Abstract
Neurobiological disorders have diverse manifestations and symptomology. Neurodegenerative disorders such as Alzheimer’s disease (AD) manifest late in life and are characterized by, among other symptoms, progressive loss of synaptic markers. Developmental disorders, such as autism spectrum, appear in childhood. Neuropsychiatric and affective disorders, such as schizophrenia and major depressive disorder, respectively, have broad ranges of age of onset and symptoms. However, all share uncertain etiologies, with opaque relationships between genes and environment. We propose a “Latent Early–life Associated Regulation” (LEARn) model, positing latent changes in expression of specific genes initially primed at the developmental stage of life. In this model, environmental agents epigenetically disturb gene regulation in a long–term fashion, beginning at early developmental stages, but these perturbations might not have pathological results until significantly later in life. The LEARn model operates through the regulatory region (promoter) of the gene, specifically through changes in methylation and oxidation status within the promoter of specific genes. The LEARn model combines genetic and environmental risk factors in an epigenetic pathway to explain the etiology of the most common, i.e., sporadic, forms of neurobiological disorders.
THE LEARn MODEL
Neurobiological disorders have diverse manifestations and symptomology. Neurodegenerative disorders, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), usually appear late in life and are characterized by progressive loss of synaptic markers and cholinergic neurons in AD and nigrostriatal projection neurons in PD, both eventually resulting in dementia.1–3 Developmental disorders, such as autism spectrum (AS), usually manifest in childhood and are typified by symptoms such as difficulty in pragmatic communication and social interaction, repetitive and stereotyped movements, and perseveration on a limited range of interests.4 Neuropsychiatric disorders such as bipolar disorder and schizophrenia tend to appear in adolescence and early adulthood. Bipolar disorder is associated with a cycling of depressive and manic/hypomanic symptoms.5 Schizophrenia has a complex of symptoms that includes lack of affect, social withdrawal, hallucinations, and disordered thinking. Other disorders include major depressive disorder (MDD) and anxiety disorders, which have a broad range of age of onset and manifest as a severe and pervasive emotional state together with behavioral and somatic symptoms.
All of these disorders share genetic risk factors that have incomplete penetrance. It is possible to have a “pathogenic” allele and not develop the associated disorder The popular current model is a polygenic/multifactorial model that renders no single contributory factor either necessary or sufficient. It is, instead, the interaction of multiple agents that are presumed to lead to a disorder, with some potential for environmental input.
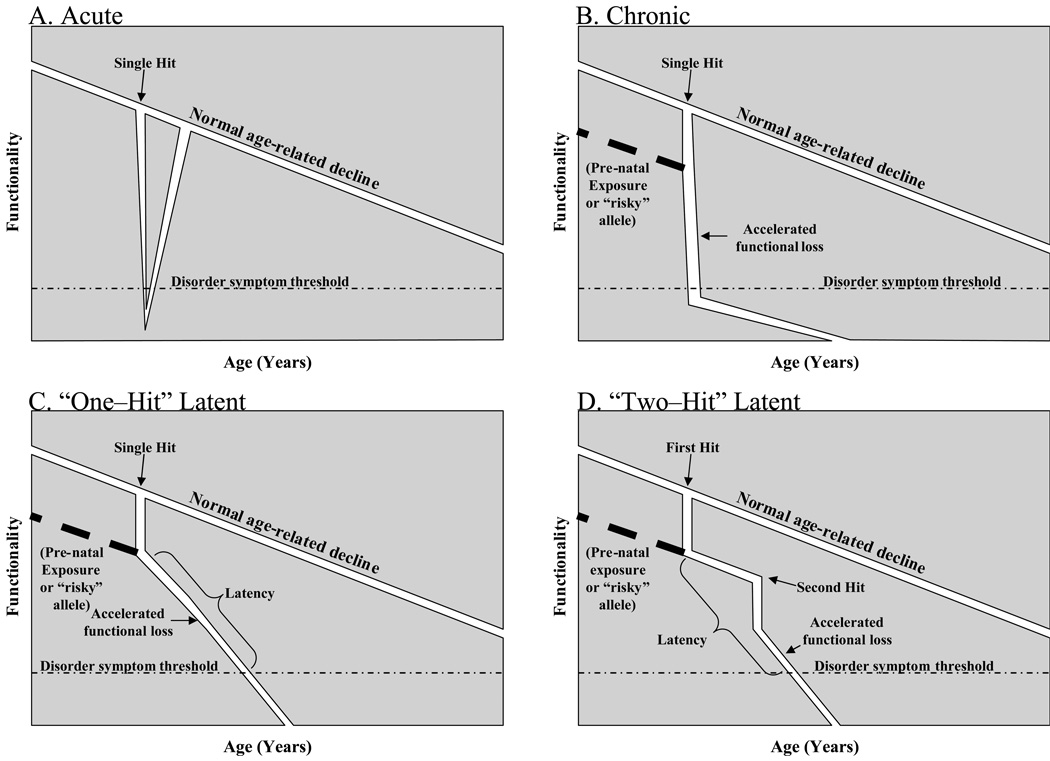
A useful categorization of disease progression divides disorders and diseases into a few general models. An acute disorder begins shortly after a trigger event has occurred (Fig. 1A) and progresses, ultimately (presuming that it is not fatal) being self–limiting, with restoration of function at end of the disease episode. Many infectious diseases, such as rubella, fall into this category. A chronic (Fig. 1B) disorder likewise clinically presents after a trigger has occurred. However, the chronic disorder remains active for an extended period, potentially throughout an organism’s lifespan. This model would likely fit several classes of disorder, such as emphysema. Latent (Fig. 1C–D) disorders do not quickly manifest symptoms after an initial etiologic “hit” occurs. Within latent disorders, two subtypes can be characterized. Long prodromal phase disorders (Fig. 1C) are characterized by a “one–hit” etiology. A single triggering event is sufficient to induce the disease state, which simply takes an appreciable amount of time to “build up” to a clinical level, this latency could be manifest as recurrent sporadic symptomatic periods, such as with herpes simplex virus infection, or a latent period followed by an extended symptomatic period, as with HIV infection “Two–hit” latency (Fig. 1D) is an etiology that begins with an initial event or condition that, in and of itself, is insufficient to produce a disease state. The first hit could be exposure to environmental toxins, heavy metals, nutritional deficiency, stress, or even inherited genetic or epigenetic substrate. Later in life, a second hit would occur that, in those organisms that had suffered the first hit, would lead to disease. However, in organisms that were not subject to the first hit, no disease would occur. This model makes the greatest allowance for “sporadic” appearance of a disorder, especially if, instead of being “two–hit”, it is actually “many–” or “n–hit”, where “n” can be any number of individual risk factors, each with the potential to instill or activate a latent alteration. The most well-characterized “n-hit” disorders would be cancers.
Fig. 1. Models of disease progression.
Disease progression usually follows one of four models. A) Acute disease. An etiologic agent or condition “single hit” (including a genetic mutation) takes immediate or near–term to exposure effect, resulting in loss of function that progresses to disease symptoms, which eventually recede. B) Immediate–trigger chronic disease. An etiologic agent or condition takes immediate or near–term to exposure effect, resulting in loss of function that progresses to disease symptoms, which continue throughout life, or at least for some considerable time. C) One–hit latent disease. An etiologic agent or condition imposes immediate or near–term effect, but the effects are subclinical for an extended period. Over time, loss of function increases to a pathological level without any further “hits” to the organism. D) Two–hit latent disease. An etiologic agent or condition affects an organism but does not result in a diseased state. This alteration is maintained through the organism’s lifespan without readily visible effect unless a second hit intrudes. This interacts with the “embedded effect” of the first to produce a disease state.
