Summary
Basal cells (BCs) are p63-expressing multipotent progenitors of skin, tracheoesophageal and urinary tracts. p63 is abundant in developing airways, however, it remains largely unclear how embryonic p63+ cells contribute to the developing and postnatal respiratory tract epithelium, and ultimately how they relate to adult BCs. Using lineage-tracing and functional approaches in vivo, we show that p63+ cells arising from the lung primordium are initially multipotent progenitors of airway and alveolar lineages, but later become restricted proximally to generate the tracheal adult stem cell pool. In intrapulmonary airways these cells are maintained immature to adulthood in bronchi, establishing a rare p63+Krt5− progenitor cell population that responds to H1N1 virus-induced severe injury. Intriguingly, this pool includes a CC10 lineage-labeled p63+Krt5− cell subpopulation required for a full H1N1-response. These data elucidates key aspects in the establishment of regionally distinct adult stem cell pools in the respiratory system, potentially with relevance to other organs.
eTOC Blurb
Yang et al. show that embryonic p63+ cells are initially multipotent progenitors of airways and alveoli. Later, however, they become proximally restricted to generate tracheal basal cells and an intrapulmonary p63+Krt5− progenitor pool that is maintained immature to adulthood. This pool contains p63+CC10Lineage+ cells and mediates H1N1 virus-induced pathological remodeling.

Introduction
Basal cells (BCs) are multipotent tissue-specific stem cells of a variety of organs, including skin, esophagus, olfactory and airway epithelia. In the respiratory tract of humans, BCs are distributed throughout the pseudostratified epithelium from the trachea to bronchioles, but in mice they are restricted to trachea and extrapulmonary airways (collectively referred here as trachea) (Rock et al., 2010). Mouse models of injury-repair demonstrate the BCs’ roles in maintaining local stem cell pools and the differentiated cell types of the adult tracheal epithelium (Rock et al., 2009). These models also reveal these cells as highly heterogeneous, appearing ectopically as part of the repair/remodeling process; BC-like cells can be found in the alveolar space after severe damage by Bleomycin or H1N1 (Influenza-A) infection (Kumar et al., 2011). BCs are broadly identified by expression of intermediate filaments (cytokeratins Krt5, Krt14) and Trp63 (transformation-related protein 63, hereafter p63), a p53 family member crucial for BC identity (Yang et al., 1999). p63 null mice lack BCs and die at birth with multiple abnormalities, including the lung (Yang et al., 1999; Daniely et al., 2004; Romano et al., 2012). In embryonic murine airways p63 expression has been reported in the pseudostratified epithelium throughout development (Que et al., 2007; Bilodeau et al., 2014). Nevertheless, p63-expressing cells have not yet acquired all features of mature BCs prenatally. Thus, it remains unclear what distinguishes them from the other progenitors when airways are forming and how they contribute to the stem-cell pool and the luminal compartment of airways in development, adulthood and in response to severe injury.
Here we combine lineage tracing and functional genetic analysis in vivo to address this issue. We show that the BC pool of the adult trachea is built largely prenatally from p63+ lineage-labeled progenitors that are initially multipotent to generate all the airway and alveolar cell types but become regionally restricted when intrapulmonary airways start to branch. Moreover, we provide lineage-tracing evidence that a rare population of embryonic progenitors in intrapulmonary bronchi is maintained immature and expressing p63 throughout adulthood. We show that in the adult lung these cells are heterogeneous and represent the source of the aberrant alveolar remodeling in response to sever injury by H1N1 viral infection. Together, our data reveal unexpected two lineage restriction events and cellular behaviors in embryonic p63-expressing cells that elucidates their contribution to the adult airway stem cells pools under homeostatic and repair/remodeling conditions.
Results
p63 labels multipotent progenitors of airways and alveoli, later becoming lineage-restricted to airways
To identify the onset of p63 expression in respiratory progenitors (marked by Nkx2.1), we searched for the earliest p63-expressing cells during initiation of trachea/lung development in Nkx2.1-GFP embryos. Immunofluorescence (IF) first detected a small population of p63+GFP+ cells at E9.0-E9.5 in tracheal primordium and scattered proximal regions of the early lung bud (Movies S1–2). A day later p63+GFP+ cells were mostly confined to the tracheal domain, where it remains abundant in subsequent stages (Figure 1A and Movies S3–4) (Bilodeau et al., 2014; Que et al., 2007). To investigate the contribution of the embryonic p63+ progenitors to the epithelial cell types of the developing respiratory tract, we performed lineage analysis of p63-CreERT2; R26-tdTomato mice, exposing embryos to Tamoxifen (TM) at various developmental stages. Lungs and tracheas were isolated and analyzed perinatally at E18.5 or at selected postnatal ages (see below and Methods for characterization and approach validation). To lineage-trace p63 at the earliest stages observed, E8.5, E9.5 or E10.5 embryos were exposed to TM (160 μg/g, maternal oral gavage). Analysis of E18.5 tracheas showed extensive tdTom labeling in the pseudostratified epithelium at these stages, confirming the contribution of these progenitors from as early as E8.5 (Figures 1B–C upper panels; Figures S1A–D). Intrapulmonary airways were nearly unlabeled in E10.5 TM-treated embryos (Figure 1C). Surprisingly, TM exposure at E9.5 or E8.5 resulted in abundant tdTom+ cells in E18.5 intrapulmonary airways extending ectopically to alveolar saccules (Figure 1B; Figure S1D). IF showed tdTom+ cells double-labeled with markers of airways (CC10: secretory; Cgrp: neuroendocrine) and alveolar (Pdpn: type I; pro-Spc: type II) cells (Figure 1B lower panels; Figures S1A–D). E8.5 TM-treated embryos showed the broader distribution of tdTom+ cells extending to the lung domain at different stages from E9.5-E18.5 (Figures S1A–D). To rule out putative TM residual effect in vivo, we isolated E8.5 foregut explants from p63-CreERT2; R26-tdTomato embryos, treated with 4-hydroxytamoxifen (4-OHT) for 2hrs and subsequently cultured for 5 days. IF showed the ectopic tdTom extending to the distal Nkx2.1+Sox9+ domain of lung buds, confirming the pattern observed with E8.5 TM administration in vivo (Figure S1E). Thus, in the early respiratory tract p63 marks multipotent progenitors able to generate potentially proximal and distal epithelial cell components, as predicted for other progenitors occupying the Nkx2.1 domain. This ability is lost by E10.5, when they become lineage-restricted to tracheal and proximal intrapulmonary airways.
Figure 1. p63 labels multipotent progenitors of airways and alveoli and later become lineage-restricted to airways.
