Abstract
Alzheimer’s disease (AD) is a major neurodegenerative disease and the most common cause of dementia. Currently, no treatment exists to slow down or stop the progression of AD. There is converging belief that disease-modifying treatments should focus on early stages of the disease i.e., the mild cognitive impairment (MCI) and preclinical stages. Making a diagnosis of AD and offering a prognosis (likelihood of converting to AD) at these early stages are challenging tasks, but possible with the help of multi-modality imaging such as MRI, FDG-PET, amyloid-PET, and recently introduced tau-PET, which provide different but complementary information. This paper is a focused review of existing research in the recent decade that used statistical machine learning/artificial intelligence methods to perform quantitative analysis of multi-modality image data for diagnosis and prognosis of AD at the MCI or preclinical stages. We review the existing work in three sub-areas: diagnosis, prognosis, and methods for handling modality-wise missing data – a commonly encountered problem when using multi-modality imaging for prediction or classification. Factors contributing to missing data include lack of imaging equipment, cost, difficulty of obtaining patient consent, and patient drop off (in longitudinal studies). Finally, we summarize our major findings and provide some recommendations for potential future research directions.
1. Introduction
Alzheimer’s disease (AD) is the most common cause of dementia and a main cause of death for people over 65 years old. Over 5.4 million Americans presently suffer from AD. By 2050, a growing number of people, estimated up to 13.8 million, will have AD (1). AD has incurred significant health care costs. In 2016, care for AD patients over 65 years old was estimated to be about $236 million. The Medicare payment for service to AD and other dementia patients was on average twice the payment for patients with other diseases. The impact of AD on patients and their families, the health care system, and society is enormous, and growing, which makes finding effective treatments to reduce the emotional, physical, and financial burdens of this disease an extremely important priority in the US and worldwide.
Although significant attention has been paid to the treatment of AD, there has been little success so far. Within the decade of 2002-2012, 244 drugs were tested in clinical trials registered with the National Institutes of Health, but only one trial completed and received FDA approval. Presently, there are only five FDA-approved AD-related drugs (2). However, these drugs only temporally relieve symptoms. No treatment is available thus far to slow down or stop the pathological damage of AD on the brain, so the disease is fatal (1).
According to recommendations from the working groups convened by National Institute of Aging (NIA) and the Alzheimer’s Association (AA) in 2011, the staging of AD includes AD dementia, the symptomatic predementia stage called mild cognitive impairment (MCI) due to AD, and the preclinical/presymptomatic stage of AD (3–6). The term “MCI due to AD” was used to denote a subgroup of MCI patients with a high likelihood of underlying AD pathology, because MCI, as a syndrome or clinical/research construct, can have other underlying causes than AD. Today, there is converging belief that effective treatment slowing down or stopping the progression of AD should focus on early stages of the disease, i.e., MCI or even the preclinical stage.
In both the recommendations by NIA-AA (known as the NIA-AA Criteria) and by the International Working Group (known as the IWG Criteria), the use of imaging for diagnosis and prognosis at all stages of AD has been significantly highlighted. It has been recognized that different modalities of imaging, including but not limited to structural MRI, FDG-PET, and amyloid-PET, play different but complementary roles.
While the use of multi-modality imaging for diagnosis and prognosis of AD in memory clinics is primarily based on dementia specialists’ trained eyes, researchers have developed and are developing various statistical models and machine learning (ML) algorithms for quantitative imaging data analysis to produce diagnostic and prognostic results. This research area is currently pacing at an unprecedented speed. We envision that in the foreseeable future, memory and aging centers will be empowered by artificial intelligence (AI), employing these automatic, computerized algorithms to assist clinicians’ decision making.
This paper focuses on reviewing existing works that perform quantitative analysis of multi-modality image data using statistical/ML/AI methods for diagnosis and prognosis of AD at the MCI or preclinical stages. There are numerous papers using a single imaging modality, which do not fall within the scope of this review. There are also many papers focusing on classification between AD, MCI, and normal controls (NC), which are also not within our scope because they do not have an “early” stage focus. We focused on papers published in the recent 10 years, and used PubMed as the search engine.
The rest of this article is organized as follows: Section 2 reviews the existing work focusing on diagnosis; Section 3 reviews the existing works focusing on prognosis; Section 4 discusses approaches for handling multi-modality imaging data in which not all the study subjects have all the modalities available, which is a common problem encountered in this type of studies and referred to as the problem of “modality-wise missing data” in this paper. Section 5 provides an overall workflow from image processing to generation of a diagnostic/prognostic result. Section 6 concludes the article and proposes some future research directions.
2. Use of multi-modality imaging for diagnosis of AD at early stages
AD pathology consists of brain amyloid deposition and neurofibrillary tangles, generally associated with significant loss of neurons and deficits in neurotransmitter systems. Diagnosis of AD at early stages requires pathological confirmation according to well-known criteria such as the NIA-AA criteria, which are briefly reviewed as follows with a focus on the role of multi-modality imaging.
In the NIA-AA terminology, “MCI due to AD” and “preclinical AD” are used to describe stages prior to development of AD dementia. MCI due to AD is the symptomatic predementia phase of AD. The NIA-AA Criteria for diagnosing MCI due to AD highlight the incorporation of biomarkers, as shown in Table 1. Preclinical AD describes a phase that individuals have evidence of early AD pathological changes but do not meet clinical criteria for MCI or dementia. NIA-AA proposed three-stage criteria to characterize preclinical AD, as shown in Table 2. These criteria are intended purely for research purposes and have no clinical utility at the present time.
Table 1.
NIA-AA Criteria for diagnosing MCI due to AD (adapted from (3))
| Diagnostic category | Aβ (PET or CSF) | Neuronal injury markers (FDG, tau, MRI) |
|---|---|---|
| MCI – core clinical criteria | Conflicting or indeterminant or untested | Conflicting or indeterminant or untested |
| MCI due to AD – intermediate likelihood | Positive | Untested |
| Untested | Positive | |
| MCI due to AD – high likelihood | Positive | Positive |
| MCI – unlikely due to AD | Negative | Negative |
Table 2.
