Abstract
Background
We conducted an exploratory study to determine the prevalence of the PNPLA3 rs78409 [G] allele in Hmong as a risk factor for nonalcoholic fatty liver disease (NAFLD). NAFLD/NASH is the world’s most common chronic liver disease and is expected to replace viral hepatitis as the leading cause of cirrhosis and a potential precursor to hepatocellular carcinoma (HCC). Of all populations in California, Hmong experience the highest risk of death from HCC and the highest prevalence of metabolic syndrome risk factors among Asians that predispose them to NAFLD. We sought a genetic explanation that might contribute to the high rates of chronic liver disease in Hmong. Literature pointed to the PNPLA3 rs738409 [G] allele as a potential genetic culprit.
Methods
From 26 samples previously collected in community settings, we isolated cell-free DNA from serum samples. Quantitative PCR-based SNP genotyping analysis was performed with a validated TaqMan SNP Genotyping Assay and analyzed with TaqMan Genotyper Software.
Results
The PNPLA3 rs738409 [C>G] variant occurred at a frequency of 0.46 (12/26, 95% CI 0.27–0.67). This carrier rate would rank the Hmong as the third highest population in the 1000 Genomes Project.
Conclusions
While this small sample size limits generalizability, the high frequency rates of this allele along with the presence of metabolic syndrome risk factors would warrant further studies as to the etiology of NAFLD in Hmong.
Keywords: PNPLA3 rs738409, non-alcoholic steatohepatitis (NASH), Hmong, carrier rate, hepatocellular carcinoma
INTRODUCTION
Chronic liver diseases, specifically non-alcoholic fatty liver disease (NAFLD) and its pathologically more advanced form, non-alcoholic steatohepatitis (NASH), along with hepatocellular carcinoma [HCC]1 are at epidemic proportions both world-wide and in the U.S. The prevalence of NAFLD is estimated to be at 30% in the United States2 and up to 45% in Asia.3 Based on the rapid increases in fatty liver disease, NAFLD/NASH is expected to replace viral hepatitis as the leading cause of cirrhosis; NAFLD is already the most common chronic liver disease worldwide.2,4 HCC can occur as a sequela to NASH or from chronic infection due to either hepatitis B (HBV) or hepatitis C viruses (HCV) or a combination of both. In the U.S., HCC is responsible for the highest annual percentage increases in mortality rates: 2.8% for males and 2.1% for females compared to all other cancer sites which have decreased by 1.8% for males and 1.4% for females.5 Concurrent NASH increases the risk of HCC among patients with chronic HBV.6 On a global scale, HCC has become the world’s second deadliest cancer in numerical terms [after lung cancer].7
According to an analysis of 33,270 cases of HCC diagnosed from 1988–2012 reported to the California Cancer Registry, Laotian/Hmong experienced the highest risk of cause-specific mortality (hazard ratio=1.50, 95% confidence interval [CI]: 1.29–1.73) among all 15 racial/ethnic groups.8 Multiple risk factors including viral hepatitis have been identified for HCC pathogenesis. There are different risk factors for NAFLD.4 Among Asians in Sacramento County, California, Hmong experience the highest prevalence of metabolic risk factors for HCC, such as diabetes, large waist circumference, and high body mass index (BMI),9 as well as chronic HBV infections.10 However, the biological basis for the disparity is ill-defined and understudied. Hence, the purpose of our study was to ascertain the potential for ethnic-specific variations as mechanisms mediating chronic liver disease and possibly being a genetic factor contributing to this disparity in the Hmong.