(A) Immunofluorescence (IF) of E9.5 and E10.5 Nkx2.1-GFP embryos; boxed areas enlarged in right panels. GFP+p63+ cells in lung (arrowheads) or tracheal (arrow) primordia. (*) GFP+p63− in lung. (B–C) p63-CreERT2; R26-tdTomato lineage labeling. E18.5 trachea (top panels) and lungs (bottom panels: intrapulmonary airways and alveolar saccules) from embryos exposed to TM at E9.5 (left) or E10.5 (right). tdTom+ double-labeling with cell type-specific differentiation marker: insets depict secretory (CC10), neuroendocrine (Cgrp), alveolar type 1 (Pdpn) and type 2 (pro-Spc) cells. Labeling pattern similar in trachea but dramatically different in lung indicative of lineage restriction. E9.5 tracing: N=11 embryos from 3 litters, all with extensive tdTom labeling in alveolar compartment; E10.5 tracing: N=7 embryos from 2 litters, all with minimal tdTom labeling in alveolar compartment. For TM exposure after E10.5: N>18 from at least 8 litters, none with alveolar labeling. Scale bars: 10μm unless noted. See also Figure S1 and Movies S1–3.
To learn whether intrapulmonary airways were lineage-restricted at comparable early stages, we performed lineage tracing of Sox9, which, like Id2, Spc and others, marks distal lung epithelial buds at E10.5 (Rawlins et al., 2009). Sox9-CreERT2; R26-tdTomato embryos were exposed to TM between E9.5-E11.5. Analysis of TM-treated at E11.5 embryos showed expected distribution of tdTom selectively in intrapulmonary (bronchioles) but not in tracheal epithelium. By contrast, TM at E9.5 resulted in tdTom extending ectopically to the tracheal epithelium (Figure S1F)
Further lineage restriction of p63+ cells establishes the basal stem cell pool in developing trachea
Analysis of p63 expression in the developing trachea showed distinct distribution of signals in the luminal and basal compartments from early to late stages. This was confirmed by profile analysis of signal intensity in representative sections of E14.5-E18.5 tracheas (Figure S2A). The distinct p63 expression patterns suggested an additional segregation event during epithelial differentiation. We performed lineage-tracing analysis targeting three distinct stages: (i) E13.5-E14.5, when endogenous p63 is expressed at variable levels in most tracheal epithelial cells (>60%); (ii) E14.5-E15.5, first evidence of the basal-luminal segregation as high-expressing p63+ cells assume a basal position and acquire Krt5; (iii) E17.5-E18.5, characterized by a well-defined basal layer of strongly labeled p63+Krt5+ (prebasal) cells occupying ~25% of the tracheal epithelium (Figure S2A–C). Exposure of p63-CreERT2 embryos to TM at E17.5 labeled 96% of the p63+Krt8− prebasal cells at E18.5 and labeling was maintained high throughout adulthood (postnatal days P7, P21) (Figure 2A left graph; Figure S2D; Table S1). Labeled luminal cells (p63−Krt8+tdTom+) increased significantly to 23% of the total tracheal epithelium at P21, confirming that the E18.5 lineage-labeled prebasal cells self-renew and generate luminal descendants, like their postnatal BC counterpart (Figure 2A right graph; Figure S2D; Table S1; Rock et al., 2009). In spite of differences in recombination from E17.5 tracing, TM at E13.5 or E14.5 both resulted in labeling of >84% of the E18.5 p63+Krt8− prebasal cells and could be ascribed to the strong p63 promoter activity in some of these cells (Figures 2A–C, left graphs; Figure S2D; Table S1). The data suggested that the pool of multipotent precursors destined to become BCs was largely established around E13.5-E14.5, even preceding the appearance of Krt5, one of the earliest markers of initiation of BC program (Figure S2A; Bilodeau et al., 2014). Consistent with this, TM exposure before E13.5 resulted in markedly reduced number of p63+tdTom+ cells at E18.5 (ex. only 57.9% ± 10.6% with TM treatment at E12.5, Table S1). Interestingly, the labeling frequency in luminal compartment analyzed at E18.5 increased significantly when mice were exposed to TM at earlier stages (TM at E14.5: 18.1% ± 1.9%; TM at E13.5: 33.7% ± 3.2%), suggesting that at least some of the p63+tdTom+ cells could have been committed to luminal differentiation before the basal compartment was established (Figures 2A–C, right graphs; Figure S2D; Table S1). TM residual effect was ruled out by results from a TM titration assay, as explained in Methods and Table S1. We asked whether the population of p63+tdTom+ cells expanded preferentially compared to the other cell types. EdU was administered simultaneously with TM to p63-CreERT2 embryos at E13.5, E14.5, and E17.5 and incorporation was quantified 24hr later in p63+tdTom+ or tdTom−, and p63− cells. As expected, EdU incorporation declined with age in all groups (Figure 2D; Table S1). Interestingly, we found no evidence of preferential EdU incorporation in any of the p63+ populations (Figure 2D; Table S1). Therefore, p63 expression does not confer proliferative advantage at least in the developing trachea. The data also suggest that our lineage labeling was not biased towards a higher proliferative p63+ subpopulation.
Figure 2. Lineage restriction of p63+ cells establishes the basal stem cell pool of developing trachea with equal contributions to ciliated and secretory lineages.
(A–C) Lineage tracing of p63-CreERT2; R26-tdTomato embryos: TM at E17.5, E14.5 or E13.5. Left graphs: % tdTom+p63+ cells in total p63+ population; right graphs: % tdTom+p63− cells in total p63− population at each stage. (D) p63-CreERT2; R26-tdTomato mice exposed to EdU and TM simultaneously at E13.5, E14.5 or E17.5. EdU incorporation and lineage-labeling analysis in tracheal sections after 24hr: p63+ (tdTom+ or tdTom−) or p63− cells (representative images and quantification). (E–F) Immunofluorescence (IF) of tracheal sections from p63-CreERT2; R26-tdTomato mice. Contribution of embryonic p63 to luminal tracheal secretory and ciliated cells pre- and postnatally. TM exposure at E13.5 or E14.5 and analysis at E18.5, P0-P21. tdTom double-labeled with differentiation marker; dotted area shown as single channel in lateral panels or insets. Graph: % tdTom+ cells in ciliated and secretory cells at E18.5. All graphs: mean ± SEM from 6–12 fields per sample, N>=3 per stage, except P7 in E14.5 tracing (N=2); details in Table S1. Statistics: one-way ANOVA, *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; n.s. non-significant. Scale bars: 10μm. See also Figure S2 and Table S1.
Thus, a second lineage-restriction event occurs in embryonic p63+ progenitors around E13.5-E14.5 that defines the basal and luminal compartments and generates the majority of the prebasal population.
Embryonic p63+ cells contribute equally to ciliated and secretory lineages
We tested whether lineage-labeled luminal cells were biased towards a cell fate choice during differentiation. Marker analysis of p63-CreERT2 embryos exposed to TM at E13.5 or E14.5 showed lineage-labeled A-tub+ or CC10+ luminal cells at late prenatal and postnatal stages (Figures 2E–F). Interestingly, E18.5 tracheas showed the proportion of lineage-labeled Foxj1+ or CC10+ luminal cells averaging 37% and 36%, respectively (Figure 2E; Table S1). Thus, p63+ cells contributed nearly equally to the ciliated and secretory cell populations. This observation was inconsistent with the reported role of p63 in preventing excessive ciliated cell differentiation (Daniely et al., 2004; Marshall et al., 2016).