NIA-AA Criteria for preclinical AD research (adapted from (2))
| Stages | A β (PET or CSF) | Neuronal injury markers (FDG, tau, MRI) | Subtle cognitive change |
|---|---|---|---|
| Stage 1 (Asymptomatic cerebral amyloidosis) | Positive | Negative | Negative |
| Stage 2 (Amyloid positivity + synaptic dysfunction and/or neurodegeneration) | Positive | Positive | Negative |
| Stage 3 (Amyloid positivity + neurodegeneration + subtle cognitive decline) | Positive | Positive | Positive |
Clearly, the NIA-AA Criteria for diagnosing MCI due to AD (Table 1) and for characterizing preclinical AD (Table 2) involve the use of multi-modality imaging such as amyloid-PET, FDG-PET, MRI, and possibly the recently introduced tau-PET. These imaging modalities can be generally classified into two types: imaging for identifying amyloid positivity and imaging for identifying neuronal injury. Existing research focuses on investigating the relationship between the two types of imaging. Some studies interrogated the relationship between amyloid deposition and glucose metabolism by FDG-PET in cognitively normal individuals. Several groups found hypometabolism in cognitively normal individuals with significant amyloid deposition (7,8). In the study by Yi et al. (9), hypermetabolism was demonstrated in frontal and anterior temporal regions in cognitive normal APOE ε4 carriers but hypometabolism in temporoparietal regions. After adjusting for amyloid deposition, most of the hypometabolic regions disappear while the hypermetabolic regions still exist. This implied that while hypometabolism may be amyloid dependent in this patient cohort, hypermetabolism was not. Knopman et al. showed that cognitively normal individuals with significant amyloid deposition at baseline demonstrate significant FDG hypometabolism at follow-up (7,10). Several studies also found that individuals with both markers of amyloid deposition and neurodegeneration were more likely to develop cognitive impairments at follow-up (10,11). On the other hand, several studies found that there were cognitively normal, elderly individuals who had at least one significant neurodegeneration marker including FDG hypometabolism but did not have detectable amyloid deposition (11–13). This group of individuals were considered to be more likely to have other preclinical pathophysiologic processes than AD, such as cerebrovascular disease, tauopathies, or synucleinopathies (12), and therefore fall into a special category called Suspected Non-Alzheimer Pathology (SNAP).
While Aβ can be measured by both CSF and PET imaging, the latter allows for cerebral spatial patterns (i.e., topographies) of amyloid deposition to be examined. Brier et al. studied the relationship between Aβ topograghy and tau topography measured by amyloid and tau PET imaging, respectively, in a cohort of 36 cognitively normal elderly and 10 with mild AD (14). Singular Vector Decompositions (SVDs) were performed on the tau and Aβ burdens of originally extracted 42 ROIs, respectively. Each SVD produced two significant components representing two important topographies contained in the imaging data. For both tau and amyloid imaging, the first topography corresponded to the mean of the image. The second PET tau topography was most strongly localized in the temporal lobe including the hippocampus. In contrast, the second PET Aβ topography was most strongly localized in frontal and parietal regions. This analysis demonstrated that PET imaging data for tau and Aβ exhibited strong autocorrelation across ROIs but that each had distinct topographies. Furthermore, this study showed that tau Aβ deposition in the temporal lobe more closely tracked dementia status and was a better predictor of cognitive performance than Aβ deposition in any region of the brain.
3. Use of multi-modality imaging for prognosis of AD at early stages
Equally important to diagnosis is prognosis which concerns quantification of disease progression such as estimation of the time to dementia onset or prediction of conversion within an interested time frame. A significant amount of existing work focuses on predicting MCI conversion to AD. In what follows, we will provide a detailed review of the existing studies. A brief summary can be found in Table 3. Comparison of the pros and cons between the studies is provided in Table 4.
Table 3.
Summary of studies using multi-modality imaging for AD prognosis at early stage (Acc.= Accuracy; Sens.=Sensitivity; Spec.=Specificity)
| Papers | Stage | Imaging modalities | Non-imaging data | Cross-sectional or longitudinal | ML model | Data source | Sample size | Acc. | Sens. | Spec. |
|---|---|---|---|---|---|---|---|---|---|---|
| Ritter et al. (16) | MCI | MRI, FDG-PET | CSF, neuropsychological testing, medical history, symptoms, neurological & physical exams, demographics | cross-sectional | SVM | ADNI | 86 MCI-c vs. 151 MCI-nc | 0.730 | 0.405 | 0.913 |
| Shaffer et al. (17) | MCI | MRI, FDG-PET | CSF, neuropsychological testing, APOE, age, education | cross-sectional | ICA and logistic regression | ADNI | 43 MCI-c vs. 54 MCI-nc | 0.716 | 0.853 | 0.862 |
| Zhang et al. (18) | MCI | MRI, FDG-PET | CSF | cross-sectional | MKL | ADNI | 51 AD, 43 MCI-c, 56 MCI-nc, 52 NC | 0.764 | 0.818 | 0.660 |
| Liu et al. (19) | MCI | MRI, FDG-PET | none | cross-sectional | LASSO & MKL | ADNI | 44 MCI-c vs. 56 MCI-nc | 0.678 | 0.649 | 0.700 |
| Zhang and Shen (20) | MCI | MRI, FDG-PET | CSF | cross-sectional | Multi-task learning & SVM | ADNI | 40 AD, 38 MCI-c, 42 MCI-nc, 47 NC | 0.739 | 0.686 | 0.736 |
| Cheng et al. (21) | MCI | MRI, FDG-PET | CSF | cross-sectional | Transfer learning & group lasso | ADNI | 51 AD, 43 MCI-c, 56 MCI-nc, 52 NC | 0.794 | 0.845 | 0.727 |
| Young et al. (22) | MCI | MRI, FDG-PET | CSF, APOE | cross-sectional | GP for AD vs. NC, applied to MCI | ADNI | 47 MCI-c vs. 96 MCI-nc | 0.741 | 0.787 | 0.656 |
| Hinrichs et al. (23) | MCI | MRI, FDG-PET | CSF, APOE, NeuroPshychological Status Exam scores | Longitudinal | MKL for AD vs. NC, applied to MCI | ADNI | 48 AD, 119 MCI, 66NC | 0.791 | N/A | N/A |
| Zhang et al. (24) | MCI | MRI, FDG-PET | CSF, MMSE, ADAS-Cog | Longitudinal | MKL | ADNI | 35 MCI-c vs. 50 MCI-nc | 0.784 | 0.79 | 0.780 |
| Wang et al. (25) | MCI | MRI, FDG-PET, flobetapir-PET | ADAS-Cog | cross-sectional | PLS | ADNI | 64 MCI-c vs. 65 MCI-nc | 0.861 | 0.813 | 0.907 |
Table 4.
