Abstract
Variations in levels of apolipoprotein E (ApoE) have been tied to the risk and progression of Alzheimer’s disease (AD). Our group has previously compared and contrasted the promoters of the mouse and human ApoE gene (APOE) promoter sequences and found notable similarities and significant differences that suggest the importance of the APOE promoter’s role in the human disease. We examine here three specific single-nucleotide polymorphisms within the human APOE promoter region, specifically at −491 (A/T), −427 (T/C), and at −219 (G/T) upstream from the +1 transcription start site. The −219 and −491 polymorphic variations have significant association with instance of AD, and −491AA has significant risk even when stratified for the APOEε4 allele. We also show significant effects on reporter gene expression in neuronal cell cultures, and, notably, these effects are modified by species origin of the cells. The −491 and −219 polymorphisms may have an interactive effect in addition to any independent activity. DNA–protein interactions differ between each polymorphic state. We propose SP1 and GATA as candidates for regulatory control of the −491 and −219 polymorphic sites. This work’s significance lies in drawing connection among APOE promoter polymorphisms’ associations with AD to functional promoter activity differences and specific changes in DNA–protein interactions in cell culture-based assays. Taken together, these results suggest that APOE expression levels are a risk factor for AD irrespective of APOEε4 allele status.
Keywords: aging, Alzheimer’s disease, dementia, genetics, gene regulation
INTRODUCTION
Alzheimer’s disease (AD) is the leading cause of dementia among the elderly [Hebert et al., 2003]. A known risk factor for AD is the apolipoprotein E (ApoE) ε4 genotype [Corder et al., 1993; Saunders et al., 1993; Lahiri, 2004a]. In addition to coding sequence polymorphisms, three promoter polymorphisms have been identified on the ApoE gene (APOE) with potential influence on sporadic AD [Bullido et al., 1998; Artiga et al., 1998b; Lambert et al., 2002]. These polymorphisms reside at −491 (rs449647, A/T), −427 (rs769446, T/C), and −219 (TH1/E47cs, rs405509, G/T) [Lambert et al., 2002, 2004]. It has been reported that the −491AA genotype confers an independent risk for developing AD [Lambert et al., 2002, 2004], as has likewise been stated for the −219T allele [Lambert et al., 2002, 2004], although the field is not unanimous, since reports that failed to find an AD association with the −491 [Thome et al., 1999; Toji et al., 1999] or the −219 [Zurutuza et al., 2000; Tycko et al., 2004] loci are also in the literature. Meta-analysis of 38 studies indicated an AD-associated OR (95% CI) of 0.73 (0.64, 0.82) for the T allele at −491; meta-analysis of 13 studies showed an OR (95% CI) of 0.86 (0.68, 1.09) for the C allele at −427; and meta-analysis of 20 studies revealed an OR (95% CI) of 0.73 (0.68, 0.78) for the G allele at −217 [Bertram et al., 2007]. While important work has been done in characterizing effects of individual single nucleotide polymorphisms (SNPs) on promoter activity [Artiga et al., 1998b; Ramos et al., 2005], evidence exists that these polymorphisms may function in vivo as haplogroups, with pathogenic influence beyond independent effects they may exert [Parra-Bonilla et al., 2003].
We have previously characterized the 5′-flanking regions of the APOE genes of mouse [Lahiri et al., 2002] and human [Du et al., 2005] and, more recently, determined important structural and functional differences between them, including the presence of functional promoter regulatory domains HuA (“human A”) through HuE and MoA (“mouse A”) through MoD [Maloney et al., 2007]. While the mouse sequence was determined to share homology at the −219 polymorphic site, no homology was found between human and mouse at either of the other two sites (−491 and −427). This notable structural difference between two species suggests an important role for the APOE promoter in the pathogenesis of AD. Therefore, we continued our work on the APOE promoter with the human sequence.
Healthy and AD-diagnosed subjects were genotyped for the APOE ε2/ε3/ε4 allele and for each of the two alleles at −491, −427, and −219. Statistically significant associations were observed with homozygosity for the A allele at the −491 promoter polymorphism and with homozygosity for the T allele at the −219 SNP. After stratification for presence of the ε4 allele, the −491AA genotype retained significance as a risk factor for AD in the ε4-negative population, whereas no such an association was detected in the ε4-positive group. Regarding the −219TT genotype, this same analysis revealed that the association was lost in both groups after stratification.