To reexamine this issue, we generated a homozygous deletion of p63 from our p63-CreERT2 knock-in line. We reasoned that this knockout (KO) model could be more suitable for correlations with the findings of our lineage analysis. Our KO animals phenocopied the severe skin and limb defects previously reported (Yang et al., 1999; Romano et al., 2012). The tracheal epithelium showed no p63+Krt5+ cells and unaffected proliferation rate, as shown by Ki67 staining and predicted from our EdU incorporation data (Figures 3A, S2E, 2D). Similar expression of CC10, Scgb3a2 and Foxj1 in E18.5 WT and KO tracheas by IF and qRT-PCR suggested that p63 did not influence secretory vs. ciliated fate specification (Figures 3A–B). This was in agreement with the comparable distribution of tdTom in secretory and ciliated cells in lineage-tracing analysis (Figure 2E). No difference in intrapulmonary airways including neuroendocrine lineage was observed (Figure 3D).
Figure 3. p63 KO mutants have preserved luminal lineage balance with altered maturation and organization of the pseudostratified epithelium.
(A–E) Tracheal differentiation in E18.5 WT and p63 null (KO) mice. (A) IF, markers of basal (p63, Krt5), luminal (Krt8), secretory (CC10, SSEA1), ciliated (Foxj1, A-tub), airway progenitor (Sox2) cells. Far right panel: maximum projection of confocal z-stack images showing colocalization of A-tub+ cilia and Foxj1+ nuclei; magenta arrows showing Foxj1+ ciliated cells without cilia. (B) qRT-PCR, p63, Foxj1, CC10, Jag2 (Notch ligand), Scgb3a2 (secretory marker), (C) Morphometric analysis of A-tub and SSEA1. (D) E18.5 intrapulmonary airways of WT and p63 KO showing similar expression of Cgrp and Sgb3a2 (neuroendocrine and secretory cell markers). (E) Alcian Blue and A-tub staining of E18.5 WT and KO tracheas: Brown arrows showing A-tub+ cilia; Cyan arrows showing Alcian Blue+ goblet cells. (F) IF showing simplified epithelial structure from the pseudostratified tracheal epithelium in the KOs at E18.5; graph: epithelial cell number per unit basement membrane length (mm). (G) p63-CreERT2 lineage tracing and IF showing Notch3+tdTom+ in tracheal epithelium at E18.5 (TM administration at E13.5); insets: single channels depicted from boxed area. (H) IF E18.5 tracheal sections: Strong Notch3 nuclear staining in suprabasal cells of WT (nuclear Sox2 in all epithelial cells), however in p63 KO Notch3 was diffusely expressed at low levels in cytoplasm and no longer in the nucleus. Graphs: mean ± SEM from 6–12 fields per sample, N>=3, details in Table S1. Statistics: Student’s t-test: *P<0.05; **P<0.01; ****P<0.0001; n.s. non-significant. Scale bars: 10μm. See also Figure S2 and Table S1.
Nevertheless, A-tub labeling was more abundant in KO epithelia, suggesting that once committed, ciliated cells matured to a greater extent to form multicilia (Figures 3A, 3C; Table S1). Moreover, expression of SSEA1, which marks immature secretory cells (Xing et al., 2010), was greatly reduced in KO tracheas both at early (E15.5) and late (E18.5) stages, while unaffected in intrapulmonary airways (reflecting the regional distribution of p63+ cells in WT) (Figures 3A, 3C, S2F; Table S1). By contrast, numerous Alcian-Blue-labeled cells in KO indicated that, without p63, secretory progenitors undergo aberrant goblet cell differentiation (Figure 3E).
As expected from the absence of BCs, the pseudostratified architecture of the tracheal epithelium was converted into a columnar epithelium with a reduction in the epithelial cell number by 36.4% in KOs (Figure 3F; Table S1). Nuclear Notch3+tdTom+ was present in E18.5 p63-CreERT2 tracheas (Figure 3G; Mori et al., 2015). However, in KO tracheas Notch3 was expressed diffusely and no longer in the nucleus, likely from the limited Jag ligand available for Notch3 activation without p63+ cells (Figure 3H). Indeed qRT-PCR showed decreased Jag2 expression (Figure 3B). Low Jag levels, presumably from ciliated cells, could still induce expression but not activation of Notch3. Altered Notch3 could potentially influence differentiation behavior but not fate choice. Thus, the phenotype we observed could be potentially explained by the loss of BC progenitors in the absence of p63, with an alternative progenitor (p63-independent) then undergoing an aberrant luminal differentiation program. This would suggest that prenatal p63 is crucial to generate not only the postnatal BC pool, but also the embryonic progenitors of luminal cells able to carry the normal differentiation of the developing airways.
Embryonic p63+ cells establish rare p63+Krt5− intrapulmonary bronchial progenitors that generate H1N1-induced alveolar Krt5+ pods
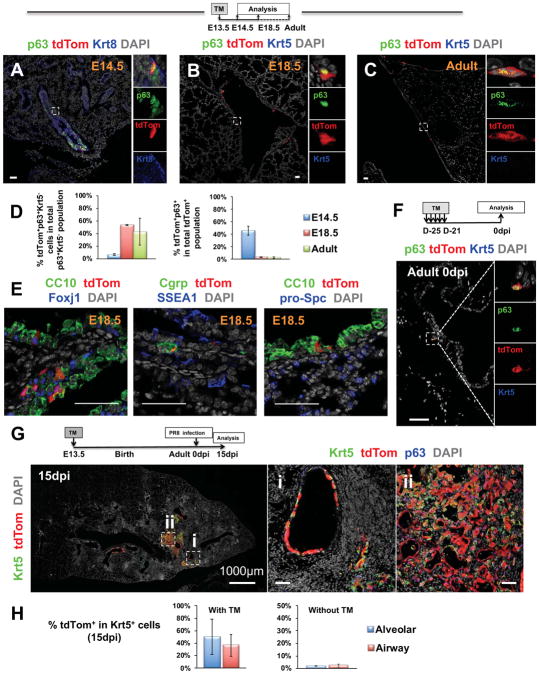
We had already evidence that p63+tdTom+ cells remained uncommitted in trachea from late gestation to adulthood (Figures 2A–C). We asked whether a similar population also existed in intrapulmonary airways, since scattered embryonic p63+ cells in main bronchi could potentially be maintained undifferentiated to adulthood (Movie S4; Figure 4A). E13.5 embryos were exposed to TM as before, and examined from E14.5 to adult. p63+tdTom+ cells could be identified at all stages; these were rare, restricted to intrapulmonary stem bronchi, often near the junction of extra- and intra-pulmonary airways. These cells never acquired Krt5, remaining immature (Figures 4A–C), similar to the p63+Krt5− intrapulmonary cells labeled in adulthood (Figure 4F). The low recombination efficiency in intrapulmonary p63+ cells compared to the tracheal p63+ cells reflected the dramatic difference in p63 promoter activities in these regionally distinct p63+ population (6.7% ± 1.4% vs. 35.7% ± 4.5%) (Figures 4D, 2C; Table S1). Interestingly, with such low labeling efficiency and the fact that more than 96% of these lineage-labeled cells committed to luminal differentiation by E18.5, we could still detect 53.9% ± 0.9% of the intrapulmonary p63+Krt5− cells bearing the tdTom label at E18.5, and 43.1% ± 21.6% in adulthood (Figure 4D; Table S1). tdTom+ cells also infrequently co-labeled with markers of airway, but not alveolar cell types (Figure 4E).