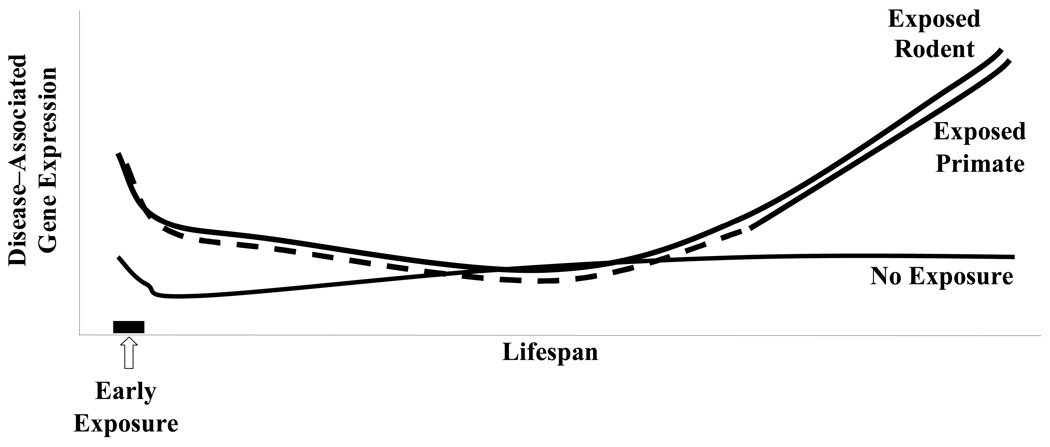
We propose that many neurobiological disorders, regardless of their symptomatic variety, share a fundamentally similar mechanistic etiology: “Latent Early–life Associated Regulation” or “LEARn”.6–8 Under LEARn, early–life stressors modify potential expression levels of disorder–associated genes in a latent fashion. Latent changes in these genes are maintained by epigenetic mechanisms such as DNA methylation, DNA oxidation, and chromatin reorganization. LEARn–modified genes may require multiple “hits” to express at pathological levels. This is similar to the “two–hit” model of oncology.9 Recent work on primates and rodents has led to the determination that early–life exposure to external stress agents, such as lead (Pb) may induce a short term upregulation of disease–associated genes, followed by a long latency period of “normal” levels of gene expression, but this period ends when disease–associated gene expression levels increase later in life (Fig. 2A, B).10, 11 In addition, oxidative DNA damage, as measured by the presence of Oxo8dG, remains low early in life but at some point is increased, potentially due to suppression of repair enzyme expression. This oxidation would contribute to overall epigenetic changes and influence expression of pathogenic genes, resulting in increase in expression levels of pathogenic genes (Fig. 2B).
Fig. 2. LEARn effected gene expression.
Diagrammatic representation of expression levels for disease–associated genes according to the LEARn model based upon rodent and primate research.10, 11 A) Susceptible genes undergo an early–life exposure to a stress such as heavy metals, inadequate maternal care, or nutritional deficit. This may result in an acute increase in expression levels that quickly returns to “normal” expression. Later in life an additional trigger may affect genes. Those organisms that have been previously subject to the initial trigger experience a significant increase in expression levels of disease–associated genes, while genes in organisms not primed in such a manner do not. B) Schematic representation of pathologically–associated genes and DNA oxidation damage in relationship to LEARn. Shortly after exposure to initial trigger, genes associated with a disorder would experience alterations such as an increase in expression level, while overall DNA oxidation would not be perturbed early in life. Later in life, DNA oxidation would be perturbed, which would lead to activation of the promoters of disease–associated genes, leading to active disease.
LEARn also postulates that in the case of disorders with distinctive ages of onset the final “hit” may be normal large–scale changes in gene expression that occur at various stages of development, such as puberty (both onset and end) or onset of senescence. However, for disorders that do not have such a characteristic age of onset, “non–shared” environmental influences would act as a temporally proximal trigger. In addition, LEARn posits two categories of target genes, “LEARNed” and “unLEARNed”, which are structurally distinct from each other. These structural differences are in the primary DNA sequences of the gene, specifically in densities of CpG and GG dinucleotides.
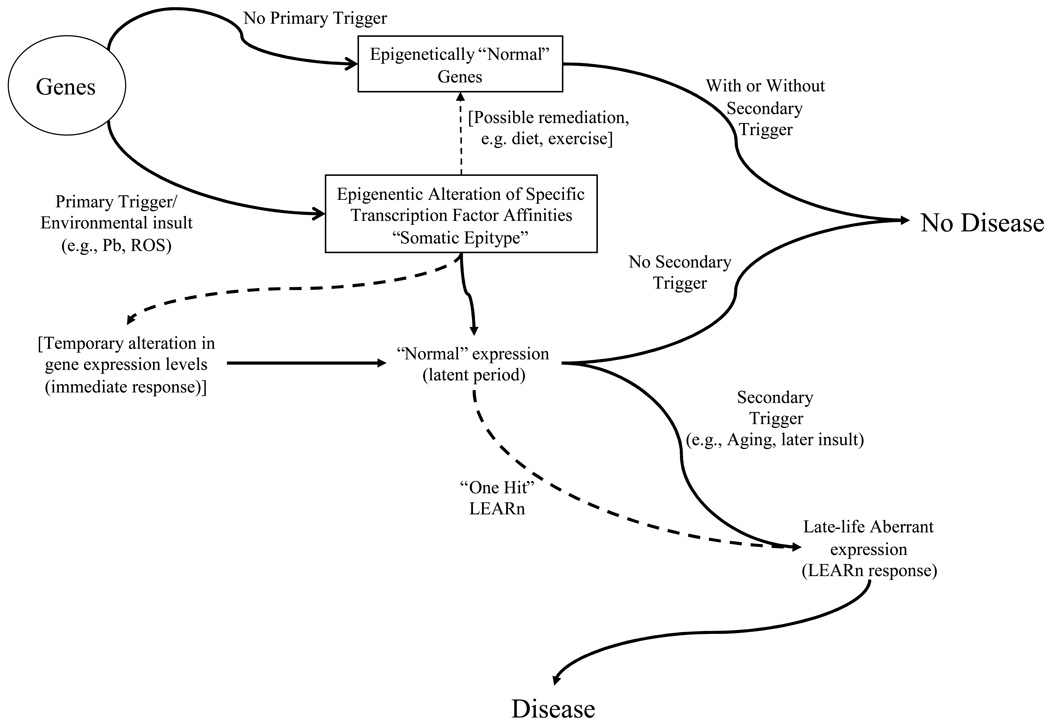
In the LEARn model, environmental agents (e.g., heavy metals), agrichemicals (e.g., pesticides-containing water), intrinsic factors (e.g., alleles or inflammatory cytokines), and dietary factors (e.g., folate and cholesterol) and lifestyle habits (e.g., physical exercise) act at early developmental stages to alter gene regulation in a long–term fashion. These disturbances would not produce disease states on their own,7, 10 and even then only after a latent period that may last up to several years (Fig. 3). Similar hypotheses were developed in the 1980s by Barker and colleagues.12. However, Barker’s model is based on low birth weight and rapid childhood weight gain, without identifying a molecular mechanism. LEARn is based on the regulatory structure common to eukaryotic genes and on epigenetic processes operating at specific sites within the promoter (regulatory) region of specific genes. LEARn is consistent with the Barker model of fetal origins of adult disease. It is also consistent with the more recently detailed “DOHaD” (developmental origins of health and disease) model.13
Fig. 3. The LEARn model.
Schematic representation of LEARn–type disease progression. A gene or genes associated with a disorder may be subject to an environmental “first trigger”, such as exposure to Pb or ROS. This results in an epigenetically marked gene, through methylation, oxidative damage of DNA, and/or chromatin rearrangement. The epigenetically marked gene may undergo a temporary change in expression levels, but this returns to “normal” levels. If a secondary trigger, such as additional environmental insult or systemic changes in gene expression patterns associated with aging, further affects the gene(s), expression levels deviate from normal, resulting in a disease state. Dotted lines represent alternate pathways.
