Abstract
Objectives
To extend the coverage of phenotypes in SNOMED CT through post-coordination.
Methods
We identify frequent modifiers in terms from the Human Phenotype Ontology (HPO), which we associate with templates for post-coordinated expressions in SNOMED CT.
Results
We identified 176 modifiers, created 12 templates, and generated 1,617 post-coordinated expressions.
Conclusions
Through this novel approach, we can increase the current number of mappings by 50%.
Keywords: phenotype, SNOMED CT, ontology, post coordination
Introduction
While the usefulness of coarse phenotyping based on electronic health record (EHR) data has been demonstrated in the context of recent genomic studies (e.g., [1]), the study of rare syndromes requires detailed phenotyping. More generally, deep phenotyping is required in order to understand how genetic variation relates to clinical manifestations [2]. Despite efforts to facilitate the adoption of standards for phenotyping across domains (e.g., PhenX project [3]), resources for phenotyping tend to vary between clinical data repositories used for translational research and in healthcare settings. For example, while somewhat overlapping, the Human Phenotype Ontology (HPO) used for annotation of research data and SNOMED CT used in EHRs have not been developed in a coordinated fashion and are only partially interoperable.
In previous work, we assessed the coverage of HPO terms in standard terminologies using simple lexical mapping through the UMLS [4]. Only 54% of HPO classes mapped to UMLS concepts and only 30% mapped to SNOMED CT. This simple approach only considered mapping to pre-coordinated terms in SNOMED CT. However, in addition to the pre-coordinated terms distributed with the terminology, SNOMED CT supports the creation of post-coordinated expressions, i.e., logical definitions based on the SNOMED CT concept model. The examination of HPO terms with no lexical mapping to SNOMED CT reveals that they can often be decomposed into simple elements, which could be mapped to SNOMED CT and aggregated into post-coordinated expressions.
For example, the HPO term “Renal Hypoplasia” [HPO:HP_0000089] maps to the (pre-coordinated) SNOMED CT concept “Congenital hypoplasia of kidney” [SCTID:32659003]), with synonym “Renal Hypoplasia”. In contrast, there is no pre-coordinated concept in SNOMED CT for the HPO term “Macular hypoplasia” [HPO:HP_0001104]. However, as shown with “Congenital hypoplasia of kidney”, the notion of congenital hypoplasia can be represented in SNOMED CT, which also provides a concept for the anatomical structure “Macula lutea structure” [SCTID:82859000]. Therefore, it is possible to create a post-coordinated expression for “Macular hypoplasia”, using the template provided by “Congenital hypoplasia of kidney”.
The main objective of this work is to extend the coverage of phenotypes in SNOMED CT through post-coordination, i.e., beyond simple lexical mapping to pre-coordinated terms. More specifically, we remove various types of modifiers in HPO terms in order to decompose them into semantic elements that can be recomposed into SNOMED CT expressions through post-coordination. We demonstrate that we can increase the current number of mappings by 50%.
Background
HPO
The Human Phenotype Ontology (HPO) is an ontology of phenotypic abnormalities, used for the annotation of databases such as OMIM (Online Mendelian inheritance in Man), Orphanet (knowledge base about rare diseases), and DECIPHER (RNAi screening project) [5]. The version of HPO used in this investigation is the (stable) OWL version downloaded on April 16, 2014 from the HPO website. It contains 10,491 classes and 16,414 names for phenotypes, including 5,923 exact synonyms, in addition to one preferred term for each class.
SNOMED CT
Developed by the International Health Terminology Standard Development Organization (IHTSDO), SNOMED CT is the world’s largest clinical terminology and provides broad coverage of clinical medicine, including findings, diseases, and procedures for use in electronic medical records [6]. It is implemented with a description logic backend and supports two types of concepts. Pre-coordinated concepts are named and defined in SNOMED CT. Post-coordinated concepts are made up of other concepts in a compositional approach [7]. The U.S. edition of SNOMED CT dated September 2013 is used in this work.
UMLS
The Unified Medical Language System (UMLS) is a terminology integration system developed by the U.S. National Library of Medicine [8]. The UMLS Metathesaurus integrates many standard biomedical terminologies, including SNOMED CT. Although the UMLS does not currently integrate HPO, it is expected to provide a reasonable coverage of phenotypes through its source vocabularies. In the UMLS Metathesaurus, synonymous terms from various sources are assigned the same concept unique identifier, creating a mapping among these source vocabularies. Terminology services provided by the UMLS support the lexical mapping of terms to UMLS concepts. Additionally, each UMLS concept is assigned one of the 15 Semantic Groups, which represent broad domains, including Disorders, Anatomy and Physiology. The 2013AB version of the UMLS is used in this work.