Pros and cons of the studies in Table 3
| Papers | Pros | Cons |
|---|---|---|
| Ritter et al. (16) | Use different data imputation and feature selection algorithms | Concatenating features from different modalities does not preserve within-modality feature integrity. |
| Shaffer et al. (17) | Use ICA for voxel-based analysis and imaging feature/component extraction | Same as above |
| Zhang et al. (18) | Encapsulate features of each modality to preserve intra-modality integrity by MKL; can handle high-dimensional features | Kernel-based classifier makes interpretation harder. |
| Liu et al. (19) | Multitask joint feature selection across different modalities | Same as above |
| Zhang and Shen (20) | Use multitask learning to exploit auxiliary data (MMSE and ADAS-Cog scores) | Same as above; lack of theoretical justification of why using auxiliary data would help despite empirically good performance. |
| Cheng et al. (21) | Use multitask learning to exploit auxiliary domain (AD vs. NC classification) | Same as above |
| Young et al. (22) | Use AD vs. NC classifier to classify MCI conversion | Based on an assumption yet to be validated that MCI converters are more like AD an non-converters more like NC |
| Hinrichs et al. (23) | Incorporate longitudinal image data | Kernel-based classifier makes interpretation harder. |
| Zhang et al. (24) | Incorporate longitudinal image data and perform feature selection | Same as above. |
| Wang et al. (25) | Use PLS to interrogate between-modality covariance structure to include as features | Hard to tell if the improved accuracy is due to the PLS method or inclusion of pathological imaging data. |
Some studies focused on understanding the roles that different imaging modalities play in predicting MCI conversion. For example, Jack et al. (15) studies the correlation of Aβ load and hippocampal volume with MCI time-to-conversion to AD. To measure Aβ load, they computed a global PiB-PET retention score formed by combining the cerebellum-adjusted standard uptake value ratios (SUVRs) of prefrontal, orbitofrontal, parietal, temporal, anterior cingulate and posterior cingulate/precuneus regions using a weighted average. For patients who did not have PiB-PET, the global scores were imputed using CSF Aβ42 and APOE ε4. Amyloid positivity was defined using a cutoff of 1.5. Then, cox proportional hazards models were fitted to estimate the effect of Aβ load and hippocampal volume on the relative hazard of progression. The major findings from this study include: (i) MCI patients with amyloid positivity were more likely to progress to AD than MCI with amyloid negativity (50% vs. 19% by two years). (ii) Among amyloid positive MCI patients, hippocampal atrophy predicted shorter time-to-conversion while amyloid load did not. (iii) In contrast, in the combined amyloid positive and negative MCI cohort, hippocampal atrophy and brain Aβ load predicted time-to-conversion with comparable power; and (iv) however, the effects of these two biomarkers differ. The risk profile is linear throughout the range of hippocampal atrophy values whereas the profile reaches a ceiling at higher values of brain Aβ load.
A typical approach adopted by relatively earlier studies for predicting MCI conversion was to concatenate features from multiple modalities into a combined feature set, which was then used to build a classifier. Because of the high dimensionality of the combined feature set, feature selection algorithms were commonly used before building a classifier. For example, Ritter et al. (16) composed a feature set that consisted of features from multiple imaging and non-imaging modalities, including MRI, FDG-PET, CSF, neuropsychological testing, medical history, medical symptoms at baseline, neurological and physical examinations, and demographic information. Feature selection was followed by a support vector machine (SVM) classifier to predict MCI converters and non-converters with three years from the ADNI datasets. Several algorithms were used to impute missing data, such as mean imputation and Expectation-Maximization (EM). Their approach achieved 73% accuracy in classifying 86 MCI converters and 151 non-converters based on 10-fold cross validation. Shaffer et al. (17) considered MRI, FDG-PET, CSF measurements, neuropsychological testing, APOE ε4, age, and education. Independent Component Analysis (ICA) was performed to extract four and nine components from MRI and FDG-PET voxel-based measurements, respectively. These components together with other non-imaging features were then used to build a logistic regression model, which achieved 71.6% accuracy in classifying 97 MCI converters and non-converters within four years from ADNI based on 10-fold cross-validation.
Different from the above-reviewed research that concatenated features from multiple modalities into a combined feature set, another line of research chose to “encapsulate” the features of each modality by themselves to better preserve intra-modality integrity and reveal inter-modality difference. Multiple kernel learning (MKL) is a commonly used approach to achieve this, in which features from the same modality were encapsulated into a kernel. The modality-wise kernels were then combined to make a classifier. Zhang et al. (18) used MKL to build a classifier for MCI and NC, which was further used to classify MCI converters and non-converters within 18 months. Two imaging modalities, MRI and FDG-PET, were used together with one non-imaging modality, CSF measurements. MRI volumetric features were extracted from 93 manually labeled ROIs. Regional FDG of these ROIs were computed and used as PET features. Using 10-fold cross validation, the MKL classifier achieved a classification accuracy of 76.4% (with a sensitivity of 81.8% and a specificity of 66%) on the ADNI datasets, while the best single-modality classifier achieved 72% accuracy.
Liu et al. (19) acknowledged that existing studies had shown evidence that combining multi-modality information improved the accuracy of AD-related classification. On the other hand, they pointed out that feature selection – an important step that warrants a good classifier – had typically been performed separately for each modality, which ignored the potential inter-modality relationship. Therefore, they proposed a multitask learning method to jointly select features from different modalities. The basic idea was to pose an additional constraint to the LASSO-based feature selection algorithm, which demanded the predictions using the features in modality A and B, ŷA and ŷB, to be similar if the features, xA and xB, are similar. They applied the proposed feature selection method together with MKL to classify 43 MCI converters and 56 non-converters within 18 months from the ADNI datasets, using gray matter volume from MRI and regional FDG of 93 manually labeled ROIs. The proposed method achieved an accuracy of 0.6783 based on 10-fold cross validation, which is better than methods using single modalities alone.