LEARn, however, enlarges upon both of these models by providing specific biochemical and molecular pathways that can be directly tested. LEARn introduces an intermediate step between genotype and phenotype, specifically the “somatic epitype”, which is an epigenotype acquired after birth.8 Evidence for the somatic epitype includes changes in DNA methylation induced by maternal behavior.14 LEARn explains maintained latency as epigenetic modification of gene regulatory sequences. LEARn is similar to the proposals of Szyf et al.15 However, LEARn is not a toxicological model. We expand upon these other models by positing structurally “LEARNed” and “unLEARNed” gene regulatory sequences and by including DNA oxidation in addition to methylation as an latent epigenetic marker. LEARn addresses the “black box” of sporadic disorder origins by expanding upon our findings that, in addition to latency and epigenetics, expresion levels of specific transcription factors, such as stimulatory protein 1 (SP1), and changes in primary DNA sequence oxidation levels interacting with neighboring methylation sites make up testable physical sites of the model’s activity. In addition, we have determined that expression of critical transcription factors, such as SP1, parallel expression levels of disease–associated genes in model disorders, such as β–amyloid precursor protein (APP).16
NEURODEGENERATIVE DISORDERS
Degenerative neurobiological disorders include diseases such as Parkinson’s disease and Alzheimer’s disease. These diseases have a significant genetic component. Longitudinal studies have shown disease concordance for PD of 77% in monozygotic twins and 22% in dizygotic twins; however, cross–sectional twin studies contradict these findings.17, 18 Twin studies of AD indicate concordance of 75%–80% in monozygotic twins and 26%–46% in dizygotic.19, 20 Nevertheless, the majority of AD and PD cases cannot be explained by autosomal inheritance, which account for less than 1%21 of PD and 5%2 of AD. Genes associated with PD include α–synuclein (SNCA),22 Catechol–O–methyl transferase (COMT),23 and microtubule–associated protein τ (MAPT),24, 25 Genes associated with AD include APP,26 apolipoprotein E (APOE),27 β–amyloid cleaving enzyme 1 (BACE1).28 MAPT,25, 29 and the presenilins, 1 and 2 (PSEN1, PSEN2).30 However, many more genes have been associated with these disorders (Suppl. Table 1).
Environmental factors, such as nutritional imbalance, stress, and exposure to risk agents such as heavy metals (for AD)31 or pesticides (for PD)32 are implicated but also fail to completely explain disease incidence. Presentation of disease symptoms usually occurs in the absence of an immediate trigger, strongly suggesting a long latent period. Finally, there is growing evidence that many of these diseases may have “interlocking”; risk factors that contribute to one of these diseases and contribute to other neurodegenerative disorders, such as the involvement of α–synuclein, β–amyloid, and microtubule–associated protein τ in both AD and PD.33, 34
NEURODEVELOPMENTAL DISORDERS
Neurodevelopmental conditions have a highly sporadic nature characterized by genetic and environmental risk factors. These include schizophrenia and autism, both of which have distinctive ages of onset.35, 36 Genetic risk factors such as neuregulin–1 (NRG1)37 for schizophrenia and reelin (RELN) or fragile–X mental retardation protein (FMR1)38 for autism, are among the many that have been implicated (Suppl. Table 1). However, as is the case for neurodegenerative disorders, genetic risk factors are not certain indicators. Environmental risk factors for schizophrenia include complications at birth,39, 40 childhood viral infections of the CNS41 and maternal exposure to severe adverse life effects such as death of a close relative during pregnancy.42 Autism has been linked to greater paternal age43 and prenatal stress,44 among other factors.
OTHER NEUROPSYCHIATRIC DISORDERS
Incomplete penetrance extends to other conditions, including bipolar disorder, major depressive disorder, and obsessive–compulsive disorder. These disorders manifest at any age or life stage. There are several gene associations (Suppl. Table 1), such as brain–derived neurotrophic factor (BDNF)45 for bipolar disorder and the serotonin transporter protein (SLC6A4) for major depressive disorder46 and obsessive–compulsive disorder.47 These associations have also been found to be “incomplete”. Likewise, strong environmental components have been recognized in the etiology of OCD, bipolar disorder, and major depressive disorder, such as advancing paternal age and bipolar disorder, parental bonding variation and OCD, and childhood parental loss and major depressive disorder.48–51
ALZHEIMER’S DISEASE AS A MODEL FOR IDIOPATHIC NEUROPSYCHIATRIC DISORDERS
From an epidemiological standpoint, AD could be considered sufficient to model many aspects of neurodegenerative, neurodevelopmental, and neuropsychiatric disorders. AD manifests late in adult life, and it is not clearly understood when the disease begins its progression, nor how long this process takes. Large longitudinal studies have concluded that significant risk for developing AD is established by early adulthood.52 While this has led some researchers to conclude that AD must be a genetic disorder, such a conclusion dismisses well–established environmental risk factors such as inflammation,53 cholesterol/diet,54 head injury,55 and reduced midlife physical activity.56 Even taking into account the very–well established genetic risk factor of the APOEε4 genotype,27 the majority of AD cases are idiopathic. A unifying hypothesis for the etiology of AD must take into account both its neuropathological features and multiple environmental factors associated with AD.
BIOLOGICAL MECHANISM OF THE LEARn MODEL
Oxidation of DNA often occurs as oxidation of d–guanosine to oxo8d–guanosine. The 5’ “G” of a “GG” dinucleotide is a hot spot of such oxidation.57 This interferes with the DNA–binding capacity of methyl CpG–binding protein (MeCP) to a methylated cytosine,58 producing an “effective demethylation” due to oxidative damage of DNA.
Methylation in mammalian DNA usually is the addition of a methyl group to cytosine residues at CpG dinucleotides.58 This reaction is catalyzed by DNA methyltransferase (DNMT) enzymes.59 Hypomethylation in the promoter region leads to elevated gene expression, whereas hypermethylation results in decreased gene expression. Environmental stressors, including exposure to metals and dietary factors, may interfere with the methylation of CpG clusters, thus altering affinity with potential transcription factors proteins, such as MeCP and stimulatory protein 1 (SP1). The activity of DNMT is reduced by heavy metal (cadmium) exposure.60 Furthermore, heavy metals such as Pb are known to induce oxidative stress,61 oxidative stress modulates DNA methylation during malignant transformation.62 In addition, trace copper in drinking water has been shown to facilitate formation of Aβ plaque in rabbits,63, 64 although the specific pathway for this effect is not determined. Aluminum has been shown to induce Z–DNA conformation at CCG repeats.65
Histone acetylation is also likely to function in the LEARn process, with differences in acetylation occurring in response to DNA methylation or demethylation.66 This, of course, would involve chromatin remodeling. However, it is our contention that changes in DNA oxidation and methylation are the initial means through which LEARn effects arise in reaction to the environment.
