. 2015:20, 73–82. doi: 10.1016/j.coph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Hardingham GE, Do KQ. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat Rev Neurosci. 2016;17(2):125–134. doi: 10.1038/nrn.2015.19. [DOI] [PubMed] [Google Scholar]
- 46.Geisheker MR, Heymann G, Wang T, Coe BP, Turner TN, Stessman HAF, Hoekzema K, Kvarnung M, Shaw M, Friend K, Liebelt J, et al. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat Neurosci. 2017;20(8):1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, Holloman CM, Golas G, Adams DR, Boerkoel CF, Gahl WA, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun. 2014;5:3251. doi: 10.1038/ncomms4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. Functional Evaluation of a De Novo GRIN2A Mutation Identified in a Patient with Profound Global Developmental Delay and Refractory Epilepsy. Mol Pharmacol. 2017;91(4):317–330. doi: 10.1124/mol.116.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, Su Z, et al. Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 2017;13(1):e1006536. doi: 10.1371/journal.pgen.1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortum F, Fritsch A, Pientka FK, Hellenbroich Y, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nature genetics. 2010;42(11):1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 51.Lemke JR, Geider K, Helbig KL, Heyne HO, Schutz H, Hentschel J, Courage C, Depienne C, Nava C, Heron D, Moller RS, et al. Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology. 2016;86(23):2171–2178. doi: 10.1212/WNL.0000000000002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Shieh C, Swanger SA, Tankovic A, Au M, McGuire M, Tagliati M, Graham JM, Madan-Khetarpal S, Traynelis SF, Yuan H, et al. GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J Hum Genet. 2017 doi: 10.1038/jhg.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullier B, Wolff C, Sands ZA, Ghisdal P, Muglia P, Kaminski RM, Andre VM. GRIN2B gain of function mutations are sensitive to radiprodil, a negative allosteric modulator of GluN2B-containing NMDA receptors. Neuropharmacology. 2017:123, 322–331. doi: 10.1016/j.neuropharm.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 54.Platzer K, Yuan H, Schutz H, Winschel A, Chen W, Hu C, Kusumoto H, Heyne HO, Helbig KL, Tang S, Willing MC, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet. 2017;54(7):460–470. doi: 10.1136/jmedgenet-2016-104509. [DOI] [PMC free article] [PubMed] [Google Scholar]