A classic approach for predicting MCI conversion, such as the afore-reviewed papers, is to train a classification model using the data of MCI converters and non-converters and then use the trained classifier to classify new MCI patients. Alternatively, researchers have tried different ways to incorporate “auxiliary data” to improve MCI classification accuracy. Zhang and Shen (20) developed a multi-modal multi-task learning method (M3T) to jointly predict MCI conversion and 2-year changes of MMSE and ADAS-Cog scores, which were treated as three tasks. Here, 2-year changes of MMSE and ADAS-Cog scores are considered “auxiliary data”. They used baseline MRI, FDG-PET, and CSF data as three modalities. Gray matter volume and regional FDG of 93 manually labeled ROIs and three CSF measurements were extracted from each modality and used as features. M3T includes a multi-task feature selection as its first step to select common features relevant to all the tasks, followed by building multi-modal SVM classification/regression models using the selected features for each task. M3T was applied to 167 subjects including 40 AD, 80 MCI (38 and 42 converters and non-converters), and 47 NC from ADNI datasets. Using 10-fold cross validation, M3T achieved 0.739 accuracy (0.686 sensitivity and 0.736 specificity) in classifying MCI converters and non-converters, which outperformed individual modality based methods and a simple concatenation method (CONCAT) which concatenated all MRI, FDG, and CSF features into a single feature vector and applied lasso for feature selection and SVM regression/classification.
Cheng et al. (21) pointed out that most existing studies classifying MCI converters and non-converters used data from MCI patients alone (called the target domain), but ignored data in other related domains such as classification of AD and NC (called the auxiliary domain). They believed that leveraging information from the auxiliary domain can improve MCI conversion prediction accuracy. Therefore, they proposed a domain transfer learning method that included three steps: First, a domain transfer feature selection algorithm was used to select features that are informative to both the target and auxiliary domains. This joint feature selection was achieved by imposing a group-lasso penalty on the weights of features in the two domains. Second, an instance transfer approach was used to select AD and NC subjects who are more separable than MCI converter vs non-converter separation to include in the target domain. Finally, a classifier was built using the features selected in the first step and the samples selected in the second step to predict MCI conversion. The proposed method was applied to 202 subjects from ADNI, including 51 AD patients, 99 MCI patients (43 converters and 56 non-converters with 24 months), and 52 NCs, using gray matter volume from MRI and regional FDG of 93 manually labeled ROIs as well as CSF measurements. Using 10-fold cross validation, the proposed method achieved 79.4% accuracy (83.5% sensitivity and 72.7% specificity), which was higher than methods not using information from the auxiliary domain.
Young et al. (22) proposed to build an AD vs. NC classifier using a Gaussian process (GP), which was then used to classify MCI converters and non-converters. Note that the difference between the work of Young et al. and the previously reviewed work by Cheng et al. is that the former did not use any MCI data to build the classifier. The proposed method was used to classify 96 MCI non-converters and 47 converters within three years from the ADNI datasets, using gray matter volume from MRI and regional FDG of 27 pre-selected ROIs as well as CSF measurements and APOE. Using leave-one-out cross validation, their model achieved an accuracy of 74%.
Most existing work as reviewed previously focused on using baseline multi-modality imaging to predict MCI conversion. A few researchers explored the use of longitudinal multi-modality image data. Hinrichs et al. (23) used multi-kernel learning (MKL) to build an AD vs. NC classifier, which was then used to compute a Multi-Modality Disease Marker (MMDM) for each MCI patient. The MMDM was used to classify MCI patients as converters, stablers, and reverters within three years. They used MRI and FDG-PET at baseline and roughly 24 months as well as non-imaging data including CSF assays of AB1-42, total Tau, and P-tau 18, NeuroPshychological Status Exam scores (NPSEs), and APOE genotype. For MRI processing, voxel-based morphometry (VBM) was used to process the scans at baseline and 24 months, and voxel intensities were used as features. Also, longitudinal MRI processing was performed using the tensor-based morphometry (TBM) approach in SPM5 to obtain the amount of volume change by taking the determinant of gradient of deformation at a single-voxel level. For FDG-PET processing, voxels of the scan at each time point were scaled to each individual’s Pons average FDG uptake value. Also, longitudinal features were calculated as voxel-wise difference and ratio between scans at the two time points. MKL was applied to build a classifier of 46 AD and 66 NC subjects from ADNI using a number of modalities. The MRI modality includes VBM features at baseline and 24 months and TBM-based longitudinal features. The FDG modality includes normalized voxel-level measurements at baseline and 24 months as well as longitudinal features of voxel-wise difference and ratio. Three non-imaging modalities include CSF, NPSE2, and APOE. This AD vs. NC classifier was applied to 119 MCI patients, giving an MMDM for each patient, which was used to predict each patient as a converter, stabler, or reverter within three years. The proposed method was demonstrated using 10-fold cross validation. Results showed that using all the imaging and non-imaging modalities as mentioned above achieved 81.40% accuracy, 79.69% sensitivity and 83.08 specificity, which outperforms separately using imaging data at baseline, longitudinal image data, NIPEs at baseline, longitudinal NIPEs, and biological measures.
Zhang et al. (24) used MRI, PET, MMSE, and ADAS-Cog at multiple time points to predict MCI conversion. The proposed method included two major steps: First, longitudinal feature selection was performed to select common brain regions across the multiple time points for MRI and PET, respectively. This was achieved by imposing a group-lasso penalty on a regression that predicted cognitive test scores using imaging data. Then, an MKL classifier was built using imaging features at each time point, longitudinal imaging features that reflected the rates of change over time, together with cognitive test scores at each time point. The proposed method was applied to 88 MCI subjects (35 converters and 50 non-converters) who had MRI, PET, MMSE, and ADAS-Cog data at five different time points (baseline, 6, 12, 18, and 24 months). MRI images were processed to produce gray matter, white matter, and CSF volumes of 93 ROIs, and PET images were processed to produce regional FDG of the 93 ROIs. Using 10-fold cross validation, the proposed method achieved a classification accuracy of 78.4%, a sensitivity of 79.0%, a specificity of 78.0%, and an AUC of 0.768, which outperformed methods using baseline data alone and using single modality alone.