BIOCHEMICAL EVIDENCE FOR THE LEARn MODEL
A specific example of LEARn activity may involve the response of Alzheimer–associated genes to early–life exposure to the heavy metal lead Pb, although this is not to be taken to mean that Pb is an exclusive operator in the LEARn model. It is presented as a model agent known to influence both DNA methylation67 and oxidation.68 When Basha and colleagues introduced Pb–acetate into the drinking water of dams of infant rats at 20 days post–birth (Pb–E) and at 19 months of age (Pb–L), levels of brain Pb when all rats were sacrificed at 20 months of age were the same between early–exposed (Pb–E and control rats but elevated in Pb–L animals. However, at the age of 20 months, the early–exposed (Pb–E rats’ APP expression levels and Aβ peptide levels increased to levels higher than those of control rats and likewise higher than levels found in Pb–L rats. Longitudinal assays of APP mRNA and Aβ peptide levels revealed that these end–of–life changes did not reflect a “chronic” condition. While there was a temporary increase in these markers, levels returned to normal levels when Pb was removed from their diet. When SP1–DNA binding was compared between Pb–E and control rats, this followed a similar trend to APP mRNA levels. SP1–DNA interaction was elevated during initial Pb exposure but fell to normal levels after Pb was removed from the diet. Late in life, SP1–DNA interaction increased significantly above that seen in control animals. In addition, DNA oxidation levels were also measured at end of life, and it was determined that oxo–d8–GTP levels were elevated in Pb–E animals.10 It was also determined that Pb treatment of cell cultures did not alter enzymatic activity of the α–, β–, or γ–secretases that process APP through the amyloidogenic or non–amyloidogenic pathways. However, Pb did increase Aβ aggregation in vitro.69 This in vitro aggregation may have an in vivo counterpart in the discovery that water quality influences Aβ aggregation in rabbits,70 although in that case copper was specifically implicated.63 Likewise, in humans, elevated concentrations of agrichemicals in surface water in April–July coincided with higher risk of birth defects in live births with last menstrual periods during those months.71
Cynomolgus monkeys were also exposed to Pb during the first year of life (Pb–E) and were sacrificed for brain tissue analysis at the age of approximate 23 years. Comparing Pb–E brains with control (non–exposed) monkeys revealed that Pb levels between the two groups were the same. On the other hand, APP and SP1 mRNA levels were significantly elevated (BACE1 levels were elevated, but p = 0.06) vs. controls. In addition, Pb–E monkeys had significantly higher levels of Aβ42 and Aβ40 in later life. Exposed monkeys also showed greater brain amyloid aggregation and more dense aggregates than did non–exposed monkeys.72 Finally, comparison of DNA methyltransferase activity in both rodent cells exposed short term to Pb and in the early–lifespan Pb–exposed monkeys showed that Pb exposure significantly reduced activity of this enzyme in both systems.72
It is particularly notable that SP1 is likely to be regulated in a LEARn fashion, given that SP1 binds to the human73–76, rhesus monkey77 and rat78 APP promoter and accelerates the production of APP mRNA.73, 74, 78 Location and frequency of SP1 sites are more similar between monkey and human than between either primate species and rodent.77 Furthermore, SP1 regulates the expression of the MAPT gene.79 In addition, SP1 and APP mRNA levels closely follow each other over the life spans of mice and monkeys.16
EPIDEMIOLOGICAL AND CLINICAL EVIDENCE FOR THE LEARn MODEL
A recent survey of twin pairs for AD with Lewy body dementia showed discordance in 90% of monozygotic twins.80 In a separate study, a pair of identical twins with discordant development of AD had been brought up in the same household and had similar career paths, both becoming chemical engineers. One twin developed AD, while the other did not. It was determined that DNA methylation levels were different from each other, with the AD twin having a lower level of methylation than the non�AD twin.81 This is, of course, not DNA methylation, but it does indicate that AD etiology may be influenced by other epigenetic pathways. Regarding disorders other than AD, an association between prenatal exposure to Pb and schizophrenia has also been reported.82, 83
In neurodevelopmental disorders, autism has been associated with DNA hypomethylation in parents.84 Schizophrenia has been associated with both hypomethylation of MB–COMT85 and with hypermethylation of RELN.86 Aberrant methylation has been implicated in bipolar disorder.85, 87 In mood and anxiety disorders, there have been several associations raised with abnormal methylation.85, 87, 88 Likewise, hypermethylation of the gene for ribosomal RNA has been implicated in suicide.89 Likewise, epigenetic dysregulation has been observed near several genes involved in neuronal development in the brain in individuals with major psychosis.90
While the above clinical data demonstrate association between aberrant methylation states and disease, they do not necessarily show that disease–associated methylation or other epigenetic states have been acquired after prenatal imprinting. Acquisition of an aberrant epigenetic state would be necessary for LEARn–style etiology to apply. It has been recently determined that methylation levels for an individual person change over a period of a decade for 29% of an Icelandic cohort.91 This raises the possibility of disease–related, environmentally induced methylation states. In addition to lifespan changes in DNA methylation states in humans, delayed neurological response has been demonstrated in rats subjected to postnatal inflammation. The treatment resulted in increased susceptibility to seizure in adulthood.92 Combination of these various elements, i) differential methylation patterns in disease, ii) potential for post–natal changes in gene methylation in individuals, and iii) demonstrated induction of lifespan–delayed neurological symptoms in an animal model, indicate a reasonable epidemiological case for further investigation of LEARn as a general model of idiopathic neurobiological diseases.
“LEARned” VS. “unLEARned” GENES