To investigate activity and potential interactions of the three APOE promoter SNPs, we constructed eight different clones containing 1.4 kb of the APOE 5′-flanking region. These clones included all currently known polymorphic variants at each of three locations fused to the chloramphenicol acetyltransferase (CAT) reporter gene. The clones were transfected into human SK–N–SH neuroblastoma (NB) and rat pheochromocytoma (PC12) neuronal cells and resulting reporter levels were analyzed. We determined that the −491 A/T polymorphism exerted independent effect on reporter protein levels in both NB and PC12 cells. The −219 G/T polymorphism had significant independent effects in NB cells but not in PC12 cells. Multiple ANOVA analysis of data indicated a significant interaction between the effects exerted by the polymorphisms at −491 and −219 in PC12 cells but not in NB cells. The −427 polymorphism did not have any significant effect on reporter gene product levels. In addition, each polymorphic state displayed differential DNA–protein interactions as revealed from electrophoretic mobility shift assay and Southwestern blotting experiments. Taken together, our work partially confirms the proposed existence [Parra-Bonilla et al., 2003] of a haplogroup of the −491 and −219 polymorphisms of the APOE promoter. Furthermore, the −491 polymorphism was confirmed as significantly altering APOE promoter activity independently of other polymorphisms in two cell lines from two different species while the −219 polymorphism only had this effect in human NB cells.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, reagents were purchased from Sigma (St. Louis, MO) and were of “molecular biology” or “analytic” quality. Enzymes were purchased from Roche (Indianapolis, IN). Cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
Cell Lines
Rat pheochromocytoma (PC12) and human SK–N–SH neuroblastoma (NB) cell cultures were acquired from ATCC and routinely cultured in our laboratory according to ATCC instructions [Ghosh et al., 2000]. Tissue culture reagents were obtained from Invitrogen.
Preparation of Nuclear Extracts
Nuclear extracts from NB and PC12 cell lines and from mouse brain tissue were obtained commercially (Active Motif, Carlsbad, CA).
Populations
The study population was recruited from the Alzheimer’s disease research center at Mayo Clinic, Rochester, MN. Samples from all subjects were collected under IRB approved protocols with informed consent signed by the individuals or next of kin. Patients were diagnosed with probable AD according to NINCDS–ADRDA criteria. DNA was extracted from peripheral blood cells using standard protocols. All sample DNA’s were plated in 96-well plates with cases and controls being randomly plated together.
The series comprised 310 sporadic late-onset AD cases (67% female, 15% autopsy confirmed) with mean age of 83.2±8.0 and mean age at onset of 77.6 ± 8.0. Controls for this cohort included 425 individuals (68% female; 5% autopsy confirmed) with mean age of 83.3 ± 7.1.
Genotyping of Patient DNA for APOε Promoter SNP Status
DNA extraction and APOE genotyping were performed according to published methods with minor modifications [Lahiri and Nurnberger, 1991; Crook et al., 1994; Lambert et al., 1998; Artiga et al., 1998b]. A summary of primers, conditions, and genotyping details can be found in Table I.
TABLE I.
Genotyping Conditions for the APOE Locus
| Polymorphic site | Primer sequencea | Product length (bp) |
Restriction enzyme |
PCR conditionsb |
Agarose (%) |
Size of fragments (bp) |
|---|---|---|---|---|---|---|
| −491 A/T (rs449647) | 5′-TGT TGG CCA GGC TGG TCT CGA-3′ | 228 | DpnII | TD65 | 3.5 | A: 147 |
| 5′-CTT CCT TTC CTG TCC CAG TCC-3′ | T: 128 + 19 | |||||
| −427 T/C (rs769446) | 5′-TGT TGG CCA GGC TGG TCT CGA-3′ | 228 | AluI | TD65 | 3.5 | T: 144 + 84 |
| 5′-CTT CCT TTC CTG TCC CAG TCC-3′ | C: 228 | |||||
| −219A/C (rs405509) | 5′-AGA ATG GAG GAG GGT GCC TG-3′ | 233 | BstNI | TD65 | 4.0 | T: 69 + 59 + 57 + 48 |
| 5′-ACT CAA GGA TCC CAG CAT TG-3′ | G: 69 + 59 + 57 + 30 + 18 | |||||
| Coding region (rs429358; codon 112) (rs7412; codon 158) |
5′-TAA GCT TGG CAC GGC TGT CCA AGG A-3′ | 244 | CfoI | TD60c | 4.5 | ε2: 91 + 81 |
| 5′-ACA GAA TTC GCC CCG GCC TGG TAC AC-3′ | ε3: 91 + 48 + 33 | |||||
| ε4: 72 + 48 + 33 + 19 |
Mismatches introduced to increase amplification specificity are indicated in bold.