In the context of NAFLD, single nucleotide variants (SNVs) in PNPLA3, the gene encoding patatin-like phospholipase domain containing A3 represent an important genetic mechanism. The most prominent variant is PNPLA3 rs738409[G], which is a nonsynonymous substitution of cytosine to guanine (C>G) that changes codon 148 from encoding isoleucine (I) to methionine (M) (I>M, I148M).11,12 This allele was identified in a genome-wide analysis of nonsynonymous variations (i.e., those likely to impact protein function) and was found to be strongly associated with hepatic fat/triglyceride content (P = 5.9 × 10−10) and hepatic inflammation (P= 3.7 × 10−4), and its frequency was concordant with the relative prevalence of NAFLD in European Americans, African Americans, and Hispanics. In a cohort of Chinese patients, the PNPLA3 rs738409 [G] allele was associated with susceptibility to NAFLD (OR 1.94, 95% CI 1.12–3.37, p = 0.018)13 and found at a frequency of 0.34 (HapMap).
The PNPLA3 protein is a triacylglycerol lipase with hydrolytic activity towards triglycerides in hepatocytes and retinyl esters in hepatic stellate cells.14 The I148M amino acid change occurs in the patatin-like phospholipase domain and leads to a loss of function promoting triacylglycerol accumulation in hepatocytes,15 as well as gain of functions including elevated lysophosphatidic acid acyltransferase and thioesterase activities.15 Taken together, heterozygous (CG) or homozygous (GG) PNPLA3 rs738409 [G] genotypes can increase the susceptibility to the development of NAFLD including fibrosis risk and progression. Thus, we hypothesized that the PNPLA3 rs738409 [G] variant could be a genetic mechanism that may partially explain the health disparity of increased rates of chronic liver disease in Hmong.
MATERIALS AND METHODS
Research participants
Twenty-six Hmong adults who participated in a community screening for viral hepatitis and cancer research in Sacramento County, CA each donated 5mL blood.10 Participants were recruited by partnering community based organizations and through in-language flyers and radio public service announcements. Participants completed an intake form with the assistance of lay bilingual community health workers as all participants preferred responding in Hmong rather than English. The intake included the following: 1) questions regarding country of birth, gender, and age; 2) research staff measuring and documenting participant’s waist circumference, height and weight; and 3) self-reported history of high blood pressure, high cholesterol and smoking. No information on alcohol intake was collected nor were any lipid profiles conducted. Our limited budget meant confining testing only to PNPLA3 even though we recognize that other mutations could have been studied. Participants also provided two additional samples for HBV and hemoglobin A1C testing. All lab tests were completed by the University of California, Davis Department of Pathology and Laboratory Medicine.
Serum samples
Whole blood samples were obtained from volunteers by venipuncture, drawn into 6.0-ml red-top Vacutainer tubes (Becton-Dickinson), and transferred to the UC Davis Comprehensive Cancer Center’s (UCDCCC) Biorepository Shared Resource. Serum was obtained by allowing the blood to clot (15–30 minutes at room temperature), and followed by centrifugation at 1,000–2,000 × g for 10 minutes at 4ºC to remove the clot. The serum was then aliquoted and stored at −80ºC.
Serum cell-free DNA isolation
Serum samples were submitted to the UCDCCC’s Genomics Shared Resource for DNA isolation and PNPLA3 SNP analysis. Circulating cell-free DNA (cfDNA) was isolated from serum samples (0.5 ml) using the QIAamp Circulating Nucleic Acid Kit (Qiagen), quantified with a NanoDrop Spectrophotometer and a Qubit 2.0 Fluorometer (dsDNA High-sensitivity Assay Kit; Thermo Fisher Scientific), and sized with an Agilent 2100 Bioanalyzer.
PNPLA3 SNP genotyping
We performed PNPLA3 SNP genotyping on the 26 Hmong participants. SNP genotyping analysis was performed on each DNA sample (5 ng DNA, >0.2 ng/μL concentration, 1,342 genome equivalents). A predesigned and validated TaqMan SNP Genotyping Assay (C______7241_10; catalog #: 4351379) (Thermo Fisher Scientific)16 was used and conducted according to the manufacturer’s standard protocols for reaction set-up and thermal cycling with endpoint readings on a StepOnePlus Real-Time PCR System (Applied Biosystems). For the PNPLA3 rs738409 [G] SNP, the context sequence utilized for assay design is: AGGCCTTGGTATGTTCCTGCTTCAT[C/G]CCCTTCTACAGTGGCCTTATCCCTC. Triplicate assays were performed for each sample as well as a no template control (NTC). Data analysis was performed for genotype calling (i.e., C/C, C/G, G/G) using TaqMan Genotyper Software (Applied Biosystems), and allelic discrimination plots were generated to visualize the genotypes of the entire cohort.