Related work
Researchers have investigated the representation of phenotypes through pre- and post-coordinated terms. Groza et al. developed an automated approach for decomposing skeletal phenotype concepts defined in HPO [9]. Oellrich et al. proposed an automated transformation of pre-coordinated phenotypes into Entity-Quality (EQ) statements for achieving interoperability between phenotype ontologies and compare their results to manually created EQ statements available for about half of the terms in HPO [10].
Besides our previous study [4], there have been a few efforts to study the coverage of phenotypes in standard terminologies such as SNOMED CT. Sollie et al. found that there are sizable gaps in SNOMED CT for metabolic disorders (and possibly for other classes of rare and genetic disorders) [11]. However, when Beck et al. assessed the suitability of HPO, ICD10, MeSH, SNOMED CT, and the Human Disease Ontology (DO) for describing GWAS traits, they concluded that, despite generally poor coverage (~20%), partial term matching to SNOMED CT is most successful [12].
These findings suggest, that there is a need for systematic improvement of the coverage of phenotypes in SNOMED CT, and that such an effort might benefit from decomposition techniques similar to prior work [9,10].
Specific contribution
The specific contribution of this work is to identify templates in SNOMED CT for the creation of post-coordinated expressions for phenotype concepts from HPO. This approach significantly extends the mapping to pre-coordinated concepts used in most mapping studies, including our earlier work on the coverage of HPO terms in SNOMED CT.
Methods
Our approach to assessing the coverage of HPO phenotypes in SNOMED CT can be summarized as follows. Starting from HPO terms with no lexical mapping to pre-coordinated SNOMED CT, we identify frequent modifiers and transformation rules in order to make HPO terms compatible with SNOMED CT. We examine the logical definition of existing phenotype concepts in SNOMED CT for templates. We then apply the transformation rules in order to decompose the HPO terms into simple elements. Finally, we combine them again into post-coordinated expressions consistent with the templates.
Establishing a list of modifiers and transformation rules for HPO terms
Starting from HPO terms with no lexical mapping to pre-coordinated SNOMED CT through the UMLS, we analyze word frequencies in order to identify frequent modifiers. More specifically, one author (FD) manually reviewed samples of terms containing words occurring more than 25 times in HPO terms (150 words). Frequent words include “abnormality [of]” (1126), “aplasia/hypoplasia” (134) and “congenital” (44). In addition, we noted frequent abbreviations for ordinals (e.g., “2nd”), while SNOMED CT typically uses the extended form (“second”).
We create four types of lexical transformations for making HPO terms compatible with SNOMED CT. These transformations are presented in increasing order of aggressiveness.
Level 1: Replace
We simply replace the abbreviations for ordinals by their expanded form. For example, the HPO term “Aplasia of the phalanges of the 4th toe” is transformed into “Aplasia of the phalanges of the fourth toe”.
Level 2: Split
HPO uses “/” for coordinating variants (e.g., “aplasia/hypoplasia”), while SNOMED CT does not. Here, we interpret “/” as disjunction, i.e., “aplasia or hypoplasia”. Therefore, we split expressions with words concatenated by “/” into two individual words, which we substitute to the corresponding expression in the term. For example, we transform the term “aplasia/hypoplasia of the thymus” into two terms, “aplasia of the thymus” and “hypoplasia of the thymus”, which both map to SNOMED CT.
Level 3: Demodification to disorder (D0)
HPO contains modifiers that specialize disorders (e.g., “congenital” specializes “bilateral cataract” to form “bilateral congenital cataract”). By removing these modifiers, we create a more general concept, which is more likely to map to SNOMED CT. For example, “bilateral cataract” maps to SNOMED CT.
Level 4: Demodification to anatomy, physiology or chemical substance (D1)
Another category of modifiers are those that denote a disorder (e.g., an abnormality) of a specific anatomical structure, physiological process or chemical substance. Examples of such terms include “Abnormality of the lip”, “Abnormality of intracranial pressure” and “Abnormality of prothrombin”. By removing “abnormality [of the]”, we extract the anatomical structure (e.g., lip), physiologic process (e.g., intracranial pressure) or chemical substance (e.g., prothrombin), which is the object of the abnormality.