Observed differences of response to early–life exposure to Pb may define a more general class of gene regulation in response to methylation and oxidation “availability”. When Wu et al performed high-throughput analysis of monkey mRNA with a human neurological microarray twenty–three different genes were determined to have altered expression between Pb-exposed and control monkeys.72 In addition, APP, BACE1, and SP1 mRNA levels were found by northern blot to be elevated in Pb–exposed monkeys. Of the 23 microarray analyzed genes, 6 were upregulated and 17 were downregulated. Sequences of these 26 total genes (table 1), were downloaded from the human genome project (build 34.3) from −3000 to +3000 bp from the +1 TSS. An additional 26 "non-responding" genes were randomly chosen from the same array (table 1) and sequences of −3000/+3000 were downloaded. Sequence groups were compared for cumulative CpG and GG content, in a sliding window of 200bp. This size of sliding window was chosen as an approximation of the length of DNA in a nucleosome plus linker length between nucleosomes.
Table 1.
LEARned and randomly selected unLEARned genes.
| Expression altered by early- life–exposure to Pb (LEARned) |
Expression not altered by early–life exposure to Pb (unLEARned) |
||
|---|---|---|---|
| Upregulated | Downregulated | ||
| aAPPa | AP2M1 | ADORA1 | HLA-C |
| BACE1a | ARGHDIA | ANXA3 | PEX5 |
| CALM3 | CAPN3 | CALR | NF2 |
| HMOX2 | CLTC | CETP | PCSK2 |
| NR4A3 | DAO | CHRNA1 | PEBP1 |
| PLA2G2A | DNM2 | CRHR1 | PITPNA |
| SDHA | DRD1 | DBH | PLCG2 |
| SP1a | GDI2 | EFNB3 | POU4F1 |
| STX7 | GNB1 | FABP5 | PRKACA |
| HTR1B | GABRA3 | RABGGTA | |
| IQGAP1 | GABRG2 | SCP2 | |
| KCNJ4 | GDNF | STXBP2 | |
| OPRD1 | HK1 | SYT1 | |
| OPRM1 | |||
| RAB32 | |||
| RAB5A | |||
| RAB5C | |||
Determined by northern blot measurement
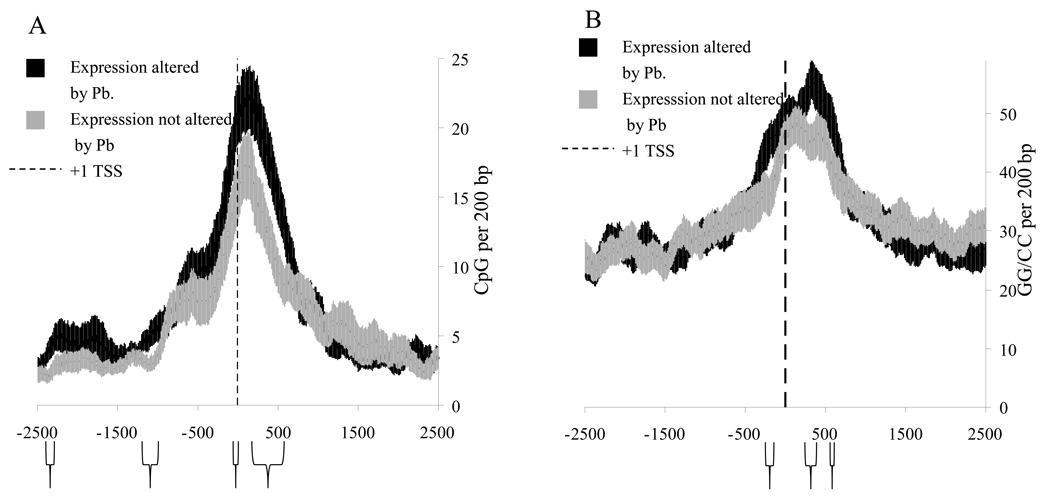
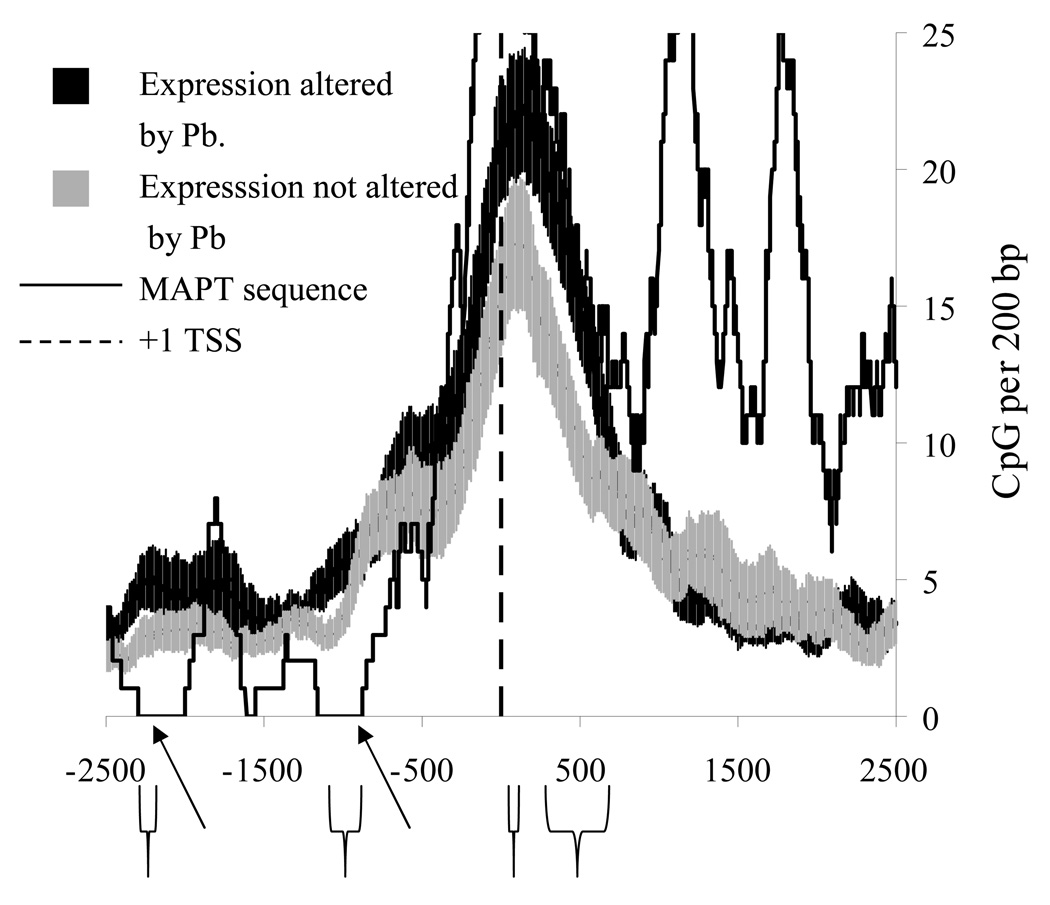
When CpG density in this 200bp window was compared, four regions of significant difference were found (Fig. 4A). Two of these regions were within 500bp of the +1 TSS, while two other regions were more distal (near −1000 and −2300). Significant differences in “GG” doublets, hotspots for DNA oxidation,93 were found at around −250/−350, +250/+350, and near +500 (Fig. 4B). The sequence derived model was tested by comparing it to the sequences of an AD–associated gene that did not show a late–life change in expression after Pb exposure, specifically MAPT (Fig. 5). While the MAPT sequence was in agreement with the LEARn model in proximal regions (between −500 and +500), it showed differences in CpG density from “LEARned” genes in a more distal region of interest, specifically around −750/−1000 and around −2300 from +1 TSS. This would suggest that the LEARn model, if further validated would structurally tie both distal and proximal elements of gene regulation.
Fig. 4. “LEARned” vs. “unLEARned” genes, CpG or GG density.
Graphs of A) CpG or B) GG density of genomic DNA sequences from 26 monkey genes, of −3000 to +3000 bases around the +1 TSS in 200bp window. Sequences were chosen after microarray of mRNA or by Northern blotting of mRNA indicated alterations in expression levels 23 years after exposure to Pb (black line).72. These were compared to 26 genes with expression levels not altered 23 years after exposure to Pb (gray line). The “non–responding” genes were selected at random from the same RNA array that determined the “responding” genes. Graphs depict standard error of the mean along the sequence of CpG or GG density. Regions where differences in CpG density in the selected window size were significant at p < 0.05 are indicated with brackets under the graphs.
Fig. 5. Comparison of “unLEARned” MAPT gene with the LEARned gene model.
The MAPT gene, which did not respond to early–life Pb, was chosen for comparison with the LEARned gene model. CpG densities in a 200bp running window were calculated. Regions where MAPT resembles “unLEARned” genes are indicated with arrows.
LEARn AND SPORADIC DISEASE: A TWO–HIT PATHWAY
APP protein, the Aβ peptide, and τ protein all appear in healthy individuals. Their mere presence is not a sign of active or incipient AD. Therefore, it stands to reason that AD and other idiopathic neurobiological diseases are likely to be “triggered” independently of the simple presence of any associated proteins. For AD, the LEARn model posits developmental triggering and latency of the APP gene until it is further triggered to express at pathological levels. What could trigger APP and Aβ peptides to be overproduced in sporadic cases of AD? LEARn proposes that the initial mechanism is primed early in life, at developmental stages. Sites of action for AD would be within the promoter of APP and other AD–associated genes. The trigger would be maintained through epigenetic means, such as DNA methylation. It is also possible that genes with products protective against AD are likely to have altered methylation patterns due to environmental stress.
AD, since it usually appears in senescence, would be a “long–latent” condition, and its etiology would be that of a “two–hit” disorder, similarly to those found in currently accepted models of cancer etiology.94 In the case of AD, this second hit would be a broad spectrum of changes in gene expression, especially upregulation of inflammatory factors, which has been shown to be a function of normal aging, as has been increased oxidative DNA damage in the aged cortex and specific targeting of genes by such oxidative damage.95 Environmental insults do not “intentionally” target AD–related genes in the brain. Instead, certain genes, by juxtaposition of CpG and GG sites with important normally active or inactive transcription factor sites, would be particularly vulnerable to the effects of environmental stresses that alter CpG methylation and G oxidation. Testing such changes could, in part, be done by microarray analysis of sporadic and familial AD. If these assays were longitudinal, individual expression profiles could be traced across a lifetime, permitting comparison of the same AD and non–AD individuals at multiple life stages.
The LEARn model lies within theories of epigenetics and the epigenome, specifically adapted to the etiology of sporadic disorders of long latency. The epigenome, the collection of epigenetic markers associated with a specific individual organism’s genome,96 has specific epigenotypes, generated by modification of DNA methylation or oxidation, by changes in histone acetylation patterns, and by variations in the physical arrangement of chromosomal material.97 It is the expression of these epigenotypes, whether they be inherited by imprinting or acquired during life as somatic epitypes8, that contributes to development of sporadic neuropsychiatric diseases.