Statistical analysis
The distribution of participant demographics (country of birth; gender; and age) and health characteristics (waist circumference; body mass index; self-reported high blood pressure, high cholesterol, and smoking status; diabetes and hepatitis B status determined by laboratory tests) were summarized among those who had the PNPLA3 rs738409 [G] allele and those who did not, and for the entire cohort. Of note, we used the Asian cut points for waist circumference17 and BMI.18 An exact 95% CI was computed for the proportion of participants with the PNPLA3 rs738409 [G] allele.
RESULTS
Twenty-six Hmong adults from community screenings for viral hepatitis donated one extra tube of blood (5mL) for cancer research in Sacramento County, CA. Demographic and HCC risk factors are displayed in Table 1. In general, participants were: foreign-born (50%); female (76.9%); had a waist circumference above the Asian cut-point18 (75%); had a BMI of 23 (Asian cut-point)18 or greater (90.5%); did not have high blood pressure (76.9%); did not have high cholesterol (80.8%); did not smoke (73.1%); were diabetic or pre-diabetic (46.1%); and 7.7% were chronically infected with HBV.18
Table 1.
Characteristics of a Sample of Hmong Americans, by PNPLA3 rs738409 [G] allele frequencies
| Yes [G] (N = 12) N (%) |
No [C] (N = 14) N (%) |
Total (N = 26) N (%) |
|
|---|---|---|---|
|
|
|||
| Country of Birth | |||
|
|
|||
| Laos | 4 (33.3) | 8 (57.1) | 12 (46.2) |
|
|
|||
| Thailand | 1 (8.3) | 0 | 1 (3.9) |
|
|
|||
| USA | 1 (8.3) | 2 (14.3) | 3 (11.5) |
|
|
|||
| Unknown | 6 (50) | 4 (28.6) | 10 (38.5) |
|
|
|||
| Gender | |||
|
|
|||
| Female | 10 (83.3) | 10 (71.4) | 20 (76.9) |
|
|
|||
| Male | 1 (8.3) | 3 (21.4) | 4 (15.4) |
|
|
|||
| Unknown | 1 (8.3) | 1 (7.1) | 2 (7.7) |
|
|
|||
| Age | |||
|
|
|||
| Mean (SD) | 42.6 (14.6) | 46.6 (11.4) | 44.8 (12.8) |
|
|
|||
| Min-Max | 24–63 | 26–70 | 24–70 |
|
|
|||
| Waist Circumference | |||
|
|
|||
| < Asian Cut-point | 2 (25.0) | 2 (25.0) | 4 (25.0) |
|
|
|||
| ≥ Asian Cut-point | 6 (75.0) | 6 (75.0) | 12 (75.0) |