All PCRs were run at 40 cycles using touchdown (TD) protocols. The starting annealing temperature is shown in the table. The PCR profile included 4 cycles at the initial annealing temperature, 20 cycles with a decrease of 0.5°/cycle of the annealing temperature and 16 cycles at the final temperature.
DMSO (5% final concentration) was added to the reaction mix to increase specificity.
Construction of APOε Promoter Polymorphism-CAT Reporter Fusion Clones
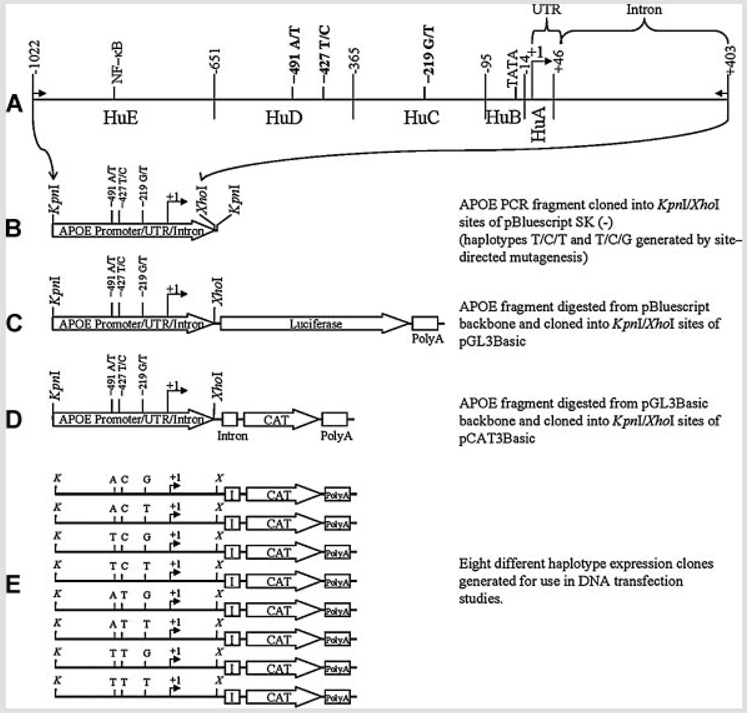
Genotyped DNA was isolated via PCR from human subjects using oligomers that inserted HindIII, KpnI, and XhoI linkers at the ends of a 1.4 kb APOE promoter/intron fragment (Fig. 1A). The fragment was inserted into the HindIII and XhoI sites of pBluescript SK (−) (Fig. 1B). Clones corresponding to six of eight possible SNP polymorphism combinations were constructed in this fashion. Two haplotypes (T/C/T and T/C/G) did not appear in our sample population (data not shown). These two clones were derived by site–directed mutagenesis of the corresponding −427T/−219T or −427T/−219 G haplotype clones using the Transformer site-directed mutagenesis kit (Clontech, Mountain View, CA). The pBluescript-backbone clones were digested with KpnI and XhoI and 1.4 kb APOE fragments were cloned into pGL3 (Promega) (Fig. 1C). Preliminary attempts to perform luciferase-based assays produced unacceptable noise due to well-to-well variation and cross talk from various neighboring wells. Therefore, the pGL3-backbone clones were digested with KpnI and XhoI. The 1.4 kb APOE promoter fragment band was purified from each and subcloned into the KpnI and XhoI sites of vector pCAT3Basic (Promega) to produce eight fusion clones, containing each possible permutation of the SNP variants, driving the CAT reporter coding sequence (Fig. 1D) to produce eight different polymorphic haplotype clones (Fig. 1E).
FIG. 1.