Identifying templates for phenotype concepts
As shown in the example presented in the introduction, some pre-coordinated SNOMED CT concepts include the very modifiers (or constructs) we have identified as generally preventing the mapping to SNOMED CT. For example, the SNOMED CT concept “Congenital hypoplasia of kidney” contains “Hypoplasia”, which we have identified as a modifier (at the D1 level). Such SNOMED CT concepts therefore suggest valuable templates for the decomposition of similar concepts. For example, the logical definition of the concept “Congenital hypoplasia of kidney” in SNOMED CT suggests a template for the modifier “Hypoplasia”.
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Hypoplasia (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some ‘Kidney structure (body structure)’)))
The template suggested by this SNOMED CT concept is “Congenital hypoplasia of <ANATOMICAL STRUCTURE>” and corresponds to the following definition:
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Hypoplasia (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some ‘<ANATOMICAL STRUCTURE> ‘)))
where <ANATOMICAL STRUCTURE> is what remains from the original term after demodification.
Applying transformation rules to HPO terms
We apply the transformation rules to HPO terms in increasing order of aggressiveness, from level 1 to level 4. Moreover, at a given level, the transformation rule is applied to not only the original terms, but also to the terms produced by all previously applied rules (at lower levels).
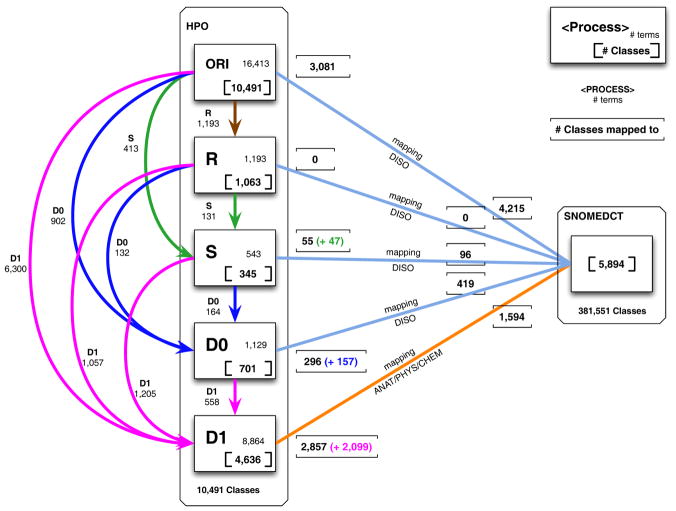
In practice, as shown in Figure 1, we first apply the Replace rule to the original terms (brown path). We then apply the Split rule to the original terms and to the terms produced by the Replace rule (green paths). The D0 rule is applied to the original terms and to the terms produced by the Replace and Split rules (blue paths). Finally, we apply the D1 rule to all the terms (pink paths).
Figure 1.
Levels of transformations for the 5,384 mappings between HPO and SNOMEDCT Classes (ORI : original HPO terms; R=Replace transformation; S=Split transformation; D0/D1= Demodification transformations)
For example, starting from the original term “Congenital adrenal gland hypoplasia”, the rules Replace and Split do not produce any results. The modifier “Congenital” is removed by rule D0, producing the demodified disorder term “adrenal gland hypoplasia”. Finally, the modifier “hypoplasia” is removed by rule D1, producing the term “adrenal gland”, which corresponds to an anatomical structure.
Mapping demodified HPO terms to SNOMED CT concepts
Having removed the modifiers according to the transformation rules, we map all transformed terms to SNOMED CT through the UMLS using simple lexical mapping techniques [4]. More specifically, we attempt an exact match, followed by a normalized match using the functions provided by the UMLS Terminology Services API.
It is important to note that the first three rules (Replace, Split and D0) are expected to produce more general terms for disorders, and yield mappings to disorder concepts in SNOMED CT (light blue links on Figure 1). In contrast, the D1 rule produces terms for anatomical structures, physiologic processes or chemical substances. These terms are expected to map to entities of these types in SNOMED CT (orange link on Figure 1). For example, the term “factor XIII” (demodified from the HPO concept “Reduced factor XIII activity” [HPO:HP_0008357]) is mapped to the SNOMED CT concept “Factor XIII” [SNCTID: 319930009].
Of note, the original term and several demodified terms that derive from it may map to SNOMED CT. For example, “Congenital adrenal gland hypoplasia” (original HPO term), “adrenal gland hypoplasia” (produced by D0) and “adrenal gland” (produced by D1) all map to SNOMED CT. In this case, precedence is given to the mapping from the original term or to the mapping derived from the less aggressive transformation.