|
|
|||
| BMI | |||
|
|
|||
| BMI < 23 | 0 (0) | 2 (20.0.) | 2 (9.5) |
|
|
|||
| BMI ≥ 23 | 11 (100.0) | 8 (80.0) | 19 (90.5) |
|
|
|||
| High Blood Pressure | |||
|
|
|||
| No | 12 (100.0) | 8 (57.1) | 20 (76.9) |
|
|
|||
| Yes | 0 (0) | 6 (42.9) | 6 (23.1) |
|
|
|||
| High Cholesterol | |||
|
|
|||
| No | 9 (75.0) | 12 (85.7) | 21 (80.8) |
|
|
|||
| Yes | 3 (25.0) | 2 (14.3) | 5 (19.2) |
|
|
|||
| Smoking Status | |||
|
|
|||
| No | 9 (75.0) | 10 (71.4) | 19 (73.1) |
|
|
|||
| Yes | 3 (25.0) | 1 (7.1) | 4 (15.4) |
|
|
|||
| Unknown | 3 (21.4) | 3 (11.5) | |
|
|
|||
| Diabetes Status | |||
|
|
|||
| None | 8 (66.7) | 6 (42.9) | 14 (53.9) |
|
|
|||
| Pre-Diabetes | 2 (16.7) | 6 (42.9) | 8 (30.8) |
|
|
|||
| Diabetic | 2 (16.7) | 2 (14.3) | 4 (15.4) |
|
|
|||
| Hep B Status | |||
|
|
|||
| No | 11 (91.7) | 13 (92.9) | 24 (92.3) |
|
|
|||
| Yes | 1 (8.3) | 1 (7.1) | 2 (7.7) |
|
|
|||
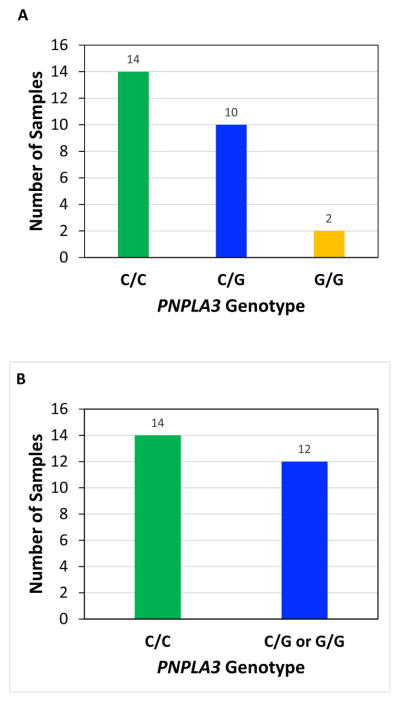
As noted in Figure 1, our results demonstrated that the PNPLA3 rs738409 [C>G] variant occurs at high frequency of 0.46 in the Hmong (12/26 samples, 95% CI 0.27–0.67), with 10 samples being heterozygous (C/G) and 2 samples being homozygous (G/G) for the allele (Figure 1, A and B). These results are quite compelling since a similarly high frequency was reported for Hispanics (0.49), who have the highest prevalence of NASH in the Dallas Heart Study and the only statistical significant association between the PNPLA3 rs738409 [G] allele and elevated serum alanine aminotransferase (ALT), compared to European Americans (0.23 frequency) and African Americans (0.17 frequency).11
Figure 1. PNPLA3 rs738409[G] Genotypes in Hmong in California.
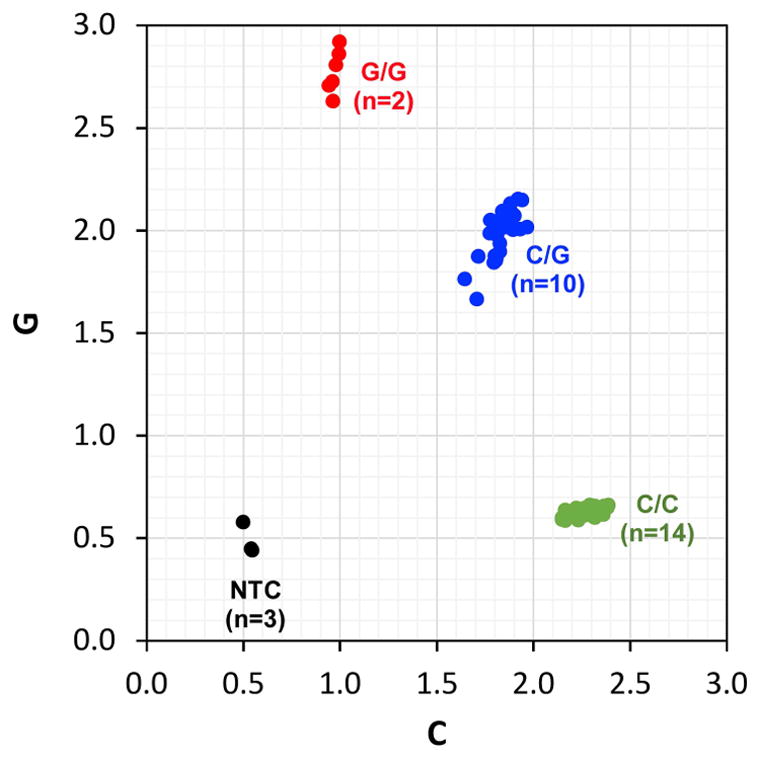
TaqMan SNP genotyping assays were performed in triplicate on serum cell-free DNA samples obtained from 26 Hmong individuals. Genotype calls (i.e., C/C, C/G, G/G) were made with TaqMan Genotyper Software (Applied Biosystems) and genotype frequencies were summarized according to A) the three possible genotypes: C/C, C/G, G/G, B) samples with (C/G, G/G) or without (C/C) the “G” allele. C) The genotypes of the entire cohort of samples (in triplicate) are visualized as an allelic discrimination plot.