The epigenome is inherently less stable than the genome. The epigenome is subject to changes in epigenetic markers over time within an individual lifespan. When individuals were followed longitudinally over average periods of 11 and 16 years, both increases and decreases in the methylation of an individual person’s genome were found in a well–characterized Icelandic sample set and in a cohort of families in Utah.91
FURTHER IMPLICATIONS OF LEARn
The LEARn model proposes environmentally–induced changes in DNA methylation and oxidative damage as a fundamental mechanism of pathogenesis in idiopathic sporadic diseases.68 These perturbations would be latent, not as immediately apparent in the same manner found in conventional toxic responses. Thus, reversal of the symptoms of acute exposure to environmental stressors, such as Pb or poor nutrition, would not necessarily entail no long–lasting repercussions for an environmental insult. Conventional anti–toxicity treatments would be insufficient, since removing the cause would not remove the effect. This could be epidemiologically tested. For example, bans enacted upon Pb in gasoline in previous decades significantly reduce levels of Pb in urban dwellers. However, the LEARn model would suggest that levels of AD would not be likely to decrease in response until 50–60 years after the bans were enacted, when individuals would begin to reach ages at risk of sporadic late–onset AD but would not have suffered higher levels of childhood Pb exposure. It should be pointed out that the possibility of “latent sequelae” to asymptomatic exposure to Pb was raised over 30 years ago, although without a specific underlying mechanism.98
However, the potential exists that the latent sequelae modeled by LEARn have a longer reach than inducing late–life disease in an exposed individual. Several groups have recently raised the possibility that acquired characteristics may very well be heritable. This, possibility, of course, brings to mind the theories of Jean–Baptiste Lamarck, who posited a connection between environmental responses and heritable traits in evolution.99 An increasing body of evidence has begun to suggest the possibility of medically important heritable traits.100 Whitelaw’s group determined that the epigenetic methylation states of the mouse Axin gene and associated phenotypes were heritable across generations.101 Anway and Skinner administered the fungicide vinclozolin to mice during gestation and determined that not only did the offspring develop latent disease but the F1 through F4 generations of non–exposed offspring developed these same late–life disorders at a significant level.102 Skinner’s group further determined that alterations in DNA methylation accompanied the pathogenic changes.103 The implication of such work is that LEARn pathogenesis operating through altered DNA methylation may permanently instill greater incidence of latent neuropsychiatric disease such as AD in the descendants of an exposed population.
Long–term latency suggests the possibility of biologically–based remediation. Changes in methylation and/or oxidative damage to DNA, lend towards potential solutions to a LEARn–type environmental exposure. For example, fruit juices, such as concentrated apple juice, have been shown to reverse acute oxidative damage and be a useful source of S–adenosyl methionine, reversing hypomethylation in rats.104 Likewise, dietary melatonin supplementation reduced levels of Aβ in rat cerebral cortex.105 This suggests investigation of the use of appropriate dietary supplementation early in life as a prophylactic or treatment measure against possible latent response to environmental insult. In addition, exercise in rats has been shown to modulate the activity of mucosal betaine–homocysteine methyltransferase 2, potentially reducing aberrant methylation.106 This suggests the possibility that those lifestyle habits suspected to protect against AD, such as physical exercise,107 may work through remediation of early–life aberrant DNA methylation. The LEARn model is not only a useful framework for the study of idiopathic neurobiological disease, but it has further implications in personal and public health practices.
Supplementary Material
Acknowledgments
Sources of Support:
This work was supported by grants from the Alzheimer’s association and NIH (AG18379 and AG18884) to DKL. The authors wish to acknowledge the critique and suggestions made by Dr. John Nurnberger for this manuscript.
References
- 1.Hardy J. A hundred years of Alzheimer's disease research. Neuron. 2006;52(1):3–13. doi: 10.1016/j.neuron.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Maslow K. 2008 Alzheimer's disease facts and figures. Alzheimers Dement. 2008;4(2):110–133. doi: 10.1016/j.jalz.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub D, Comella CL, Horn S. Parkinson's disease--Part 3: Neuropsychiatric symptoms. Am J Manag Care. 2008;14(2 Suppl):S59–S69. [PubMed] [Google Scholar]
- 4.Steyaert JG, De la Marche W. What's new in autism? Eur J Pediatr. 2008;167(10):1091–1101. doi: 10.1007/s00431-008-0764-4. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez JM, Bowden CL, Katz MM, Thompson P, Singh V, Prihoda TJ, et al. Development of the Bipolar Inventory of Symptoms Scale: concurrent validity, discriminant validity and retest reliability. Int J Methods Psychiatr Res. 2008;17(4):198–209. doi: 10.1002/mpr.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahiri DK, Zawia NH, Greig NH, Sambamurti K, Maloney B. Early-life events may trigger biochemical pathways for Alzheimer's disease: the "LEARn" model. Biogerontology. 2008 doi: 10.1007/s10522-008-9162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahiri DK, Maloney B, Basha MR, Ge YW, Zawia NH. How and when environmental agents and dietary factors affect the course of Alzheimer’s disease: the “LEARn” model (Latent Early Associated Regulation) may explain the triggering of AD. Curr Alzheimer Res. 2007;4(2):219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- 8.Lahiri DK, Maloney B. Genes are not our destiny: the somatic epitype bridges between the genotype and the phenotype. Nat Rev Neurosci. 2006;7 [Google Scholar]
- 9.Scrable H, Cavenee W, Ghavimi F, Lovell M, Morgan K, Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci USA. 1989;86(19):7480–7484. doi: 10.1073/pnas.86.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basha MR, Wei W, Bakheet SA, Benitez N, Siddiqi HK, Ge YW, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J Neurosci. 2005;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, McPherson CA, et al. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2(8663):577–580. doi: 10.1016/s0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- 13.Gluckman PD, Hanson MA, Beedle AS. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol. 2007;19(1):1–19. doi: 10.1002/ajhb.20590. [DOI] [PubMed] [Google Scholar]
- 14.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 15.Szyf M. The dynamic epigenome and its implications in toxicology. Toxicol Sci. 2007;100(1):7–23. doi: 10.1093/toxsci/kfm177. [DOI] [PubMed] [Google Scholar]
- 16.Dosunmu R, Wu J, Adwan L, Maloney B, Basha MR, McPherson CA, et al. Lifespan profiles of Alzheimer’s disease-associated genes and their products in monkeys and mice: Common gene regulatory mechanisms but different pathways to Aβ production. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1138. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross OA, Farrer MJ. Pathophysiology, pleiotrophy and paradigm shifts: genetic lessons from Parkinson's disease. Biochem Soc Trans. 2005;33(Pt 4):586–590. doi: 10.1042/BST0330586. [DOI] [PubMed] [Google Scholar]
- 18.Wirdefeldt K, Gatz M, Schalling M, Pedersen NL. No evidence for heritability of Parkinson disease in Swedish twins. Neurology. 2004;63(2):305–311. doi: 10.1212/01.wnl.0000129841.30587.9d. [DOI] [PubMed] [Google Scholar]
- 19.Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, et al. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52(2):M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- 20.Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54(3):264–270. doi: 10.1001/archpsyc.1997.01830150090013. [DOI] [PubMed] [Google Scholar]
- 21.Foundation NP. A Primer on Parkinson Disease. 2009. [cited 2009 February 3]; Available from: http://www.parkinson.org/NETCOMMUNITY/Page.aspx?pid=226&srcid=198.