Creating post-coordinated expressions for HPO terms
In this preliminary work, we focus on expressions for D1 level modifiers, because D1 transformation rules tend to be more productive compared to D0 rules.
Using the templates suggested by existing SNOMED CT concepts, we generate post-coordinated expressions by inserting into the template the anatomical structure, physiologic process or chemical substance extracted by the transformation rule D1. (An occasional D0 modifier may also have been removed from the term, but the final mapping happens at level D1.)
For example, the term “Macular” is extracted from the original HPO term “Macular hypoplasia” [HPO:HP_0001104] by the D1 rule. It is mapped to the SNOMED CT concept “Macula lutea structure (body structure)” [SNCTID:362517001]. The modifier “hypoplasia” is associated with the template “Congenital hypoplasia of <ANATOMICAL STRUCTURE>”, into which we insert the anatomical structure concept “Macula lutea structure (body structure)”. The resulting logical definition for “Macular hypoplasia” is as follows.
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Hypoplasia (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some Macula lutea structure (body structure)’)))
Results
Establishing a list of modifiers and transformation rules for HPO terms
As mentioned earlier, we identified four transformation rules (Replace, Split, D0 and D1). As shown in Table 1, each rule is associated with a list of substitution patterns for replacement (Replace, Split) or modifiers to be removed (D0, D1). The number of patterns/modifiers per transformation rule ranges from 5 (Replace) to 69 (Split). The most frequent modifiers in HPO terms are those from the D1 list (65 modifiers found in 8,864 HPO terms).
Table 1.
List and count of modifiers organized in 4 levels: R (REPLACE), S (SPLIT), D0 and D1.
| Description | n |
|---|---|
|
Level R: Modifiers preventing terms from being mapped to disorders Modifiers for substitution patterns (one-to-one) 1st => first, 2nd => second, 3rd => third, 4th => fourth, 5th => fifth |
5 (present in 1,193 HPO terms) |
|
Level S: Modifiers preventing terms from being mapped to disorders Modifiers for substitution patterns (one-to-two) Terms containing a “/” are split into two terms, for example: Aplasia/Hypoplasia of kidney => Aplasia of kidney; Hypoplasia of kidney |
69 (present in 543 HPO terms) |
|
Level D0: Modifiers preventing terms from being mapped to disorders asymmetric, asymmetrical, bilateral, complete, congenital, cutaneous, generalized, lethal, marked, mildly, multiple, nearly, osseous, partial, patchy, severely, symmetric, symmetrical, unilateral, Alcohol-induced, Aminoglyco-side-induced, Anesthetic-induced, Aspirin-induced, Cold-induced, Drug-induced, Effort-induced, Exercise-induced, Fava bean-induced, Heparin-induced, Radiation-induced, Stress-induced, infection-induced, Viral infection-induced, Warfarin-induced, “, recurrent”, “, acute”, “, chronic” |
37 (present in 1,129 HPO terms) |
|
Level D1: Modifiers preventing terms from being mapped to anatomical structures/physiological processes/chemical substances abnormal, abnormality, absence, absent, activity, agenesis, aplasia, aplastic, atresia[s], atrophy, bracket, broad, bullet shaped, bullet-shaped, chevron shaped, chevron-shaped, cone shaped, cone-shaped, contractures, curved, decreased, deficiency, degeneration, delayed, duplication, dystrophy, eeg, elevated, enlarged, fragmentation, hypoplasia, hypoplasic, hypoplastic, impaired, increased, irregular, ivory, loss, lytic defect[s], malrotation, number[s], osteolytic defect[s], prominent, pseudoepiphysis, reduced, rhomboid shaped, rhomboid-shaped, rudimentary, sclerosis, shortened, shortening, small, sparse, stippling, symphalangism, synostosis, thin, triangular, triangular shaped, triangular-shaped, wedge shaped, wedge-shaped, widened, widening |
65 (present in 8,864 HPO terms) |
Identifying templates for phenotype concepts
As shown in Table 2, we identified 12 templates corresponding to modifiers at the D1 level (with an occasional D0 modifier), corresponding to five distinct logical definitions. Each template has the form:
Table 2.