If the Hmong prevalence of 0.46 were considered as part of the 1000 Genome Project, they would rank third highest (Figure 2). All other Asian ancestral populations in the 1000 Genomes Project exhibited comparable, but lower, PNPLA3 rs738409 [G] allele frequencies (Figure 2), including Han Chinese in Beijing (CHB, frequency = 0.383), Southern Han Chinese (CHS, frequency = 0.390), Vietnamese (KHV, frequency = 0.308), and Japanese (JPT, frequency = 0.423).19
Figure 2. PNPLA3 rs738409 [G] variant frequency in Hmong compared to that in other populations worldwide.
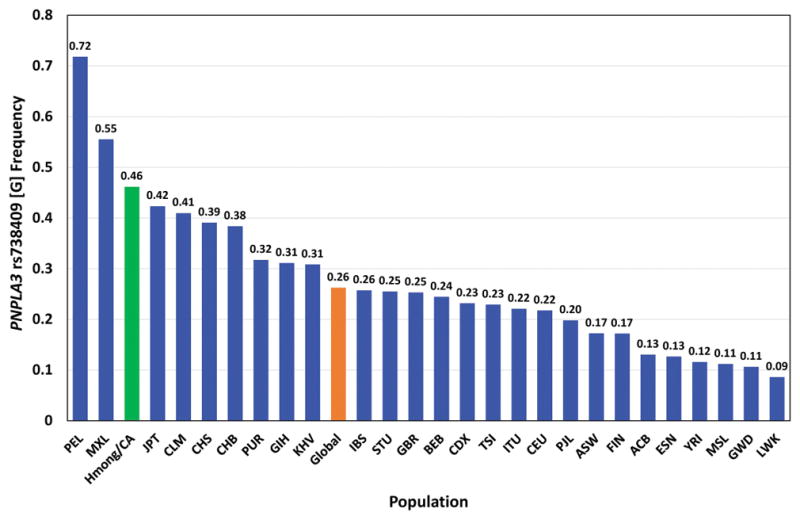
The mean global frequency of the PNPLA3 rs738409 [G] variant and its frequencies in 26 populations worldwide were sourced from the 1000 Genomes Project (NCBI 1000 Genomes Browser). Comparison of the frequency defined for the Hmong in California (Hmong/CA) to the other populations is depicted in the bar graph. The populations are abbreviated as follows: PEL (Peruvians from Lima, Peru), MXL (Mexican Ancestry in Los Angeles, CA), Hmong/CA (Hmong in California), JPT (Japanese in Tokyo, Japan), CLM (Colombian in Medellin, Colombia), CHS (Southern Han Chinese), CHB (Han Chinese in Beijing, China), PUR (Puerto Ricans from Puerto Rico), GIH (Gujarati Indian from Houston TX), KHV (Kinh in Ho Chi Minh City, Vietnam), IBS (Iberian Population in Spain), STU (Sri Lankan Tamil from the UK), GBR (British in England and Scotland), BEB (Bengali from Bangladesh), CDX (Chinese Dai in Xishuangbanna, China), TSI (Toscani in Italia), ITU (Indian Telugu from the UK), CEU (Utah residents (CEPH) with Northern and Western European ancestry), PJL (Punjabi from Lahore, Pakistan), ASW (African Ancestry in Southwest US), FIN (Finnish in Finland), ACB (African Carribbeans in Barbados), ESN (Esan in Nigeria), YRI (Yoruba in Ibadan, Nigeria), MSL (Mende in Sierra Leone), GWD (Gambian in Western Divisions in the Gambia), LWK (Luhya in Webuye, Kenya).