In terms of imaging modalities, a vast majority of existing research, including all reviewed previously, used MRI and FDG-PET. The accuracy of classifying MCI converters and non-converters, even with combining CSF, cognitive scores, demographics, and longitudinal data, is below 80%. We were able to find only one paper that used florbetapir-PET together with MRI and FDG-PET, which achieved significantly improved classification accuracy. This paper is reviewed in more details as follows: Wang et al. (25) proposed two PLS-based approaches to classify MCI converters and non-converters using MRI, FDG-PET, and florbetapir-PET. The first approach, called informed PLS, worked by concatenating features from three modalities into a combined feature set and then using the combined set to train a PLS classifier. The second approach, called agnostic PLS, used PLS as a feature extractor not a classifier. Specifically, it used PLS to extract latent variables between two sets of imaging modalities. PLS is separately performed on MRI and FDG, MRI and florbetapir, FDG and florbetapir, MRI and FDG & florbetapir together, FDG and MRI & FDG and florbetapir together, florbetapir and MRI & FDG together, respectively. The extracted latent variables were used to train a Fisher’s LDA classifier. Both approaches were applied to 64 MCI converters and 65 non-converters from ADNI based on voxel-wise gray matter measurements of MRI, voxel-wise FDG normalized to the average FDG of the cerebrum (as the reference region), and voxel-wise standardized uptake value ratio (SUVR) normalized to the cerebellum (as the reference region for florbetapir-PET). Using leave-one-out cross validation, results showed that the informed PLS approach achieved 81.40% classification accuracy (79.69% and 83.08% sensitivity and specificity); agnostic PLS achieved 82.17% classification accuracy (81.25% and 83.03% sensitivity and specificity), both outperforming single-modality models. Moreover, by adding ADAS-cog scores, agnostic PLS achieve a better accuracy of 86.05% classification accuracy (81.25% and 90.77% sensitivity and specificity).
4. Multi-modality models with modality-wise missing data
In using multi-modality data to build a prediction or classification model, a commonly encountered challenge is that not all the subjects in a study cohort have all the modalities available. This is due to various reasons such as cost, availability of imaging equipment, lack of patient consent, and patient drop off (in longitudinal studies). For example, although MRI is used in almost every medical institute/clinic for AD-related diagnosis and prognosis, not many places have FDG-PET. The availability of amyloid-PET imaging is even more limited, especially that Medicare currently does not reimburse the cost of amyloid-PET imaging. Also, due to patient drop off, some patients may only have baseline MRI (or other imaging modalities) but not at later time points. Note that we call this problem “modality-wise missing data”, which is different from conventional missing data problems. The latter can typically be handled by imputation algorithms, while the former cannot because 1) there can be a substantial portion of patients who miss at least one modality, 2) it is possible that few patients have all the modalities available, and 3) the mechanism of missing data is clearly not “missing-at-random” which is a fundamental assumption that most imputation algorithms assume.
All the studies reviewed in the previous sections used datasets containing subjects with all modalities available, or with a small portion of missing data that can be imputed. No paper has been found that handled modality-wise missing data for diagnosis or prognosis at early stages of AD. However, there are a few papers developing methods to handle modality-wise missing data in classifying AD, MCI, and NC. Although this type of classification is outside the scope of this review, we would like to discuss these papers for purpose of raising awareness for the problem of modality-wise missing data and encouraging future research to apply the methods in these papers to the study of early stages of AD. In what follows, we discuss each paper in detail. Comparison of the pros and cons between these papers is provided in Table 5.
Table 5.
Pros and cons of the studies in Section 4
| Papers | Pros | Cons |
|---|---|---|
| Yuan et al. (26) | Allow for joint feature selection across cohorts with different missing modality patterns | Assume little correlation between modalities so same features are selected for each cohort; cannot do out-of-sample prediction |
| Xiang et al. (27) | Use two separate weights to achieve feature and modality selection; can do out-of-sample prediction | Many parameters to be estimated; assume a product form for modality-wise and feature-wise coefficients |
| Thung et al. (28) | Use multitask learning to reduce features and samples making computation easier | Conventional missing data imputation algorithms are used on reduced dataset; same features are selected for each cohort. |
| Liu et al. (29) | Exploiting subject relationship by multi-view hypergraph representation and fusion naturally gets around the issue of missing modalities. | Many parameters to be estimated; hard to identify important features; model needs to be re-learned using all the data every time new test data is available. |
| Li et al. (30) | Use DL to create “pseudo” PET from MRI; raw images are used for classification not features. | Creating PET from MRI needs justification from imaging physics; Black-box DL model is hard to interpret. |
Yuan et al. (26) proposed an incomplete multi-source feature learning method (iMSF) to classify AD, MCI, and NC using baseline MRI, FDG-PET, CSF, and proteomics as four modalities. Data of 780 subjects were downloaded from ADNI with 172 AD, 397 MCI, and 211 NC. Each subject had at least one modality. FreeSurfer was used to extract 305 features of MRI falling into five categories: average cortical thickness, standard deviation in cortical thickness, volumes of cortical parcellations, volumes of specific white matter parcellations, and total surface area of the cortex. 116 FDG-PET features were extracted using Automated Anatomical Labeling (AAL) corresponding to 116 ROIs. iMSF worked by first separating study subjects into multiple blocks, with each block having the same available modality/modalities. Then, one classifier was built for each block. Instead of building the classifiers separately, iMSF adopted an L21 penalty to force blocks sharing the same modality to use a common subset of features in that modality when building block-wise classifiers. iMSF was combined with four other methods to construct a classifier ensemble, which outperformed commonly used imputation methods, methods using single modalities, and the method that threw out missing data. This comparison was based on a training-test split of the dataset with the training set containing from 50% to 75% of the data.
Xiang et al. (27) proposed an Incomplete Source-Feature Selection model (ISFS), which was applied to the same datasets as Yuan et al (26). The advantages of ISFS over iMSF included that the former could select modalities that were most relevant to the classification and could be used to classify new subjects whose modality availability was different from the training set (called “out-of-sample” classification), while iMSF did not have these two capabilities. The basic idea of ISFS was to estimate the weights β that combined features to produce a classification result as well as the weights α that combined the modalities. By penalizing β and α, both feature and modality selections could be achieved. Although such a bi-level learning model has been studied in the machine learning literature, Xiang et al. innovated it by further considering modality-wise missing data and allowing α to vary across different blocks. This paper showed that ISFS outperformed iMSF and other competing methods especially when the training sample size was small. This comparison was based on a training-test split of the dataset with the training set containing 10% or 50% of the data.