Templates and logical definitions for post-coordinated expressions (D1 level).
| Template | Logical definition | # Classes (terms) |
|---|---|---|
| {abnormal, abnormality of, abnormality of the, abnormality involving the }<monospace><ANATOMICAL STRUCTURE></monospace> <monospace><ANATOMICAL STRUCTURE></monospace>{abnormal} |
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Developmental anomaly (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some <monospace><ANATOMICAL STRUCTURE></monospace>))) |
618 (714) |
| {aplastic, aplasia of, aplasia of the, aplasia involving the, absence of}<monospace><ANATOMICAL STRUCTURE></monospace> <monospace>ANATOMICAL STRUCTURE></monospace>{agenesis, aplasia, absent} {congenital absence of, congenital aplasia of }<monospace><ANATOMICAL STRUCTURE></monospace> |
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Congenital absence (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some <monospace><ANATOMICAL STRUCTURE></monospace>))) |
415 (977) |
| {hypoplastic, hypoplasia of, hypoplasia of the, hypoplasia involving, hypoplasia involving the, hypoplasia affecting the}<monospace><ANATOMICAL STRUCTURE></monospace> <monospace><ANATOMICAL STRUCTURE></monospace>{hypoplasia} {congenital hypoplasia of}<monospace><ANATOMICAL STRUCTURE></monospace> |
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Hypoplasia (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some <monospace><ANATOMICAL STRUCTURE></monospace>))) |
409 (853) |
| {duplication of, duplication of the, duplication involving}<monospace><ANATOMICAL STRUCTURE></monospace> <monospace><ANATOMICAL STRUCTURE></monospace>{duplication} {complete duplication of, complete duplication of the}<monospace><ANATOMICAL STRUCTURE></monospace> |
‘Disease (disorder)’ and (‘Role group (attribute)’ some ((‘Associated morphology (attribute)’ some ‘Double structure (morphologic abnormality)’) and (‘Occurrence (attribute)’ some ‘Congenital (qualifier value)’) and (‘Finding site (attribute)’ some <monospace><ANATOMICAL STRUCTURE></monospace>))) |
140 (232) |
| {bilateral, D1} <monospace><ANATOMICAL STRUCTURE></monospace> examples : {bilateral aplasia}<monospace><ANATOMICAL STRUCTURE></monospace> {bilateral}<monospace><ANATOMICAL STRUCTURE></monospace>{aplasia} {bilateral absence of}<monospace><ANATOMICAL STRUCTURE></monospace> … |
‘Disease (disorder)’ and (‘Role group (attribute)’ some … D1 logical definition here … and (‘Finding site (attribute)’ some ‘left <monospace><ANATOMICAL STRUCTURE></monospace>’)) and (‘Role group (attribute)’ some … D1 logical definition here … and (‘Finding site (attribute)’ some ‘right <monospace><ANATOMICAL STRUCTURE><monospace>’)) |
35 (63) |
| TOTAL = 1,617 (2,839) | ||
{modifier1, modifier2, …, modifiern} <ENTITY TYPE>
(e.g., {absence of}<ANATOMICAL STRUCTURE>)
Or : <ENTITY TYPE>{modifier1, modifier2, …, modifiern}
(e.g., <ANATOMICAL STRUCTURE>{absent})
Each template is associated with a logical definition. Multiple templates can share the same logical definition. For example, the two templates listed above have the same logical definition. One of the authors (JTC) familiar with the SNOMED CT concept model inspected the templates for validity.
Applying transformation rules to HPO terms
The results of the successive application of transformation rules to HPO terms are depicted in Figure 1. The number of terms generated by each rule is listed next to the corresponding arrow. For example, 1,057 terms result from transformation D1 applied to the 1,193 unique terms resulting from the Replace transformation (pink path R D1).
Mapping demodified HPO terms to SNOMED CT concepts
As we showed in our previous work [4], the mapping of the 16,413 original HPO terms to SNOMED CT through UMLS results in 3,081 HPO classes mapped to 4,215 SNOMED CT disorder concepts. Additionally, a total of 2,865 modified HPO terms (2,109 unique HPO classes) results from the three transformation processes (Replace, Split, D0). These disorder terms map to 515 SNOMED CT classes. The Replace transformation does not yield any new mappings. The Split transformation yields 55 mappings, of which 47 are new (i.e., not produced earlier). Transformation D1 yields 2,857 mappings to anatomical structure, physiologic process or chemical substance concepts in SNOMED CT, 2,099 of which are new. The details of the mapping results can be found in Figure 1. Note that these mappings to pre-coordinated concepts in SNOMED CT only contribute to the post-coordination strategy developed in this paper.