DISCUSSION
The purpose of this study was to evaluate whether the PNPLA3 rs738409 [C>G] variant could be a genetic factor contributing to the disproportionately high rates of chronic liver disease, specifically NAFLD, in California Hmong. With a sample size of 26, we recognize that this study should be considered exploratory and the findings interpreted with caution, since a larger cohort would be required to be more conclusive. For instance, the sample sizes for all populations in the 1,000 Genomes Project were ≥100 samples. Because of the small sample size, our 95% confidence interval for the prevalence of this gene variant is very wide (0.27–0.67), indicating that the estimated prevalence of 0.46 is quite imprecise and must be interpreted with caution. Nevertheless, our findings are evidence that this gene variant occurs in Hmong with above average frequency. The sample size was too small to provide adequate power for a statistical comparison of participants with and without the [G] allele which would be necessary to interpret any differences. The sample size was also too small to assess a correlation between the prevalence of the mutation and gender—in particular, only 4 participants were identified as male. Other limitations included the fact that our sample of Hmong was derived from a community screening, rather than a clinical setting. Those who participated may have been motivated by their interest in liver health and hence not representative of the Hmong at-large. On the other hand, being a community-derived sample could also mean that the true prevalence is under-estimated whereas a clinically-derived sample could mean an over-estimation of the true prevalence.
Despite these known constraints, this study has several strengths. Since our original study included collection of primary data intended to characterize HCC risk factors, we were able to report findings characterizing diabetes using Asian cut-off values for each participant. Demographic characteristics included country of birth, gender, and age. HCC risk factors included waist circumference, BMI, blood pressure, cholesterol, smoking status, diabetes status, and HBV status. While the Hmong are a relatively small Asian group, their distinctiveness as the population experiencing the lowest liver cancer survival rates among all Californians,8 means that adding a genetic dimension to our prior data on their considerable behavioral risk factors9 for chronic liver disease expands our insights into contributing etiologies. Having these findings suggest that a high rate of the PNPLA3 SNV should be examined further. Our literature review indicated that PNPLA3 rs738409 [G] has been implicated as a genetic influence in other populations such as the Hispanic population in the Dallas Heart Study.11 Based on our analysis, the very high frequency of PNPLA3 in Hmong is potentially pathologically significant. To the best of our knowledge, we are the first to report the finding that a Hmong community sample has a high prevalence of PNPLA3 rs738409 [G]. At the same time, we realize that this study is exploratory. In future studies, we need to recruit additional participants to adequately assess statistical and pathological significance. In addition, since the Hmong community is small, highly annotated samples (e.g., family history, relatives) will be obtained in order to determine the contribution of heritability to the higher frequency of this variant. Whole-exome sequencing is needed to identify candidate and/or novel genetic variants that could contribute to the health disparity either independently, or through functional linkage. Other genetic variants associated with susceptibility to NAFLD, such as GCKR rs780094[T], PPP1R3B rs4240624[A], NCAN rs2228603[T], LYPLAL1 rs12137855, TM6SF2 rs58542926[G], and MBOAT7-TMC4 rs641738[T] should be investigated.20–231
The gold standard for determining a diagnosis of NAFLD is through a liver biopsy or MR elastography which is not practical in community settings. Nevertheless, assessing the extent of the prevalence of the PNPLA3 rs738409 [G] allele, alone or in combination with other variants, could help in estimating genetic predisposition. Associating genetic predisposition with known non-genetic and modifiable risk factors, e.g., healthier diet and more exercise, could be used to identify those with higher risks and be the basis for precise population-based or clinical interventions to mitigate disease onset or progression.