Thung et al. (28) developed a matrix completion method for classification of AD, MCI, and NC, and prediction of three cognitive test scores (CDR global, CDR-SB2 and MMSE). The basic idea of the proposed method is to apply multitask learning twice, one on features and the other on samples, in order to reduce the original matrix composed of samples and features into a much smaller size. The reduced matrix was more “imputation-friendly” as it contained a smaller number of missing values, and therefore can be imputed using existing imputation algorithms. Specifically, in applying multitask learning on features, the original matrix was partitioned into small overlapping sub-matrices, each containing samples having at least one complete modality. Within each sub-matrix, a group-lasso based multitask learning algorithm was used to select a common subset of features across four tasks (one AD/MCI/NC classification task and three prediction tasks for the tree cognitive scores). In combining results from the sub-matrix-wise multitask learnings, features selected for at least one sub-matrix were kept for subsequent analysis. Next, multitask learning was applied to samples similar to the way that it was applied to features, but with samples in the test set treated as multiple output targets. The proposed method was applied to baseline MRI, FDG-PET, and CSF data of 807 subjects from ADNI (186 AD, 395 MCI and 226 NC). All of subjects had MRI, 397 had FDG-PET, and 406 had CSF. Gray matter volumes and regional FDG of 93 ROIs were used as MRI and FDG-PET features, respectively. Compared with conventional imputation algorithms and two state-of-the-art methods (iMSF, Ingalhaiikar’s algorithm), the proposed method achieved higher classification accuracy based on 10-fold cross validation.
Liu et al. (29) proposed a view-aligned hypergraph learning method (VAHL), which worked by first dividing the dataset into several views based on the availability of different modalities. Then, they computed the distances between subjects using a sparse representation model and constructed one hypergraph for each view. Coherence among different views was captured by a proposed view-aligned regularizer, which considered that if one subject is represented by two feature vectors in two views, the estimated class labels for such two feature vectors should be similar because they correspond to the same subject. Furthermore, they integrated the view-aligned regularizer into a classification framework. The proposed method was applied to baseline MRI, FDG-PET, and CSF data of 807 subjects from ADNI (186 AD, 169 pMCI, 226 sMCI, and 226 NC). A baseline MRI patient was called pMCI if converting to AD within 24 months and sMCI if not. Gray matter volumes and regional FDG of 90 ROIs were used as MRI and FDG-PET features, respectively. By comparing VAHL with four conventional imputation algorithms as well as a number of state-of-the-art methods (two ensemble based methods and the previously reviewed iMSF, ISFS, and matrix shrinkage completion methods), VAHL outperformed all the other methods in classification of MCI vs. NC and pMCI vs. sMCI based on 10-fold cross validation.
Li et al. (30) proposed a deep learning (DL) framework where the input and output are two imaging modalities, MRI and FDG-PET. A 3-D convolutional neural network (CNN) was built using a training set of patients for whom both MRI and PET were available. The trained 3-D CNN was then used to predict missing FDG-PET using MRI for patients who only have MRI. Furthermore, a classifier was built using MRI and predicted or real FDG-PET whichever is available. In image preprocessing, gray matter density maps from MRI and FDG-PET images rigidly aligned to the respective MRI images were smoothed using Gaussian kernels. Note that different from classic ML approaches, the 3-D CNN did not need feature extraction from the MRI and FDG-PET images but can directly use the 3-D images. The proposed method was applied to baseline MRI, FDG-PET, and CSF data of 830 subjects from ADNI (198 AD, 167 pMCI, 236 sMCI, and 229 NC). A baseline MRI patient was called pMCI if converting to AD within 18 months and sMCI if not. All the subjects had MRI but more than half did not have PET. Using a half-half training-test split, the proposed method achieved better accuracy in MCI vs. NC, AD vs. NC and sMCI vs. pMCI classifications compared with conventional imputation algorithms.
Finally, we would like to stress that all the aforementioned methods have strengths and limitations, as summarized in Table 5. Despite their respective unique strength, the common advantage of using these methods is to maximally utilize the available data. On the other hand, since each method is based on some assumption that may or may not be true for a real-world dataset, there also exist risks that there may be a mismatch between the assumption of a method and the characteristics of the dataset. Nevertheless, one could always opt for not using any of these methods but eliminating samples with missing modalities and including only samples with all modalities available for training a predictive model. This could be a viable approach if the amounts of samples with missing modalities are not considerably large.
5. Overall workflow – from image processing to decision support
The previous sections focus on reviewing statistical models and machine learning algorithms for building a diagnostic or prognostic model with imaging features available. This is only part of the workflow. Another important part of the workflow, upstream to diagnostic/prognostic model building, is image processing and feature computation. In this section, we summarize the major steps of this upstream building block and provide the entire workflow so that interested researchers and practitioners could follow through with their specific data.
The major steps included in MRI processing include 1) anterior commissure-posterior commissure (AC-PC) correction; 2) skull-striping followed by intensity inhomogeneity correction; 3) segmentation into grey matter (GM), white matter (WM), and CSF; 4) registration to a common anatomical space; 5) parcellation of GM into ROIs using an anatomically defined template. After these steps, volumetric measurement of each ROI is taken and used as features for subsequent machine learning models. PET processing primarily includes rigid transformation with respect to the corresponding MRI from the same subject. Then, the average intensity (e.g., FDG or SUVR depending on the type of PET imaging) within each ROI is computed. Normalization using the whole brain or a reference region is typically done for each PET ROI measurement. The normalized scores are used as features for the subsequent machine learning algorithms. Figure 1 provides a schematic diagram of the overall workflow.
Figure 1.
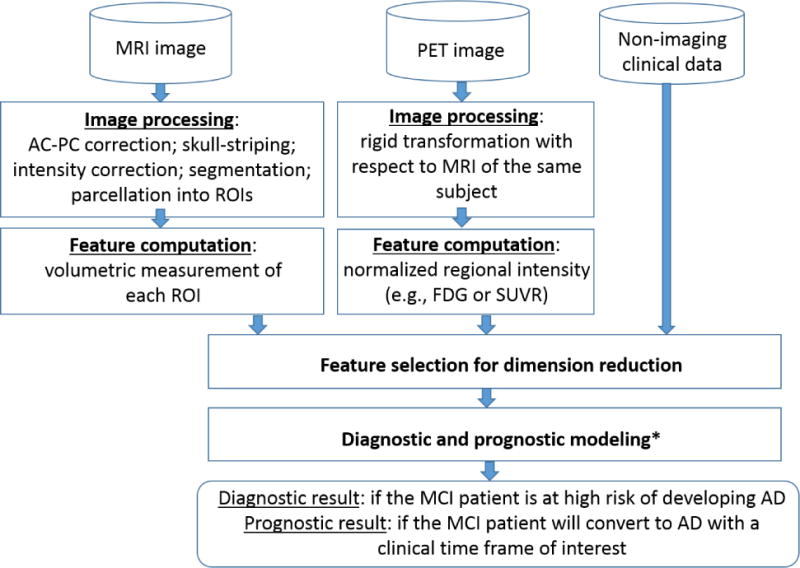
Overall workflow from image processing to diagnostic/prognostic decision support (Module with * has been reviewed in Sections 2-4; Module with * is combined with feature selection for dimension reduction in some approaches and may also be a separate step.)