Figure 4. Embryonic intrapulmonary p63+ progenitors are maintained immature throughout adulthood and mediate the H1N1 aberrant alveolar remodeling.
(A–E) Lineage tracing of p63+ descendants in intrapulmonary airways at E14.5, E18.5 and adult lung: TM in E13.5 p63-CreERT2; R26-tdTomato mice. IF tdTom double-labeled with p63 (A–C) or markers of airway/alveolar differentiation (E) Left graph: % tdTom+p63+Krt5− cells in total intrapulmonary p63+Krt5− cells; right graph: % tdTom+p63+ cells in total intrapulmonary tdTom+ cells. (F) p63+Krt5−tdTom+ cells in adult intrapulmonary bronchi in mice exposed to TM in adulthood (compare to C). (G) Adult lung from mice exposed to TM at E13.5 and infected with H1N1 at 8 weeks. IF at 15dpi: p63+tdTom+Krt5+ cells in intrapulmonary bronchi (i) and as alveolar clusters (ii. pods); boxed areas magnified in right panels. Graphs: % tdTom+Krt5+ cells in the H1N1-induced ectopic Krt5+ cells in the mice with TM treatment at E13.5 (left) or without TM exposure (right). Graphs: mean ± SEM, N>=3 except E18.5 in E13.5 tracing (N=2), details in Table S1. Scale bars: 50μm unless noted. See also Figure S3, Movie S4 and Table S1.
We then investigated the participation of these intrapulmonary p63+Krt5−tdTom+ progenitors labeled prenatally in the adult lung response to injury. From the basal-like morphology and p63 lineage, we tested their involvement in the reported H1N1-induced aberrant alveolar remodeling (Xi et al. 2017). p63-CreERT2; R26-tdTomato embryos exposed to TM at E13.5 were infected in adulthood and examined after 15 days (15dpi). p63+Krt5+tdTom+ cells were found extensively in intrapulmonary airways (36.7% ± 17.6% labeling frequency) and alveoli (50.4% ± 28.3% labeling frequency) as clusters (pods) or honeycomb-like lesions (Figures 4G–H; Table S1). Their morphology and distribution around lobar bronchi was similar to that found in these mice exposed to TM in adulthood and infected with H1N1 after 3 weeks (Figure S3A). The stepwise acquisition of Krt5 and their noticeable trail from large airways (where we identified p63+Krt5−tdTom+ cells in uninfected lungs) to areas undergoing remodeling, support their short-range migratory behavior and the idea that progenitors for these lesions reside in the main bronchi (Figure S3B). PBS-mock infection showed none of these changes (Figure S3C). Consistent with previous reports, the tdTom+Krt5+ cysts persisted with minimal contribution to alveolar regeneration in long term (Figures S3D, S4A; Vaughan et al., 2015; Xi et al., 2017).
p63+Krt5− intrapulmonary bronchial progenitors include a subpopulation of CC10 lineage-labeled cells responsible for H1N1 induction of pods
A comparison of H1N1-infected WT and p63-CreERT2 mice suggested a more attenuated response of the p63-CreERT2 knock-in mice with less pods. We asked if p63 haploinsufficiency could be influencing this response. Thus, we used another reporter line in which both p63 alleles are functional. Previous evidence of H1N1 induction of Krt5+ pods in CC10 lineage-labeled cells was deemed artefactual due to residual TM activity (Zheng et al., 2014; Vaughan et al., 2015). To circumvent this issue, we exposed adult CC10-CreERT2; R26-tdTomato mice to H1N1 21 days after the last TM-gavage administration. Analysis of 15dpi lungs showed robust induction of tdTom+Krt5+ cell clusters partially overlapping with a second population of Krt5+ clusters, however tdTom−, consisting of 30.3% ± 12.0% of the alveolar Krt5+ and 45.1% ± 11.8% of the airway Krt5+ cells (Figures 5A–C; Table S1). Collectively, all expressed p63, indicating that CC10 lineage-labeled clusters were a subpopulation of the total H1N1-responding p63 lineage-derived cells (Figure 5B). The extensive labeling pattern in the H1N1-induced ectopic Krt5+ cells was not observed in other Cre lines (Spc, Upk3a, Notch3-driven) using the same TM regimen (Figures S5A–C). Notably, the H1N1-infected CC10 reporter mice showed Krt5+ pods at a significantly higher frequency (100%), some slightly less proximal compared to p63-CreERT2 mice at similar dpi (Figures 5A, 5D; Table S2). We tested the possibility that CC10 lineage-labeled p63+Krt5− cells were already present in the adult lung prior to H1N1 infection. Remarkably, IF of TM-treated uninjured adult CC10-CreERT2 mice revealed about 31.9% ± 4.3% of the p63+Krt5− cells scattered in the main intrapulmonary bronchi labeled by tdTom (Figure 5E; Table S1). These cells could induce Krt5+ pods and thus mount a full response to H1N1, unlike those in p63-CreERT2 mice due to p63 haploinsufficiency (Figures S4C–E; Table S2). Specifically, in average 20 p63+Krt5− cells per lung section could be identified from the adult CC10-CreERT2 mice during homeostasis, while this number decreased to 3 in p63-CreERT2 mice, suggesting an allelic function of p63 in generating or maintaining this progenitor pool in intrapulmonary airways (Table S1). Overall, the pool of p63+Krt5− progenitors that responds to H1N1, although, restricted to main bronchi, seems to be more diverse and include cells that share features with other facultative progenitors, such as Club cells as shown here.
Figure 5. Intrapulmonary p63+Krt5− progenitors include a subpopulation of CC10 lineage-labeled cells responsible for H1N1 induction of pods.
(A) CC10-CreERT2 adult mice exposed to H1N1 after 21days of TM exposure. IF: lung sections at 15dpi with CC10 lineage-labeled airways and Krt5+ clusters overlapping partially with lineage-negative Krt5+ clusters (green). (B) IF: tdTom+ “trail” expanding to alveolar compartment. (C) Graphs: % tdTom+Krt5+ cells in the H1N1 induced ectopic Krt5+ cells in the mice with TM treatment in adulthood (left) or without TM exposure (right). (D) p63-CreERT2 adult lung at 15dpi: IF: less pods compared to CC10-CreERT2 suggestive of a more attenuated response to H1N1. (E) Uninjured adult CC10-CreERT2 mice 21 days after Tm: rare CC10 lineage-labeled p63+Krt5− progenitors in intrapulmonary main bronchi (i) near unlabeled p63+Krt5− cells (ii). Graph: % tdTom+p63+Krt5− cells in total intrapulmonary p63+Krt5− cells. Graphs: mean ± SEM, N>=3, details in Table S1. Scale bars: 50μm unless noted. See also Figures S4–5 and Tables S1–2.