- 22.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 23.Wu RM, Cheng CW, Chen KH, Lu SL, Shan DE, Ho YF, et al. The COMT L allele modifies the association between MAOB polymorphism and PD in Taiwanese. Neurology. 2001;56(3):375–382. doi: 10.1212/wnl.56.3.375. [DOI] [PubMed] [Google Scholar]
- 24.Dumanchin C, Camuzat A, Campion D, Verpillat P, Hannequin D, Dubois B, et al. Segregation of a missense mutation in the microtubule-associated protein tau gene with familial frontotemporal dementia and parkinsonism. Hum Mol Genet. 1998;7(11):1825–1829. doi: 10.1093/hmg/7.11.1825. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez F, Perez M, Gomez de Barreda E, Goni-Oliver P, Avila J. Tau as a molecular marker of development, aging, and neurodegenerative disorders. Curr Aging Sci. 2008;1:56–61. doi: 10.2174/1874609810801010056. [DOI] [PubMed] [Google Scholar]
- 26.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 27.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 28.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 29.Poorkaj P, Kas A, D'Souza I, Zhou Y, Pham Q, Stone M, et al. A genomic sequence analysis of the mouse and human microtubule-associated protein tau. Mamm Genome. 2001;12(9):700–712. doi: 10.1007/s00335-001-2044-8. [DOI] [PubMed] [Google Scholar]
- 30.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2(8):864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 31.Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock DB, Martin ER, Mayhew GM, Stajich JM, Jewett R, Stacy MA, et al. Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol. 2008;8:6. doi: 10.1186/1471-2377-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suh YH, Checler F. Amyloid precursor protein, presenilins, and alpha-synuclein: molecular pathogenesis and pharmacological applications in Alzheimer's disease. Pharmacol Rev. 2002;54(3):469–525. doi: 10.1124/pr.54.3.469. [DOI] [PubMed] [Google Scholar]
- 34.Forman MS, Schmidt ML, Kasturi S, Perl DP, Lee VM, Trojanowski JQ. Tau and alphasynuclein pathology in amygdala of Parkinsonism-dementia complex patients of Guam. Am J Pathol. 2002;160(5):1725–1731. doi: 10.1016/s0002-9440(10)61119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hare E, Glahn DC, Dassori A, Raventos H, Nicolini H, Ontiveros A, et al. Heritability of age of onset of psychosis in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30959. [DOI] [PubMed] [Google Scholar]
- 36.Nicholls J. A guide to autism spectrum disorders. Practitioner. 2006;250(1687):4–6. 9, 12. [PubMed] [Google Scholar]
- 37.Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12(7):824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 38.Hallmayer J, Pintado E, Lotspeich L, Spiker D, McMahon W, Petersen PB, et al. Molecular analysis and test of linkage between the FMR-1 gene and infantile autism in multiplex families. Am J Hum Genet. 1994;55(5):951–959. [PMC free article] [PubMed] [Google Scholar]
- 39.Boksa P, El-Khodor BF. Birth insult interacts with stress at adulthood to alter dopaminergic function in animal models: possible implications for schizophrenia and other disorders. Neurosci Biobehav Rev. 2003;27(1–2):91–101. doi: 10.1016/s0149-7634(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 40.Dalman C, Allebeck P, Cullberg J, Grunewald C, Koster M. Obstetric complications and the risk of schizophrenia: a longitudinal study of a national birth cohort. Arch Gen Psychiatry. 1999;56(3):234–240. doi: 10.1001/archpsyc.56.3.234. [DOI] [PubMed] [Google Scholar]
- 41.Dalman C, Allebeck P, Gunnell D, Harrison G, Kristensson K, Lewis G, et al. Infections in the CNS during childhood and the risk of subsequent psychotic illness: a cohort study of more than one million Swedish subjects. Am J Psychiatry. 2008;165(1):59–65. doi: 10.1176/appi.ajp.2007.07050740. [DOI] [PubMed] [Google Scholar]
- 42.Khashan AS, Abel KM, McNamee R, Pedersen MG, Webb RT, Baker PN, et al. Higher risk of offspring schizophrenia following antenatal maternal exposure to severe adverse life events. Arch Gen Psychiatry. 2008;65(2):146–152. doi: 10.1001/archgenpsychiatry.2007.20. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya KJ, Matsumoto K, Miyachi T, Tsujii M, Nakamura K, Takagai S, et al. Paternal age at birth and high-functioning autistic-spectrum disorder in offspring. Br J Psychiatry. 2008;193(4):316–321. doi: 10.1192/bjp.bp.107.045120. [DOI] [PubMed] [Google Scholar]
- 44.Kinney DK, Munir KM, Crowley DJ, Miller AM. Prenatal stress and risk for autism. Neurosci Biobehav Rev. 2008;32(8):1519–1532. doi: 10.1016/j.neubiorev.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geller B, Badner JA, Tillman R, Christian SL, Bolhofner K, Cook EH., Jr Linkage disequilibrium of the brain-derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am J Psychiatry. 2004;161(9):1698–1700. doi: 10.1176/appi.ajp.161.9.1698. [DOI] [PubMed] [Google Scholar]
- 46.Ogilvie AD, Battersby S, Bubb VJ, Fink G, Harmar AJ, Goodwim GM, et al. Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet. 1996;347(9003):731–733. doi: 10.1016/s0140-6736(96)90079-3. [DOI] [PubMed] [Google Scholar]
- 47.Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, et al. Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry. 2003;8(11):933–936. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- 48.Frans EM, Sandin S, Reichenberg A, Lichtenstein P, Langstrom N, Hultman CM. Advancing paternal age and bipolar disorder. Arch Gen Psychiatry. 2008;65(9):1034–1040. doi: 10.1001/archpsyc.65.9.1034. [DOI] [PubMed] [Google Scholar]
- 49.Kaymaz N, Krabbendam L, de Graaf R, Nolen W, Ten Have M, van Os J. Evidence that the urban environment specifically impacts on the psychotic but not the affective dimension of bipolar disorder. Soc Psychiatry Psychiatr Epidemiol. 2006;41(9):679–685. doi: 10.1007/s00127-006-0086-7. [DOI] [PubMed] [Google Scholar]
- 50.Wilcox HC, Grados M, Samuels J, Riddle MA, Bienvenu OJ, 3rd, Pinto A, et al. The association between parental bonding and obsessive compulsive disorder in offspring at high familial risk. J Affect Disord. 2008;111(1):31–39. doi: 10.1016/j.jad.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 51.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Childhood parental loss and adult psychopathology in women. A twin study perspective. Arch Gen Psychiatry. 1992;49(2):109–116. doi: 10.1001/archpsyc.1992.01820020029004. [DOI] [PubMed] [Google Scholar]
- 52.Ashford JW, Mortimer JA. Non-familial Alzheimer's disease is mainly due to genetic factors. J Alzheimers Dis. 2002;4(3):169–177. doi: 10.3233/jad-2002-4307. [DOI] [PubMed] [Google Scholar]
- 53.Bales KR, Du Y, Holtzman D, Cordell B, Paul SM. Neuroinflammation and Alzheimer's disease: critical roles for cytokine/Abeta-induced glial activation, NF-kappaB, and apolipoprotein E. Neurobiol Aging. 2000;21(3):427–432. doi: 10.1016/s0197-4580(00)00143-3. [DOI] [PubMed] [Google Scholar]
- 54.Sambamurti K, Granholm AC, Kindy MS, Bhat NR, Greig NH, Lahiri DK, et al. Cholesterol and Alzheimer's disease: clinical and experimental models suggest interactions of different genetic, dietary and environmental risk factors. Curr Drug Targets. 2004;5(6):517–528. doi: 10.2174/1389450043345335. [DOI] [PubMed] [Google Scholar]
- 55.Mortimer JA, van Duijn CM, Chandra V, Fratiglioni L, Graves AB, Heyman A, et al. EURODEM Risk Factors Research Group. Head trauma as a risk factor for Alzheimer's disease: a collaborative re-analysis of case-control studies. Int J Epidemiol. 1991;20(Suppl 2):S28–S35. doi: 10.1093/ije/20.supplement_2.s28. [DOI] [PubMed] [Google Scholar]
- 56.Friedland RP, Fritsch T, Smyth KA, Koss E, Lerner AJ, Chen CH, et al. Patients with Alzheimer's disease have reduced activities in midlife compared with healthy control-group members. Proc Natl Acad Sci USA. 2001;98(6):3440–3445. doi: 10.1073/pnas.061002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall DB, Holmlin RE, Barton JK. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382(6593):731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 58.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 60.Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286(2):355–365. doi: 10.1016/s0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 61.Fowler BA, Whittaker MH, Lipsky M, Wang G, Chen XQ. Oxidative stress induced by lead, cadmium and arsenic mixtures: 30-day, 90-day, and 180-day drinking water studies in rats: an overview. BioMetals. 2004;17(5):567–568. doi: 10.1023/b:biom.0000045740.52182.9d. [DOI] [PubMed] [Google Scholar]