6. Conclusion
In this paper, we reviewed research in the past decade focusing on using multi-modality imaging data for diagnosis and prognosis of AD at early stages. Based on the review, we have the following major findings:
All the works we have found and reviewed focused on MCI, while there is little research on the preclinical stage. It would be of great interest to perform and review studies focusing on the progression from normal aging to MCI, which would provide even earlier alarming and effective preventative strategies.
In the study of MCI, most existing research focuses on building classifiers that used multi-modality imaging and non-imaging data to predict MCI conversion to AD. The accuracy is generally below or barely above 80% with MRI and FDG-PET, even with inclusion of longitudinal imaging data. There is only one paper that additionally included florbetapir-PET and obtained over 80% classification accuracy. This provides evidence that including pathologic imaging helps the prognosis.
Almost all the existing research on prognosis formulates the problem into a binary classification problem of MCI conversion by a certain time of interest. Disease progression is indeed on a continuous spectrum. Even for two patients who convert to AD within two years, their paths of progression could be much different. Prognostic models that predict the path of progression would provide great clinical value for properly intervening or managing the disease. This would need new model development that goes beyond binary classification to, for example, multi-class that represents different progression trajectories.
Modality-wise missing data is a common problem when multi-modality imaging is used to make a prediction. However, little research has been found to address the problem of modality-wise missing data in developing a diagnostic or prognostic models at early stages of AD, though there are several recent studies focusing on classification of AD, MCI, and NC.
Classic machine learning algorithms are still the mainstream methodologies used in the existing studies. DL-based algorithms have only been limited applied to early AD diagnosis and prognosis using multi-modality imaging, despite their popularity in other areas of computer vision.
Driven by these findings, we provide some recommendations on future research directions:
First, future research can gear more toward using multi-modality imaging for diagnosis and prognosis of AD at the preclinical stage. It has been found that more than one third of cognitively healthy individuals over 65 have moderate to high levels of brain β-amyloidosis, who have a higher risk of developing AD (4). Optimally, treatment trials should target the presymptomatic or preclinical stage, i.e., before significant cognitive impairment, to lower Aβ burden or decrease neurofibrillary tangle in order to prevent subsequent neurodegeneration and eventual cognitive decline.
Second, imaging that provides pathologic biomarkers of AD such as amyloid PET should be combined with imaging modalities providing neuronal injury biomarkers such as MRI and FDG-PET in order to achieve a better prognosis at the early stages. An immediate, related challenge to obtaining amyloid imaging is that the modality is not presently widely available in clinics, which makes it very important to develop prognostic models that can make use of multi-modality images with modality-wise missing data. These models have greater, broader clinical utility than models that have to assume the availability of complete data or that are based on conventional missing data assumptions.
Last but not least, with the rapid growth of DL research in the AI societies and the proven effectiveness of DL applications in a variety of areas of computer vision including medical imaging, early-stage diagnosis and prognosis of AD using multi-modality imaging can benefit from DL development. More research is expected along this line.
Acknowledgments
This research is supported by the National Institute on Aging of the National Institutes of Health under award number R41AG053149. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Also, all authors have read the journal’s authorship agreement. All authors have read the journal’s policy on disclosure of potential conflicts of interest. The authors do not have anything to disclose.
Abbreviations
- AD
Alzheimer’s disease
- MCI
mild cognitive impairment
- MRI
magnetic resonance imaging
- FDG
fluorodeoxyglucose
- PET
positron emission topography
- FDA
food and drug administration
- NIA
national institute of aging
- AA
Alzheimer’s association
- IWG
international working group
- ML
machine learning
- AI
artificial intelligence
- NC
normal control
- CSF
cerebrospinal fluid
- SNAP
suspected non-Alzheimer pathology
- SVDs
singular vector decompositions
- SUVRs
standard uptake value ratios
- SVM
support vector machine
- ICA
independent component analysis
- MKL
multiple kernel learning
- ROI
region of interest
- ADNI
Alzheimer’s disease Neuroimaging Initiative
- MMSE
Mini-Mental State Examination
- ADAS-Cog
Alzheimer’s disease Assessment Scale-Cognitive subscale
- GP
Gaussian Process
- NPSEs
Neuropsychological Status Exam Score
- VBM
voxel-based morphometry
- TBM
tensor-based morphometry
- AUC
area under the curve
- PLS
partial least square
- CDR
clinical dementia rating
- CNN
convolutional neural network
- DL
deep learning
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Association A. 2016 Alzheimer’s disease facts and figures. Alzheimer’s Dement [Internet] 2016;12(4):459–509. doi: 10.1016/j.jalz.2016.03.001. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1552526016000856. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association. FDA-approved t reatments for Alzheimer’s. 2017 Available from: https://www.alz.org/dementia/downloads/topicsheet_treatments.pdf.