Moreover, the labeling pattern of H1N1-exposed p63-CreERT2 and CC10-CreERT2 differ dramatically in the trachea. Each reporter labels its respective cell type in TM-treated uninjured trachea (Figure S4B). By contrast, after injury p63-CreERT2 mice labeled almost all the epithelial cells in both basal and luminal compartments but none in CC10-CreERT2 mice (Figure S4B). This confirmed the stem cell identity of the adult tracheal p63+Krt5+tdTom+ in post-injury repair and further supports the idea that the progenitor cells sharing both CC10 and p63 lineages reside in intrapulmonary airways, not the trachea.
Discussion
Here we show that when airways are still forming, two lineage-restriction events occur in p63-expressing cells, which ultimately establish regionally distinct adult multipotent progenitor pools: the BCs in trachea and the p63+Krt5− cells in intrapulmonary airways.
In the developing trachea, cells expressing variable levels of p63 self-renewed and populated the luminal compartment, activating a differentiation program distinct from the aberrant maturation program of p63-independent progenitors. In contrast to previous studies, we found no evidence of p63 influencing the proliferative status or cell fate choices at least during development (Senoo et al., 2007; Daniely et al., 2004; Marshall et al., 2016). Given the different parameters assessed here, it was unclear the extent to which our p63 lineage-labeled cells overlapped with the previously reported Itgb4HI prebasal cells from Nkx2.1-Cherry embryonic lungs (Bilodeau et al., 2014).
Our study also provides evidence that during lung specification p63 marks multipotent progenitors for both airway and alveolar lineages, while these cells adopt a proximally restricted fate mainly for extrapulmonary airways by E10.5. This stage coincides with the emergence of distal Id2+Sox9+Spc+ progenitors that are expanded and patterned to form the bronchial tree (Morrisey et al., 2009). Intrapulmonary airways have been shown to originate from Id2 lineage-labeled distal bud progenitors (Rawlins et al., 2009). Based on our p63 and Sox9 lineage models, we propose that these two complementary lineages are specified from multipotent epithelial progenitors at distal and proximal domains early in trachea/lung development, merging to form the conducting airways of the respiratory tract.
The appearance of large areas of p63+Krt5+ BC-like cells in H1N1-injured alveoli is intriguing, as these cells become no longer restricted to airways. Recent studies ascribed their origin to Sox2+ airway progenitors (Ray et al., 2016), DASCs (p63+Krt5+ distal airway stem cells. Kumar et al., 2011; Zuo et al., 2015), or LNEPs (lineage-negative epithelial progenitors, non-CC10Lineage/non-Foxj1Lineage-derived EpCAM+Sox2+Itgb4+Krt5−. Vaughan et al., 2015; Xi et al., 2017). Here we show that these BC-like cells arise from a pool of p63+Krt5− p63 lineage-labeled intrapulmonary progenitors distinct from the tracheal prenatal p63+ cells we described. They can be traced back from the embryonic p63+ cells in main bronchi, which maintain p63 expression throughout life.
Our data support the idea that this population originates entirely from the airway compartment and thus, it is contained in the previously described Sox2 lineage-labeled pool (Ray et al., 2016). Similar to the findings in the Sox2-CreERT2 knock-in line, we also identified an attenuated response of our p63-CreERT2 mice to H1N1 challenge due to haploinsufficiency, suggesting the functional importance of these regulator genes in injury response. However, our study provides a more precise information about the nature of these progenitors, as we show that they arise from embryonic p63+ cells in the developing airway epithelium. In addition, our CC10-CreERT2 lineage-tracing analysis suggests an unexpected diversity in the progenitor pool that gives origin to the Krt5+ lesions.
The progenitor cells we describe share several features with LNEPs, including their common p63 lineage, lack of Krt5, and response to H1N1. Yet LNEPs differ in a number of aspects. The adult progenitors we identified maintained p63 expression regardless their labeling during embryonic or adult life or H1N1 challenge. By contrast, immunostaining demonstrating that p63 lineage-labeled LNEPs actively expressed p63 at the time of H1N1 infection was not available (Xi et al., 2017). Moreover, LNEPs (and DASCs) were reported in distal airways, unlike the p63+Krt5−tdTom+ cells here, which were restricted to large airways. During LNEP isolation, the p63-enriched proximal airways and CC10 lineage-labeled cells were deliberately excluded, thus differing substantially from the cells reported here. Lastly, all H1N1-induced Krt5+ cells were p63 lineage-labeled, making unlikely the contribution of p63− progenitors to the pods.
Taken together, our data provides insights into the origin and diversity of the adult stem cells in the respiratory tract, and the lineage relationships arising from responses to environmental agents.
Star Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wellington V. Cardoso (wvc2104@cumc.columbia.edu)
Experimental model and subject details
Mice
p63-CreERT2 mice were generated and characterized as described in Lee et al. (2014). This mouse line was originally in FVB background, with the CreERT2 coding sequence knocked into exon 4 which is shared by both deltaNp63 (major isoform in airway system) and TAp63 transcripts. Sox9-CreERT2, Spc-CreERT2, Upk3A-CreERT2, N3-CreERT2 knock-in lines have been previously reported and were currently used for lineage tracing of embryonic distal lung epithelial progenitors (Sox9; Soeda et al., 2010), adult alveolar type II cells (Spc; Desai et al., 2014), adult secretory cells associated with neuroendocrine bodies and terminal bronchioles (Upk3A; Guha et al., 2017), Notch3-expressing mesenchymal and rare scattered epithelial cells (N3; Fre et al., 2011), respectively. The CC10-CreERT2 knock-in line was purchased from the Jackson Laboratory (B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J) (Rawlins et al., 2009), and was currently used to follow the fate of adult club and Spc+CC10+bronchioalveolar progenitor cells. All knock-in lines were bred into R26-tdTomato (Jackson Laboratory, B6.Cg-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J) for lineage-tracing analyses. All studies were approved by Columbia University Institutional Animal Care and Use committees (IACUC).
For time-pregnancy experiments, male mice from CreERT2 driver lines were mated with female mice from the R26-tdTomato reporter line. Noon of the day when the vaginal plug was identified was determined as E0.5. The developmental stages of TM administration for each lineage-tracing experiment were detailed below.
For adult lineage-tracing experiments, 6–8 week-old mice were used and both male and female were included. Details about TM administration and H1N1 infection were described below.
Method details
Lineage tracing analysis in embryonic development
Tamoxifen (TM) was dissolved in sunflower seed oil (Sigma, T5648 and S5007) and administered by gavage to pregnant mice. To allow survival and analyses of postnatal stages, E18.5 embryos were transferred to adult CD1 foster mothers.