Construction of expression-cassette clones containing three single–nucleotide polymorphisms in the APOε promoter sequence. A: Schematic diagram of a 1.4 kb PCR fragment of the APOε gene (small arrows indicate locations of PCR primers), including the 5′-UTR and 1 kb of promoter, and a fragment of the first intron of the gene, as produced by PCR from human genomic DNA samples. Three single-nucleotide polymorphic sites [Bullido et al., 1998; Artiga et al., 1998b; Lambert et al., 2002] are indicated (−491, −427, and −219) in relationship to the transcription start site (+1). An active NF-κB site [Du et al., 2005] and promoter activity domains [Maloney et al., 2007] (HuA-“human A”-through HuE) are also indicated. B: The 1.4 kb fragment was cloned into the KpnI and XhoI sites of pBluescript SK (−). C: The pBluescript-backbone clone was digested with KpnI and XhoI, and the 1.4 kb APOε fragment was subcloned into the KpnI/XhoI sites of pGL3Basic. D: The pGL3Basic-backbone clone was digested with KpnI and XhoI, and the 1.4 kb APOε fragment was cloned into the KpnI/XhoI sites of pCAT3Basic. E: Eight different haplotype APOε promoter-CAT expression gene clones were generated and subsequently used for DNA transfection studies.
DNA Transfection of APOε Promoter Polymorphism-CAT Reporter Constructs in NB and PC12 Cell Cultures
NB and PC12 cell cultures were transfected with empty vector pCAT3Basic or one of eight polymorphic APOE promoter-CAT reporter constructs by Lipofectamine and the associated Plus Reagent (Invitrogen), as described previously [Ghosh et al., 2000]. Transfection was carried out in 2–3 × 106 cells per 60mm plate in triplicate, with 2.7 µg of CAT reporter clone plasmid DNA. To monitor transfection efficiency, cells were cotransfected with 0.3µg pSVβGAL (Promega) under the same conditions. Following transfection, cells were harvested, extracts prepared, protein concentration determined, CAT reporter protein levels were measured by enzyme-labeled immunosorbent assay (ELISA) and β-galactosidase (β-GAL) activity was assayed colorimetrically [Ghosh et al., 2000]. Presence of β-GAL levels above background was taken as indicative of successful transfection.
Reporter Gene Expression and Data Analysis
Activity of the reporter gene for all fusion clones was checked by measuring reporter protein levels by ELISA, using a commercial kit (Roche). Reporter protein level was adjusted to total protein in extract. Assays were done in linear range from three transfection experiments. Results from adjusted reporter gene activity were statistically analyzed with the SAS System 9.1 statistical analysis package (SAS Institute, Cary, NC) via three-way ANOVA followed by Waller–Duncan multiple range test or Student’s t-test.
Electrophoretic Mobility Shift Assay (EMSA) of APOε Polymorphisms
Oligomers reflecting the −491 A/T and −219 G/T SNPs, along with complementary oligomers, (Table II) were designed according to Artiga et al. [1998b] and commercially synthesized (Invitrogen) as single stranded oligomers. Oligomers were annealed with complements and labeled with [γ32P]-ATP (GE Healthcare, Piscataway, NJ) via polynucleotide kinase (Roche). All radiolabeled probes used in subsequent experiments were double stranded (ds). The assay was carried out with 10,000 cpm of probe (20–50 ng) and 10 µg of nuclear extracts. Nuclear protein extracts from NB and PC12 cells were incubated in 19 µl of EMSA binding buffer (10mM Tris–HCl, pH 7.5; 50mM NaCl; 0.5mM DTT, 5% glycerol; 0.05% Triton X-100; 100 µg/ml poly dI:dC) at 8°C for 15 min. In the “Competition-EMSA” assay, excess (150× molar concentration) unlabeled ds-oligomer was added and reactions were incubated at 8°C for 15 min. Radioactive probe was added (10,000 CPM) and reactions further incubated at 8°C for 30 min. The samples were mixed with loading dye (50% glycerol, 1mM EDTA), and the products of the binding reaction were separated on a nondenaturing polyacrylamide gel (5%) in 1 × TGE buffer (50mM Tris-base, 380mM glycine, and 2mM EDTA). The gel was dried and exposed to X-ray film with intensifying screen for fluorography at −70°C. Free unbound oligonucleotides ran at the bottom of the gel; different protein–DNA complexes were detected as mobility–retarded bands. Experiments were repeated with duplicate ds-oligomers obtained commercially (IDT Technology, Coralville, IA). Labeling of bands with Roman numerals was done according to migration rates of DNA-protein complexes.
TABLE II.