Conclusion
The high frequency of PNPLA3 among the Hmong suggests that a genetic predisposition to NAFLD is plausible though not proven. While this is consistent with our hypothesis that this variant is an etiologic factor in the disparity of increased NAFLD/NASH in Hmong, a recent study suggests that it would not be the sole determinant, but rather the pathogenesis is influenced through the interplay of genetic variants with environmental or physiological factors, such as individual adiposity.24 Taken together, this provides support for pursuing research into genetic and lifestyle determinants for the chronic liver disease disparities affecting the Hmong.
Table 2.
Results of PNPLA3 rs738409 [C>G] SNP Assays for Individual Samples
| CCB STUDY ID | Specimen Type | Gender | Age |
PNPLA3 rs738409 [C>G] SNP Calls
|
|
|---|---|---|---|---|---|
| Heterozygous, Homozygous | Genotype | ||||
| 96 | Serum | Male | 43 | Heterozygous | C/G |
| 95 | Serum | Female | 60 | Heterozygous | C/G |
| 88 | Serum | Female | 51 | Heterozygous | C/G |
| 104 | Serum | Female | 56 | Heterozygous | C/G |
| 90 | Serum | Female | 63 | Heterozygous | C/G |
| 138 | Serum | Female | 25 | Heterozygous | C/G |
| 110 | Serum | Unknown | 45 | Heterozygous | C/G |
| 107 | Serum | Unknown | 47 | Heterozygous | C/G |
| 97 | Serum | Female | 30 | Heterozygous | C/G |
| 150 | Serum | Female | 24 | Heterozygous | C/G |
| 93 | Serum | Female | 42 | Homozygous | C/C |
| 143 | Serum | Female | 26 | Homozygous | C/C |
| 140 | Serum | Male | 50 | Homozygous | C/C |
| 91 | Serum | Female | 70 | Homozygous | C/C |
| 109 | Serum | Female | 59 | Homozygous | C/C |
| 94 | Serum | Male | 45 | Homozygous | C/C |
| 89 | Serum | Female | 57 | Homozygous | C/C |
| 141 | Serum | Female | 41 | Homozygous | C/C |
| 139 | Serum | Male | 28 | Homozygous | C/C |
| 103 | Serum | Female | 48 | Homozygous | C/C |
| 92 | Serum | Female | 42 | Homozygous | C/C |
| 108 | Serum | Female | 50 | Homozygous | C/C |
| 111 | Serum | Female | 50 | Homozygous | C/C |
| 124 | Serum | Male | 44 | Homozygous | C/C |
| 105 | Serum | Unknown | Not reported | Homozygous | G/G |
Acknowledgments
Funding. We are grateful to the 26 California Hmong who participated in this study and to Ms. Irmgard M. Feldman, UC Davis Comprehensive Cancer Center Biorepository for her assistance with accessing these samples and Mr. Duke LeTran for his assistance in the graphic arts. This work was funded by U54CA153499 and by the Francis Yee Fund for Cancer Disparities Research. The Biorepository and Genomics Shared Resources are supported by Cancer Center Support Grant P30CA093373. However, the views expressed are those of the authors and do not necessarily reflect the views of the funders.
Footnotes
Author contributions: All authors made substantive contributions to the study design and interpretation of the data, were involved in drafting the article or critically reviewing it, provided final approval of the version submitted, and agree to be accountable for all aspects of the work.