- 3.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement [Internet] 2011;7(3):280–92. doi: 10.1016/j.jalz.2011.03.003. Available from: http://dx.doi.org/10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement [Internet] 2011;7(3):270–9. doi: 10.1016/j.jalz.2011.03.008. Available from: http://dx.doi.org/10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrillo MC, Dean RA, Nicolas F, Miller DS, Berman R, Khachaturian Z, et al. Revisiting the framework of the National Institute on Aging-Alzheimer’s Association diagnostic criteria. Alzheimer’s Dement. 2013;9(5):594–601. doi: 10.1016/j.jalz.2013.05.1762. [DOI] [PubMed] [Google Scholar]
- 6.Visser PJ, Vos S, Van Rossum I, Scheltens P. Comparison of international working group criteria and national institute on aging-alzheimer’s association criteria for alzheimer’s disease. Alzheimer’s Dement [Internet] 2012;8(6):560–3. doi: 10.1016/j.jalz.2011.10.008. Available from: http://dx.doi.org/10.1016/j.jalz.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe VJ, et al. Selective Worsening of Brain Injury Biomarker Abnormalities in Cognitively Normal Elderly Persons With β-Amyloidosis. JAMA Neurol [Internet] 2013;70(8):1030. doi: 10.1001/jamaneurol.2013.182. Available from: http://archneur.jamanetwork.com/article.aspx?doi=10.1001/jamaneurol.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe VJ, Kemp BJ, Jack CR, Senjem M, Weigand S, Shiung M, et al. Comparison of 18F-FDG and PiB PET in Cognitive Impairment. J Nucl Med [Internet] 2009;50(6):878–86. doi: 10.2967/jnumed.108.058529. Available from: http://jnm.snmweb.org/cgi/doi/10.2967/jnumed.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi D, Lee DY, Sohn BK, Choe YM, Seo EH, Byun MS, et al. Beta-Amyloid Associated Differential Effects of APOE ε4 on Brain Metabolism in Cognitively Normal Elderly. Am J Geriatr Psychiatry [Internet] 2014 Oct;22(10):961–70. doi: 10.1016/j.jagp.2013.12.173. [cited 2017 Sep 9] Available from: http://linkinghub.elsevier.com/retrieve/pii/S1064748114000025. [DOI] [PubMed] [Google Scholar]
- 10.Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe V, et al. Short-term clinical outcomes for stages of NIA-AA preclinical Alzheimer disease. Neurology [Internet] 2012 May 15;78(20):1576–82. doi: 10.1212/WNL.0b013e3182563bbe. [cited 2017 Sep9] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22551733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth M, Villeneuve S, Haase CM, Madison CM, Oh H, Landau SM, et al. Associations Between Alzheimer Disease Biomarkers, Neurodegeneration, and Cognition in Cognitively Normal Older People. JAMA Neurol [Internet] 2013 Oct 28;77(17):1619–28. doi: 10.1001/jamaneurol.2013.4013. [cited 2017 Sep 9] Available from: http://archneur.jamanetwork.com/article.aspx?doi=10.1001/jamaneurol.2013.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack CR, Knopman DS, Weigand SD, Wiste HJ, Vemuri P, Lowe V, et al. An operational approach to National Institute on Aging-Alzheimer’s Association criteria for preclinical Alzheimer disease. Ann Neurol [Internet] 2012 Jun 1;71(6):765–75. doi: 10.1002/ana.22628. [cited 2017 Sep 9] Available from: http://doi.wiley.com/10.1002/ana.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe VJ, et al. Brain injury biomarkers are not dependent on β-amyloid in normal elderly. Ann Neurol [Internet] 2013 Apr 1;73(4):472–80. doi: 10.1002/ana.23816. [cited 2017 Sep 9] Available from: http://doi.wiley.com/10.1002/ana.23816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brier MR, Gordon BA, Friedrichsen K, McCarthy J, Stern A, Christensen J, et al. Tau and Aβ imaging, CSF measures, and cognition in Alzheimer’s disease. Sci Transl Med [Internet] 2016;8(338):338ra66. doi: 10.1126/scitranslmed.aaf2362. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27169802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR, Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133(11):3336–48. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritter K, Schumacher J, Weygandt M, Buchert R, Allefeld C, Haynes J-D. Multimodal prediction of conversion to Alzheimer’s disease based on incomplete biomarkers∗This work was supported by the Bernstein Computational Program of the German Federal Ministry of Education and Research (01GQ1001C, 01GQ0851, GRK 1589/1), the Europe. Alzheimer’s Dement Diagnosis. Assess Dis Monit [Internet] 2015;1(2):206–15. doi: 10.1016/j.dadm.2015.01.006. Available from: http://linkinghub.elsevier.com/retrieve/pii/S2352872915000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Coleman RE, et al. Predicting Cognitive Decline in Subjects at Risk for Alzheimer Disease by Using Combined. Radiology. 2013;266(2):583–91. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage [Internet] 2011;55(3):856–67. doi: 10.1016/j.neuroimage.2011.01.008. Available from: http://dx.doi.org/10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s Disease and mild cognitive impairment identification. Neuroimage [Internet] 2014;84:466–75. doi: 10.1016/j.neuroimage.2013.09.015. Available from: http://dx.doi.org/10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage [Internet] 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. Available from: http://dx.doi.org/10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng B, Liu M, Suk H, II, Shen D, Zhang D, Munsell BC, et al. Domain Transfer Learning for MCI Conversion Prediction. Brain Imaging Behav. 2015;9(4):1805–17. doi: 10.1109/TBME.2015.2404809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. NeuroImage Clin [Internet] 2013;2(1):735–45. doi: 10.1016/j.nicl.2013.05.004. Available from: http://dx.doi.org/10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: An analysis of MCI progression in the ADNI population. Neuroimage [Internet] 2011;55(2):574–89. doi: 10.1016/j.neuroimage.2010.10.081. Available from: http://dx.doi.org/10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, Chen K, Yao L, Hu B, Wu X, Zhang J, et al. Multimodal classification of mild cognitive impairment based on partial least squares. J Alzheimer’s Dis. 2016;54(1):359–71. doi: 10.3233/JAD-160102. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. Neuroimage [Internet] 2012;61(3):622–32. doi: 10.1016/j.neuroimage.2012.03.059. Available from: http://dx.doi.org/10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang S, Yuan L, Fan W, Wang Y, Thompson PM, Ye J. Bi-level multi-source learning for heterogeneous block-wise missing data. Neuroimage [Internet] 2014;102(P1):192–206. doi: 10.1016/j.neuroimage.2013.08.015. Available from: http://dx.doi.org/10.1016/j.neuroimage.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thung KH, Wee CY, Yap PT, Shen D. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. Neuroimage [Internet] 2014;91:386–400. doi: 10.1016/j.neuroimage.2014.01.033. Available from: http://dx.doi.org/10.1016/j.neuroimage.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Zhang J, Yap PT, Shen D. View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Med Image Anal [Internet] 2017;36:123–34. doi: 10.1016/j.media.2016.11.002. Available from: http://dx.doi.org/10.1016/j.media.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Zhang W, Suk H, Wang L, Li J, Shen D, et al. Deep learning Based imaging data completion for Improved Brain Disease Diagnosis. 2014:305–12. doi: 10.1007/978-3-319-10443-0_39. [DOI] [PMC free article] [PubMed] [Google Scholar]