Oligomers for Electrophoretic Mobility Shift and Southwestern Assays
| Oligomer | Sequence |
|---|---|
| −491A-F | 5′-GCTGGTCTCAAACTCCTGACCTTAA-3′ |
| −491A-R | 5′-TTAAGGTCAGGAGTTTGAGACCAGC-3′ |
| −491T-F | 5′-GCTGGTCTCAATCTCCTGACCTTAA-3′ |
| −491T-R | 5′-TTAAGGTCAGGAGATTGAGACCAGC-3′ |
| −219G-F | 5′-GGAGGAGGGTGTCTGGATTACTGGGCG-3′ |
| −219G-R | 5′-CGCCCAGTAATCCAGACACCCTCCTCC-3′ |
| −219T-F | 5′-GGAGGAGGGTGTCTGTATTACTGGGCG-3′ |
| −219T-R | 5′-CGCCCAGTAATACAGACACCCTCCTCC-3′ |
Southwestern Blotting of Multiple Nuclear Extracts With Polymorphic ds-Oligomers
Nuclear extracts from NB and PC12 cells, 10 µg total protein, each, were run in duplicate sets on 10% SDS–PAGE in a MiniProtean gel apparatus (BioRad) as described previously [Lahiri et al., 1994]. Samples were transferred via tank blotting to 0.45 µm nitrocellulose membranes in 25mM of Tris–HCl, pH 7.4, 190mM glycine, 1mM EDTA and 0.01% SDS. Membrane was cut into two parts and each was probed individually by Southwestern blotting [Lahiri, 1998]. Briefly, proteins bound on the filter were renatured by incubation at 4°C for 24 h in 15ml of 10mM HEPES, pH 7.9, 50mM NaCl, 0.1mM EDTA, 10mM MgCl2, 1mM DTT, 10% glycerol and 5% milk powder (Carnation Non-fat). For DNA binding, the blocking solution was replaced by 15ml binding buffer (same composition as above except only 0.25% milk powder was used) with 10 µg poly dI:dC and 20 ng of each probe (specific activity 0.8 to 1.0 × 106 - CPM/ng) and was gently shaken at 4°C for 18 h. Filters were washed twice at room temperature in 50ml of the binding buffer (without probe and poly dI:dC) for 15 min each and exposed to X–ray film.
RESULTS
Genotype Frequencies of APOε Polymorphisms and AD Risk
The distribution of alleles and genotypes was consistent with that expected under Hardy–Weinberg equilibrium (−491: P>0.5; −427: P>0.9; −219: P>0.2; coding region: P>0.6). Table III summarizes the results of the genetic analysis. As expected, the strongest association was found with the polymorphism at the coding region, possession of at least one copy of the ε4 allele significantly increased the risk for the disease (OR=4.29; CI95% = [3.16–5.82]). Additionally the reported protective effect of the ε2 allele was also seen (, P = 5.43 × 10−5; OR = 0.40 CI95% = [0.25–0.63]; ε2 allele carriers vs. non-carriers). In addition to this, statistically significant associations of similar magnitude were observed with homozygosity for the A allele at the −491 promoter polymorphism (, P = 0.006; OR = 1.55 CI95% = [1.13–2.12]) and with homozygosity for the T allele at the −219 SNP (, P = 0.003; OR = 1.64, CI95% = [1.18–2.18]).
TABLE III.
Allelic and Genotypic Distributions for the Promoter and Coding Region Polymorphisms at the APOε Locus
| Allele (n (proportion)) | Genotype (n (proportion)) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | n | A | T | A/A | A/T | T/T | ||||
| −491a | ||||||||||
| Controls | 491 | 790 (0.80) | 192 (0.20) | 320 (0.65) | 150 (0.31) | 21 (0.04) | ||||
| AD | 316 | 544 (0.86) | 88 (0.14) | 235 (0.75) | 74 (0.23) | 7 (0.02) | ||||
| Allele (n (proportion)) | Genotype (n (proportion)) | |||||||||
| Polymorphism | n | T | C | T/T | T/C | C/C | ||||
| −427b | ||||||||||
| Controls | 474 | 839 (0.89) | 109 (0.11) | 371 (0.83) | 97 (0.21) | 6 (0.01) | ||||
| AD | 311 | 560 (0.90) | 62 (0.10) | 252 (0.81) | 56 (0.18) | 3 (0.01) | ||||
| Allele (n (proportion)) | Genotype (n (proportion)) | |||||||||
| Polymorphism | n | G | T | G/G | G/T | T/T | ||||
| −219c | ||||||||||
| Controls | 488 | 527 (0.54) | 449 (0.46) | 136 (0.28) | 255 (0.52) | 97 (0.20) | ||||
| AD | 315 | 292 (0.46) | 338 (0.54) | 68 (0.22) | 156 (0.49) | 91 (0.29) | ||||
| Allele (n (proportion)) | Genotype (n (proportion)) | |||||||||
| Polymorphism | n | ε2 | ε3 | ε4 | ε2/ε2 | ε2/ε3 | ε2/ε4 | ε3/ε3 | ε3/ε4 | ε4/ε4 |
| Coding regiond | ||||||||||
| Controls | 493 | 96 (0.10) | 772 (0.78) | 118 (0.12) | 5 (0.01) | 73 (0.15) | 13 (0.03) | 301 (0.60) | 97 (0.20) | 4 (0.01) |
| AD | 316 | 26 (0.04) | 394 (0.62) | 212 (0.34) | 13 (0.04) | 13 (0.04) | 125 (0.40) | 131 (0.41) | 34 (0.11) | |
Genotypic distribution ; P = 0.016.