To determine the efficiency of recombination at different TM doses and developmental stages we treated p63-CreERT2; R26-tdTomato mothers with TM by oral gavage at gestation days 13.5 (160 μg/g body weight), 14.5 (80 μg/g), 17.5 (70 μg/g) and analyzed lungs after 24hr. IF and quantitative analysis showed efficient recombination with the percentage of lineage-labeled p63+ tracheal cells averaging 35.7% ± 4.5%, 54.3% ± 1.6%, and 95.6% ± 1%, respectively (post hoc Tukey’s test: E13.5 vs. E14.5 **adjusted P<0.01; E14.5 vs. E17.5 ***adjusted P<0.001; E13.5 vs. E17.5 ****adjusted P<0.0001. Table S1). Higher recombination efficiency was achieved at later developmental stages in spite of the lower TM doses, consistent with the overall increase in p63 expression levels perinatally.
TM-independent recombination in mice treated with vehicle at E13.5 and examined at E18.5 and postnatal P21 averaged <0.1% and 1.3% ± 0.5 of the p63+tdTomato+ tracheal cells, respectively (Table S1).
To test for potential TM residual effects, we treated p63-CreERT2; R26-tdTomato E13.5 mothers with decreasing TM doses (160 μg/g, 10 μg/g and 4 μg/g body weight), looking for changes in the proportion of lineage-labeled cells in the basal versus luminal compartments at E18.5. Analysis of E14.5 tracheas confirmed the major dose-dependent decrease in recombination efficiency in p63+ cells, from 35.7% to 6.3% and 0.6%, respectively (Table S1). By E18.5 the significantly higher proportions of p63+tdTom+ in total tdTom+ cells were found at lower doses (43%, 56%, 57%, respectively; post hoc Tukey’s test: 160 μg/g vs. 10 μg/g **adjusted P<0.01; 160 μg/g vs. 4 μg/g **adjusted P<0.01; 10 μg/g vs. 4 μg/g non-significant P>0.05. Table S1). This scenario would be unlikely in the presence of sustained recombination and thus argued against a TM residual effect.
For labeling p63+ cells before E14.5, we treated p63-CreERT2; R26-tdTomato mothers with 160 μg/g Tm by oral gavage (TM exposure at E8.5, E9.5, E10.5, E12.5 or E13.5).
To label Sox9+ multipotent lung tip cells, we exposed Sox9-CreERT2; R26-tdTomato mothers with 160 μg/g TM by oral gavage at E9.5 or E11.5. Lungs were examined at E15.5 or E17.5.
Lineage tracing analysis in adult for H1N1 injury response
For lineage tracing in adult animals in H1N1-infected lungs, 6–8 -week-old p63-CreERT2, CC10-CreERT2, p63+/+, Spc-CreERT2, Upk3A-CreERT2, N3-CreERT2 (male and female, all with R26-tdTomato reporter allele) mice were given 240 μg/g body weight TM in 5 sequential days via oral gavage. A chase period of 3 weeks was used to prevent artefactual tamoxifen residual activity before viral infection.
Fluorescence intensity profile analysis
For E14.5, 150 cells from epithelium were randomly picked, circled around nuclear shape (DAPI). Fluorescent intensity and area (μm2) for each cell was measured by Zen 2.3 lite software. 9 cells from non-epithelial tissue without non-specific signals were picked and measured as negative control. The intensity value of p63 channel per unit area for each epithelial cell was calculated, and adjusted background by subtracting the averaged p63 intensity value per unit area of negative controls. The normalized values of p63 intensity per unit area lower than 3.6 were scored as p63 negative; values between 3.6 to 20.0 were scored as p63 low; values higher than 20.0 were scored as p63 high. The percentiles of cells with different p63 fluorescent intensity were shown in the graph.
Quantification of epithelial cell number per unit basement membrane length
20 lines along the basement membrane on cartilage side from each E18.5 WT and KO samples were drawn with their length recorded in Zen 2.3 lite software. The numbers of epithelial cells above the basement membrane lines were quantified. The bar graph showing the averaged number of epithelial cells per unit basement membrane length (mm). N=3 for both WT and KO. In total, cells along 7.13mm basement membrane were counted in WT; cells along 6.91mm basement membrane were counted in KO (Table S1).
EdU Incorporation
Analyses of cell proliferation in lineage-labeled cells was performed by exposing p63-CreERT2 pregnant mice to tamoxifen at designated stages through oral gavage and simultaneous intraperitoneal (i.p.) injection of EdU solution (1mg, 5mg/ml stock solution in PBS). EdU incorporation was assessed 24hr later in frozen tissue sections using the Click-iT EdU Alexa Fluor 647 Imaging kit (Thermo Fisher, C10340). Sections were then double-labeled with antibodies against selected cell markers using immunofluorescence.
Quantitative Real-Time PCR
Whole tracheas were isolated (below the cricoid cartilage to carina) from p63-CreERT2/P63-CreERT2 KO and p63+/+ WT animals at E18.5. RNA was extracted using QIAcube and QIAGEN RNeasy Mini Kit. cDNA was synthesized using the SuperScript IV First-Strand synthesis system (Thermo Fisher). Gene expression was assessed using TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and analyzed on a Step-One Plus instrument (Applied Biosystems). At least 4 tracheas were analyzed for each group. qRT-PCR in whole tracheal homogenates ensured that conclusions from analysis of tissue sections were not influenced by sampling biases due to regional distribution of these cell types.
Foregut culture
Foreguts were dissected from E8.5 p63-CreERT2; R26-tdTomato embryos (8 to 13 somites), exposed to 4-OHT (Sigma-Aldrich, H7904) for 2hrs and cultured on 6-well transwells (Costar 3450-Clear) in serum-free differentiation medium supplemented with 0.02% Ascorbic Acid at 37°C with 5% CO2. Cultures were monitored in a Zeis s Live Imaging System for 5 days and subsequently fixed (4% paraformaldehyde) for IF analysis.
H1N1 (PR8) Viral preparation and Infection
The influenza-A (H1N1) mouse-adapted PR8 viral stock was prepared and amplified in 10-day-old embryonated chicken eggs. Allantoic fluid containing the virus was collected two days after egg inoculation, cleared from cellular debris by centrifugation, aliguoted and stored at −80°C for further use. The viral titer was determined as 1.2×109 pfu/ml by plaque assay on Madin Darbin Canine Kidney cells using tenfold dilutions of allantoic fluid containing the virus. LD50 was initially determined in Balb/c mice as 450pfu. Different doses were tested in pilot experiments using WT C57Bl/6 mice, and 120pfu was found to achieve maximal generation of Krt5+ pods with minimal mortality rates in both WT and p63-CreERT2 mice.
Adult mice (2–3 months old) were anesthetized by isofluorane, and then intranasally administered with 120pfu of H1N1 virus diluted in 30μl PBS. Mock-infected animals received 30μl PBS. Successful infection was verified by weight loss and microscopy of lung frozen sections (massive infiltration of neutrophils and severe destruction of alveolar and airway structures).