Invited paper: by Dr. Fadlo R. Khuri, Editor-in-Chief
Conflict of interest All of the authors: declare no conflict of interest.
Ethical approval for research involving human participants All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study. This study was performed in accordance with the Institutional Review Board Protocol #128204 approved by the University of California, Davis for “UC Davis Pathology Biorepository: Tissue, Blood, Urine and Other Biological Materials”.
Author contributions:
All authors substantially contributed to the conception, design, data collection and analyses and provided critical analyses of the content. All approved the final version of the paper and agree to be accountable for all aspects of the work.
References
- 1.Rahman R, Hammoud GM, Almashhrawi AA, Ahmed KT, Ibdah JA. Primary hepatocellular carcinoma and metabolic syndrome: An update. World Journal Gastrointestinal Oncology. 2013;5(9):186–94. doi: 10.4251/wjgo.v5.i9.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le MH, Devaki P, Ha NB, et al. Prevalence of non-alcoholic fatty liver disease and risk factors for advanced fibrosis and mortality in the United States. PloS One. 2017;12(3):e0173499. doi: 10.1371/journal.pone.0173499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrell GC, Wong VW, Chitturi S. NAFLD in Asia—as common and important in in the West. Nat Rev Gastroenterol Hepatol. 2013;10:307–318. doi: 10.1038/nrgastro.2013.34. [DOI] [PubMed] [Google Scholar]
- 4.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313(22):2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA J Cancer Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 6.Chan AW, Wong GL, Chan HY, et al. Concurrent fatty liver increases risk of hepatocellular carcinoma among patients with chronic hepatitis B. J Gastroenterol Hepatol. 2017;32:667–676. doi: 10.1111/jgh.13536. [DOI] [PubMed] [Google Scholar]
- 7.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Intl J Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 8.Stewart SL, Kwong SL, Bowlus CL, et al. Racial/ethnic disparities in hepatocellular carcinoma treatment and survival in California, 1988–2012. World J Gastroenterol. 2016a;22:8584–8595. doi: 10.3748/wjg.v22.i38.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart SL, Dang JH, Chen MS., Jr Diabetes prevalence and risk factors in four Asian American communities. J Comm Health. 2016b Dec;41(6):1264–1273. doi: 10.1007/s10900-016-0214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang JHT, Chen MS., Jr Findings from “Increasing Hepatitis B testing and linkage to care for Sacramento’s foreign-born Asians”, 2012–2013. Public Health Reports. 2016;131(Suppl 2):119–131. doi: 10.1177/00333549161310S218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature genetics. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu R, Tao A, Zhang S, Deng Y, Chen G. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and Nonalcoholic Fatty Liver Disease: A HuGE review and meta-analysis. Scientific Reports. 2015;5:9284. doi: 10.1038/srep09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zain SM, Mohamed R, Mahadeva S, et al. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet. 2012;131:1145–1152. doi: 10.1007/s00439-012-1141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014;23:4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122:4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 17.Lear SA, James PT, Ko GT, Kumanyika S. Appropriateness of waist circumference and waist-to-hip ratio cut-points for different ethnic groups. Eur J Clin Nutr. 2010;64:42–61. doi: 10.1038/ejcn.2009.70. [DOI] [PubMed] [Google Scholar]
- 18.Araneta MR, Kanaya AM, Hsu WC, et al. Optimum BMI cut points to screen Asian Americans for type 2 diabetes. Diabetes Care. 2015;38:814–820. doi: 10.2337/dc14-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browning JD. Common genetic variants and nonalcoholic Fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:1191–1193. doi: 10.1016/j.cgh.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Hernaez R, McLean J, Lazo M, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11:1183–1190. e1182. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology. 2016;150:1219–1230. e1216. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stender S, Kozlitina J, Nordestgaard BG, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nature Genetics. 2017;49(6):842–847. doi: 10.1038/ng.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]