Creating post-coordinated expressions for HPO terms
In this proof-of-concept investigation, we identified 12 templates involving 10 D1 modifiers (and 3 additional D0 modifiers removed when necessary). The instantiation of the logical definitions associated with these templates covers 1,617 HPO classes. Compared to the 3,081 mappings to pre-coordinated SNOMED CT concepts, this approach increases the coverage of HPO classes in SNOMED CT by roughly 50% through post-coordination.
Discussion
Our approach is very productive, yielding substantial gain in terms of number of mappings. More specifically, we developed post-coordinated expressions for 1,617 HPO classes that were not previously mapped to SNOMED CT classes, increasing the total number HPO classes mapped to SNOMED CT by 50%.
A high level of implicit knowledge was observed in HPO. For example, the notion of congenitality is usually assumed, rather than stated in HPO terms. While understandable in the context of clinical genetic phenotypes, it hinders the mapping to other terminologies. In contrast, SNOMED CT uses the qualifier value “congenital” to explicitly denote congenitality. Moreover, the finer-grained definitions supported by post-coordination make it possible to represent laterality (unilateral, bilateral, left, right), which is generally not represented in pre-coordinated terminologies.
Some of the templates we created do not fit the SNOMED CT concept model (e.g., {EEG with} <PHYSIOLOGICAL PROCESS> for electroencephalogram waveforms description) or lack specific pre-coordinated classes (e.g., for specific enzymes {decreased activity of} <CHEMICAL SUBSTANCE>). Additional work is needed to explore how these templates could be represented in SNOMED CT.
In future work, we want to identify templates for D0 modifiers. We also want to assess the logical validity of the post-coordinated expressions we created by classifying them with the description logics classifier used by SNOMED CT. Finally, we would like to test the applicability of this method for mapping other specialized terminologies to SNOMED CT.
Conclusion
In this preliminary study, we explored the automatic mapping of HPO terms to SNOMED CT through post-coordination. Through this novel approach, we were able to increase the current number of mappings by 50%.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine, the French Gynecology and Obstetrics Association (Collège National des Gynécologues et Obstétriciens Français), and the Philippe Foundation.
References
- 1.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc JAMIA. 2013 Jun;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennekam RCM, Biesecker LG. Next-generation sequencing demands next-generation phenotyping. Hum Mutat. 2012 May;33(5):884–6. doi: 10.1002/humu.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton CM, Strader LC, Pratt JG, Maiese D, Hendershot T, Kwok RK, et al. The PhenX Toolkit: get the most from your measures. Am J Epidemiol. 2011 Aug 1;174(3):253–60. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winnenburg R, Bodenreider O. Coverage of Phenotypes in Standard Terminologies. Proceedings of the Joint Bio-Ontologies and BioLINK ISMB’2014 SIG session “Phenotype Day.”; Boston, USA. 2014; pp. 41–4. [Google Scholar]
- 5.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014 Jan;42(Database issue):D966–74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud Health Technol Inform. 2006;121:279–90. [PubMed] [Google Scholar]
- 7.Rector A, Iannone L. Lexically suggest, logically define: Quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. J Biomed Inform. 2012 Apr;45(2):199–209. doi: 10.1016/j.jbi.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groza T, Hunter J, Zankl A. Decomposing Phenotype Descriptions for the Human Skeletal Phenome. Biomed Inform Insights. 2013 Feb 4;6:1–14. doi: 10.4137/BII.S10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oellrich A, Grabmüller C, Rebholz-Schuhmann D. Automatically transforming pre- to post-composed phenotypes: EQ-lising HPO and MP. J Biomed Semant. 2013 Oct 16;4:29. doi: 10.1186/2041-1480-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sollie A, Sijmons RH, Lindhout D, van der Ploeg AT, Rubio Gozalbo ME, Smit GPA, et al. A new coding system for metabolic disorders demonstrates gaps in the international disease classifications ICD-10 and SNOMED-CT, which can be barriers to genotype-phenotype data sharing. Hum Mutat. 2013 Jul;34(7):967–73. doi: 10.1002/humu.22316. [DOI] [PubMed] [Google Scholar]
- 12.Beck T, Free RC, Thorisson GA, Brookes AJ. Semantically enabling a genome-wide association study database. J Biomed Semant. 2012;3(1):9. doi: 10.1186/2041-1480-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]