Histology and Immunostaining
Embryonic and neonatal lungs were fixed in 4% paraformaldehyde in PBS at 4°C for 1 hour (earlier than E14.5), 4 hours (E14.5, E15.5), overnight (E18.5 and older; whole embryos). Adult lungs were inflated with 4% paraformaldehyde (25cm water column pressure) through trachea and fixed overnight. Samples were processed for frozen or paraffin-embedding.
Immunofluorescence (IF) was performed in tissue sections (6–8μm) blocked with 10% horse serum and 0.3% TritonX-100 (Sigma) for 1 hour at room temperature (rt). Primary antibodies were incubated in 1% bovine serum albumin (Sigma) and 0.3% TritonX-100 at 4°C overnight or 2 hours at rt. Sections were then washed with PBS and incubated with Alexa Fluor-conjugated secondary antibodies (1:500) and NucBlue Live Cell ReadyProbes Reagent (DAPI) (Life Technology) for 1 hr. After washing, samples were mounted with ProLong Gold antifade reagent (Life Technology). When necessary, antigen unmasking was done using Citric Based solution (Vector Labs H-3300) heated in microwave. Mouse primary antibody staining was done using M.O.M kit (Vector Labs BMK-2202). Whole-mount IF was performed using same reagents with elongated antibody incubation time. Confocal Microscopy was performed using a Zeiss LSM 710 confocal microscope.
For visualization of labeled antibodies in immunohistochemistry (IHC), Mouse on Mouse Elite Peroxidase kit (Vector Labs, PK-2200) and Vectastain Elite ABC-HRP kit (Vector Labs, PK-6100) were used. To block endogenous enzyme activity, 0.3% hydrogen peroxide in 0.3% horse serum was used. Following DAB staining, Alcian Blue staining was performed. Images were acquired on a Nikon Labophot 2 microscope equipped with a Nikon Digital sight DS-Ri1 charge-coupled device camera.
The following primary antibodies were used: rabbit anti-p63α (1:400, CST, 13109); chicken anti-Krt5 (1:500, Biolegend, 905901); rabbit anti-Krt5 (1:500, Biolegend, 905501); chicken anti-GFP (1:1000, Abcam, ab13970); chicken anti-Krt8 (1:500, Abcam, ab107115); goat anti-CC10 (1:150, Santa Cruz, sc-9772); mouse anti-Ki67 (1:300, BD Biosciences, 550609); mouse anti-Ecad (1:100, BD Biosciences, 610181); mouse anti-Foxj1 (1:100, eBioscience, 14-9965); mouse anti-acetylated α-tubulin (1:2000, Sigma, T7451); rabbit anti-acetylated α-tubulin (1:500, CST, 5335); rat anti-Scgb3a2 (1:100, R&D, MAB3465); rabbit anti-Cgrp (1:1000, Sigma-Aldrich, C8198); rabbit anti-Sox9 (1:500, Millipore, AB5535); goat anti-Sox9 (1:200, R&D, AF3075); rat anti-Sox2 (1:200, eBioscience, 14-9811-82); hamster anti-Pdpn (1:50, Developmental Studies Hybridoma Bank); rabbit anti-proSpc (1:1000, Seven Hills, WRAB9337); mouse anti-SSEA1 (1:50, Santa Cruz, sc21702); rabbit anti-Nkx2.1 (1:100, Abcam, ab76013); rabbit anti-Notch3 (1:50, CST, 5276).
The following secondary antibodies were used: donkey anti-rabbit (conjugated with Alexa Fluor 488, 568, 647); donkey anti-chicken (conjugated with Alexa Fluor 488); goat anti-chicken (conjugated with Alexa Fluor 488, 647); donkey anti-goat (conjugated with Alexa Fluor 488, 568, 647); donkey anti-mouse (conjugated with Alexa Fluor 488, 568, 647); donkey anti-rat (conjugated with Alexa Fluor 488, 647); goat anti-hamster (conjugated with Alexa Fluor 647). All secondary antibodies were purchased from Thermo Fisher Scientific or Jackson ImmunoReseach.
Quantification and statistical analysis
Quantification and statistical analysis have already been detailed in the methods section above, associated with each experiment, as well as in the figure legends. All quantification for colocalization and marker analyses were performed in Adobe Photoshop CS6. Details about the quantified cell numbers and numbers of mice for each experiment were provided in Table S1. Statistical analyses were performed in Microsoft Excel or GraphPad Prism 7. As shown in figure legends, data in graphs were shown in mean ± SEM. Statistical significance of differences in three or more groups of data were determined by one-way ANOVA and post hoc Tukey’s test (Figures 2A–D and methods). Statistical significance of differences between two groups of data were determined by unpaired two-tailed t test (Figures 2E, 3B–C, 3F, S2E–F).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-p63α | CST | Cat#13109; RRID:AB_2637091 |
| chicken anti-Krt5 | Biolegend | Cat#905901; RRID:AB_2565054 |
| rabbit anti-Krt5 | Biolegend | Cat#905501; RRID:AB_2565050 |
| chicken anti-GFP | Abcam | Cat#ab13970; RRID:AB_300798 |
| chicken anti-Krt8 | Abcam | Cat#ab107115; RRID:AB_10976462 |
| goat anti-CC10 | Santa Cruz | Cat#sc-9772; RRID:AB_2238819 |
| mouse anti-Ki67 | BD Biosciences | Cat#550609; RRID:AB_393778 |
| mouse anti-Ecad | BD Biosciences | Cat#610181; RRID:AB_397580 |
| mouse anti-Foxj1 | eBiosciences | Cat#14-9965; RRID:AB_1548835 |
| mouse anti-acetylated α-tubulin | Sigma | Cat#T7451; RRID:AB_609894 |
| rabbit anti-acetylated α-tubulin | CST | Cat#5335; RRID:AB_10544694 |
| rat anti-Scgb3a2 | R&D | Cat#MAB3465; RRID:AB_2183548 |
| rabbit anti-Cgrp | Sigma-Aldrich | Cat#C8198; RID:AB_259091 |
| rabbit anti-Sox9 | Millipore | Cat#AB5535; RRID:AB_2239761 |
| goat anti-Sox9 | R&D | Cat#AF3075; RRID:AB_2194160 |
| rat anti-Sox2 | eBioscience | Cat#14-9811-82; RRID:AB_11219471 |
| hamster anti-Pdpn | Developmental Studies Hybridoma Bank | NA |
| rabbit anti-proSpc | Seven Hills | Cat#WRAB9337; RRID:AB_2335890 |
| mouse anti-SSEA1 | Santa Cruz | Cat#sc21702; RRID:AB_626918 |
| rabbit anti-Nkx2.1 | Abcam | Cat#ab76013; RRID:AB_1310784 |
| rabbit anti-Notch3 | CST | Cat#5276; RRID:AB_10560515 |
| donkey anti-rabbit; Alexa Fluor 488 | Thermo Fisher | Cat# A-21206; RRID:AB_2535792 |
| donkey anti-rabbit; Alexa Fluor 568 | Thermo Fisher | Cat#A10042; RRID:AB_2534017 |
| donkey anti-rabbit; Alexa Fluor 647 | Thermo Fisher | Cat# A-31573; RRID:AB_2536183 |
| donkey anti-chicken; Alexa Fluor 488 | Jackson ImmunoResearch | Cat# 703-545-155; RRID:AB_2340375 |
| goat anti-chicken; Alexa Fluor 488 | Thermo Fisher | Cat# A-11039; RRID:AB_2534096 |
| goat anti-chicken; Alexa Fluor 647 | Thermo Fisher | Cat# A-21449; RRID:AB_2535866 |