- 62.Campos AC, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D'Almeida V, et al. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia. 2007;9(12):1111–1121. doi: 10.1593/neo.07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sparks DL, Schreurs BG. Trace amounts of copper in water induce {beta}-amyloid plaques and learning deficits in a rabbit model of Alzheimer's disease. Proc Natl Acad Sci USA. 2003;100(20):11193–11194. doi: 10.1073/pnas.1832769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sparks DL, Friedland R, Petanceska S, Schreurs BG, Shi J, Perry G, et al. Trace Copper Levels in the Drinking Water, but not Zinc or Aluminum Influence CNS Alzheimer-Like Pathology. J Nutr Health Aging. 2006;10(4):247–254. [PMC free article] [PubMed] [Google Scholar]
- 65.Latha KS, Anitha S, Rao KS, Viswamitra MA. Molecular understanding of aluminum-induced topological changes in (CCG)12 triplet repeats: relevance to neurological disorders. Biochim Biophys Acta. 2002;1588(1):56–64. doi: 10.1016/s0925-4439(02)00133-3. [DOI] [PubMed] [Google Scholar]
- 66.Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life Sci. 2001;58(5–6):721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rossiello MR, Aresta AM, Prisco M, Kanduc D. DNA hypomethylation during liver cell proliferation induced by a single dose of lead nitrate. Boll Soc Ital Biol Sper. 1991;67(12):993–997. [PubMed] [Google Scholar]
- 68.Bolin CM, Basha R, Cox D, Zawia NH, Maloney B, Lahiri DK, et al. Exposure to lead and the developmental origin of oxidative DNA damage in the aging brain. FASEB J. 2006;20(6):788–790. doi: 10.1096/fj.05-5091fje. [DOI] [PubMed] [Google Scholar]
- 69.Basha MR, Murali M, Siddiqi HK, Ghosal K, Siddiqi OK, Lashuel HA, et al. Lead (Pb) exposure and its effect on APP proteolysis and Abeta aggregation. FASEB J. 2005;19(14):2083–2084. doi: 10.1096/fj.05-4375fje. [DOI] [PubMed] [Google Scholar]
- 70.Sparks DL, Lochhead J, Horstman D, Wagoner T, Martin T. Water quality has a pronounced effect on cholesterol-induced accumulation of Alzheimer amyloid beta (Abeta) in rabbit brain. J Alzheimers Dis. 2002;4(6):523–529. doi: 10.3233/jad-2002-4609. [DOI] [PubMed] [Google Scholar]
- 71.Winchester PD, Huskins J, Ying J. Agrichemicals in surface water and birth defects in the United States. Acta Paediatr. 2009;98(4):664–669. doi: 10.1111/j.1651-2227.2008.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Basha MR, Brock B, Maloney B, Cox D, Harry J, et al. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pollwein P, Masters CL, Beyreuther K. The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 1992;20(1):63–68. doi: 10.1093/nar/20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Docagne F, Gabriel C, Lebeurrier N, Lesne S, Hommet Y, Plawinski L, et al. Sp1 and Smad transcription factors co-operate to mediate TGF-beta-dependent activation of amyloid-beta precursor protein gene transcription. Biochem J. 2004;383(Pt 2):393–399. doi: 10.1042/BJ20040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lahiri DK, Robakis NK. The promoter activity of the gene encoding Alzheimer beta-amyloid precursor protein (APP) is regulated by two blocks of upstream sequences. Brain Research Molecular Brain Research. 1991;9(3):253–257. doi: 10.1016/0169-328x(91)90009-m. [DOI] [PubMed] [Google Scholar]
- 76.La Fauci G, Lahiri DK, Salton SR, Robakis NK. Characterization of the 5'-end region and the first two exons of the beta-protein precursor gene. Biochem Biophys Res Commun. 1989;159(1):297–304. doi: 10.1016/0006-291x(89)92437-6. [DOI] [PubMed] [Google Scholar]
- 77.Song W, Lahiri DK. Molecular cloning of the promoter of the gene encoding the Rhesus monkey beta-amyloid precursor protein: structural characterization and a comparative study with other species. Gene. 1998;217(1–2):151–164. doi: 10.1016/s0378-1119(98)00337-0. [DOI] [PubMed] [Google Scholar]
- 78.Hoffman PW, Chernak JM. DNA binding and regulatory effects of transcription factors SP1 and USF at the rat amyloid precursor protein gene promoter. Nucleic Acids Res. 1995;23(12):2229–2235. doi: 10.1093/nar/23.12.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heicklen-Klein A, Ginzburg I. Tau promoter confers neuronal specificity and binds Sp1 and AP-2. J Neurochem. 2000;75(4):1408–1418. doi: 10.1046/j.1471-4159.2000.0751408.x. [DOI] [PubMed] [Google Scholar]
- 80.Wang CS, Burke JR, Steffens DC, Hulette CM, Breitner JC, Plassman BL. Twin pairs discordant for neuropathologically confirmed Lewy body dementia. J Neurol Neurosurg Psychiatry. 2009;80(5):562–565. doi: 10.1136/jnnp.2008.151654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ryu H, Barrup M, Kowall NW, McKeel AC. Epigenetic modification in a monozygotic twin with Alzheimer's disease. Alzheimers & Dementia. 2008;4(4):T598. [Google Scholar]
- 82.Guilarte TR. Prenatal lead exposure and schizophrenia: a plausible neurobiologic connection. Environ Health Perspect. 2004;112(13):A724. doi: 10.1289/ehp.112-a724a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Opler MG, Buka SL, Groeger J, McKeague I, Wei C, Factor-Litvak P, et al. Prenatal exposure to lead, delta-aminolevulinic acid, and schizophrenia: further evidence. Environ Health Perspect. 2008;116(11):1586–1590. doi: 10.1289/ehp.10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jill James S, Melnyk S, Jernigan S, Hubanks A, Rose S, Gaylor DW. Abnormal transmethylation/transsulfuration metabolism and DNA hypomethylation among parents of children with autism. J Autism Dev Disord. 2008 doi: 10.1007/s10803-008-0614-2. [DOI] [PubMed] [Google Scholar]
- 85.Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 87.Rosa A, Picchioni MM, Kalidindi S, Loat CS, Knight J, Toulopoulou T, et al. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(4):459–462. doi: 10.1002/ajmg.b.30616. [DOI] [PubMed] [Google Scholar]
- 88.Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, Kundakovic M, et al. The human reelin gene: transcription factors (+), repressors (−) and the methylation switch (+/−) in schizophrenia. Pharmacol Ther. 2006;111(1):272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 89.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3(5):e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galic MA, Riazi K, Heida JG, Mouihate A, Fournier NM, Spencer SJ, et al. Postnatal inflammation increases seizure susceptibility in adult rats. J Neurosci. 2008;28(27):6904–6913. doi: 10.1523/JNEUROSCI.1901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanvah S, Schuster GB. The sacrificial role of easily oxidizable sites in the protection of DNA from damage. Nucleic Acids Res. 2005;33(16):5133–5138. doi: 10.1093/nar/gki801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 96.Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum Mol Genet. 2006;15:R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- 97.van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007;64(12):1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De la Burde B, Choat MS. Does asymptomatic lead exposure in children have latent sequelae? J Pediatr. 1972;81(6):1088–1091. doi: 10.1016/s0022-3476(72)80236-1. [DOI] [PubMed] [Google Scholar]
- 99.Liu Y. Like father like son. A fresh review of the inheritance of acquired characteristics. EMBO Rep. 2007;8(9):798–803. doi: 10.1038/sj.embor.7401060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gorelick R. Neo-Lamarckian medicine. Med Hypotheses. 2004;62(2):299–303. doi: 10.1016/S0306-9877(03)00329-3. [DOI] [PubMed] [Google Scholar]
- 101.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100(5):2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147(6 Suppl):S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 103.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chan A, Shea TB. Supplementation with apple juice attenuates presenilin-1 overexpression during dietary and genetically-induced oxidative stress. J Alzheimers Dis. 2006;10(4):353–358. doi: 10.3233/jad-2006-10401. [DOI] [PubMed] [Google Scholar]
- 105.Lahiri DK, Chen D, Ge YW, Bondy SC, Sharman EH. Dietary supplementation with melatonin reduces levels of amyloid beta-peptides in the murine cerebral cortex. J Pineal Res. 2004;36(4):224–231. doi: 10.1111/j.1600-079X.2004.00121.x. [DOI] [PubMed] [Google Scholar]
- 106.Buehlmeyer K, Doering F, Daniel H, Kindermann B, Schulz T, Michna H. Alteration of gene expression in rat colon mucosa after exercise. Ann Anat. 2008;190(1):71–80. doi: 10.1016/j.aanat.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 107.Kivipelto M, Solomon A. Alzheimer's disease - the ways of prevention. J Nutr Health Aging. 2008;12(1):89S–94S. doi: 10.1007/BF02982595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.