| donkey anti-goat; Alexa Fluor 488 | Thermo Fisher | Cat# A-11055; RRID:AB_2534102 |
| donkey anti-goat; Alexa Fluor 568 | Thermo Fisher | Cat# A-11057; RRID:AB_2534104 |
| donkey anti-goat; Alexa Fluor 647 | Thermo Fisher | Cat# A-21447; RRID:AB_2535864 |
| donkey anti-mouse; Alexa Fluor 488 | Thermo Fisher | Cat# A-21202; RRID:AB_141607 |
| donkey anti-mouse; Alexa Fluor 568 | Thermo Fisher | Cat# A10037; RRID:AB_2534013 |
| donkey anti-mouse; Alexa Fluor 647 | Thermo Fisher | Cat# A-31571; RRID:AB_162542 |
| donkey anti-rat; Alexa Fluor 488 | Jackson ImmunoResearch | Cat#712-546-153; RRID:AB_2340686 |
| donkey anti-rat; Alexa Fluor 647 | Jackson ImmunoResearch | Cat#712-606-153; RRID:AB_2340696 |
| goat anti-hamster; Alexa Fluor 647 | Thermo Fisher | Cat# A-21451; RRID:AB_2535868 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | Cat#T5648; |
| Sunflower seed oil | Sigma | Cat#S5007 |
| 4-OHT | Sigma | Cat#H7904 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 imaging kit | Thermo Fisher | Cat#C10340 |
| SuperScript IV First-Strand synthesis system | Thermo Fisher | Cat# 18091050 |
| Experimental Models: Organisms/Strains | ||
| Mouse: p63-CreERT2 | Lee et al., 2014 | NA |
| Mouse: Sox9-CreERT2 | Soeda et al., 2010 | NA |
| Mouse: Spc-CreERT2 | Desai et al., 2014 | NA |
| Mouse: Upk3A-CreERT2 | Guha et al., 2017 | NA |
| Mouse: N3-CreERT2 | Fre et al., 2011 | NA |
| Mouse: CC10-CreERT2 | Rawlins et al., 2009 | B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J; RRID:IMSR_JAX:016225 |
| Mouse: R26-tdTomato | Jackson Lab | B6.Cg-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J; RRID:IMSR_JAX:007914 |
Supplementary Material
Highlights.
Embryonic p63+ cells before E10.5 generate both airway and alveolar descendants
Further p63 lineage restriction at E13.5 defines the tracheal basal progenitor pool
H1N1-induced Krt5+ pods originate from the embryonic intrapulmonary p63+ progenitors
Intrapulmonary CC10 lineage-labeled p63+ cells are required for a full H1N1 response
Acknowledgments
We thank Jun Qian, Jun Li, Jingshu Huang, Xiaoqing Zhang, Maria Stupnikov for their help with the experiments and technical advice, Darrel Kotton and Laertis Oikonomou (Boston University, CReM) for generously sharing the Nkx2.1-GFP mice. We also thank, Virginia Papaioannou, Michael Shen and members of the CCHD, Jianwen Que and Hans Snoeck for thoughtful discussions. This work was supported by NIH-NHLBI R35-HL135834-01 to WVC and NIH CA112403 and CA193455 to JX; and CRIP (Center for Research on Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract #HHSN272201400008C) to AG-S.
Footnotes
Author Contributions
Experimental design, Y.Y., J.L., W.V.C.; Investigation, Y.Y., P.R., M.S.; Resources, M.S., D-K.L., A.G-S., J.X.; Writing & Editing, Y.Y., M.M., W.V.C.; Supervision, J.L., A.G-S., J.X., W.V.C.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bilodeau M, Shojaie S, Ackerley C, Post M, Rossant J. Identification of a proximal progenitor population from murine fetal lungs with clonogenic and multilineage differentiation potential. Stem Cell Reports. 2014;3:634–649. doi: 10.1016/j.stemcr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, Kissel H, Louvi A, Greve J, Louvard D, Artavanis-Tsakonas S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE. 2011;6(10):e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhar A, Deshpande A, Jain A, Sebastiani, Cardoso WV. Uroplakin 3a+ cells are a distinctive population of epithelial progenitors that contribute to airway maintenance and post-injury repair. Cell Reports. 2017;19:246–254. doi: 10.1016/j.celrep.2017.03.051. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CB, Mays DJ, Beeler JS, Rosenbluth JM, Boyd KL, Guasch GLS, Shaver TM, Tang LJ, Liu Q, Shyr Y, et al. p73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Reports. 2016;14:2289–2300. doi: 10.1016/j.celrep.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BLM. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2009;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Liu Y, Liao L, Wang F, Xu J. The prostate basal cell (BC) heterogeneity and the p63-positive BC differentiation spectrum. Int J Biol Sci. 2014;10:1007–1017. doi: 10.7150/ijbs.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Mahoney JE, Stupnikov MR, Paez-Cortez JR, Szymaniak AD, Varelas X, Herrick DB, Schwob J, Zhang H, Cardoso WV. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 2015;142:258–267. doi: 10.1242/dev.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BLM. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BLM. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasgawa H, Wang F, Hogan BLM. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chiba N, Yao C, Guan X, McConnell AM, Brockway B, Que L, McQualter JL, Stripp BR. Rare Sox2+ airway progenitor cells generate Krt5+ cells that repopulate damaged alveolar parenchyma following influenza virus infection. Stem Cell Reports. 2016;7:1–9. doi: 10.1016/j.stemcr.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BLM. Basal cells as stem cells of the mouse trachea and human airway epithelium. PNAS. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Randell S, Hogan BLM. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Models Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, Sinha S. DNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Soeda T, Deng JM, Crombrugghe B, Behringer RR, Nakamura T, Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendon. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee DK, Gotts JE, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Li C, Li A, Sridurongrit S, Tiozzo C, Bellusci S, Borok Z, Kaartinen V, Minoo P. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development. 2010;137:825–833. doi: 10.1242/dev.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Zheng D, Yin L, Chen J. Evidence for Scgb1a1+ cells in the generation of p63+ cells in the damaged lung parenchyma. Am J Respir Cell Mol Biol. 2014;50:595–604. doi: 10.1165/rcmb.2013-0327OC. [DOI] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. p63+ Krt5+ distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.