Genotypic distribution ; P = 0.634.
Genotypic distribution ; P = 0.007.
Genotypic distribution ; P < 10−20.
After stratification for presence of the ε4 allele, the −491AA genotype was still a risk factor for AD in the ε4-negative population (, P = 0.032; OR = 1.58, CI95% = [1.04–2.40]) whereas no such an association was still detected in the ε4-positive group. Regarding the −219TT genotype, this same analysis reveals that the association was lost in both groups after stratification.
Finally, logistic regression was used to detect interactions between the different polymorphisms, as well as age or sex. In this case, only the coding region polymorphism was still associated with the risk for AD (P < 10−4; OR = 1.90, CI95% = [1.64–2.22]).
Sequencing of APOε Promoter With SNP Regions
PCR of human genomic DNA samples with specific primers produced a 1.4 kb fragment (Fig. 1). Excepting for the specific locations of the three SNPs, DNA sequences derived within our sample were 100% homologous to GenBank sequence #M10065.
Reporter Expression Levels of APOε Promoter Polymorphisms in NB and PC12 Cells
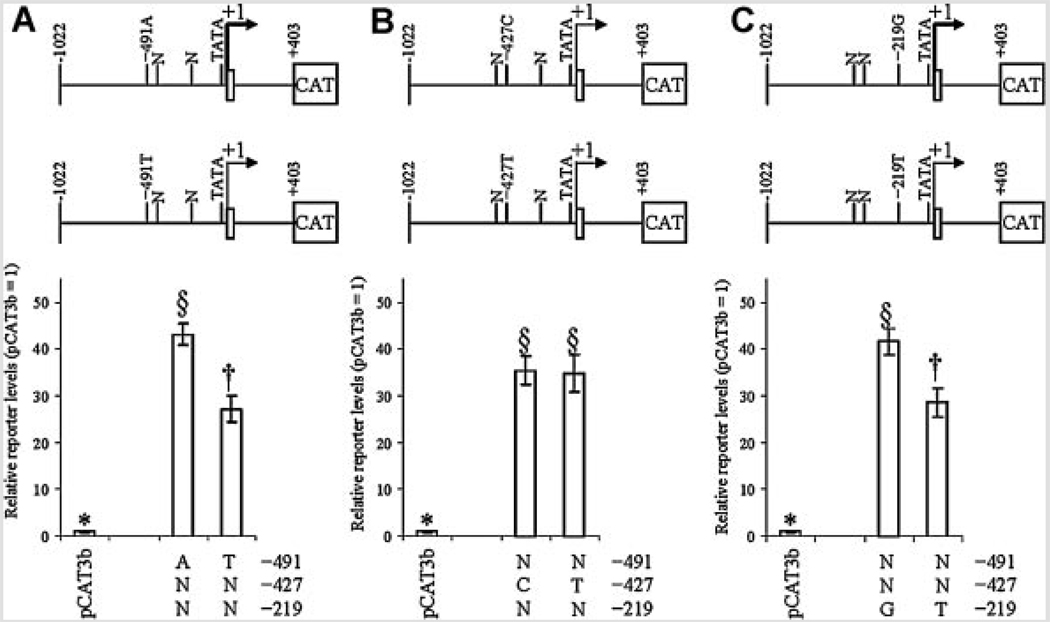
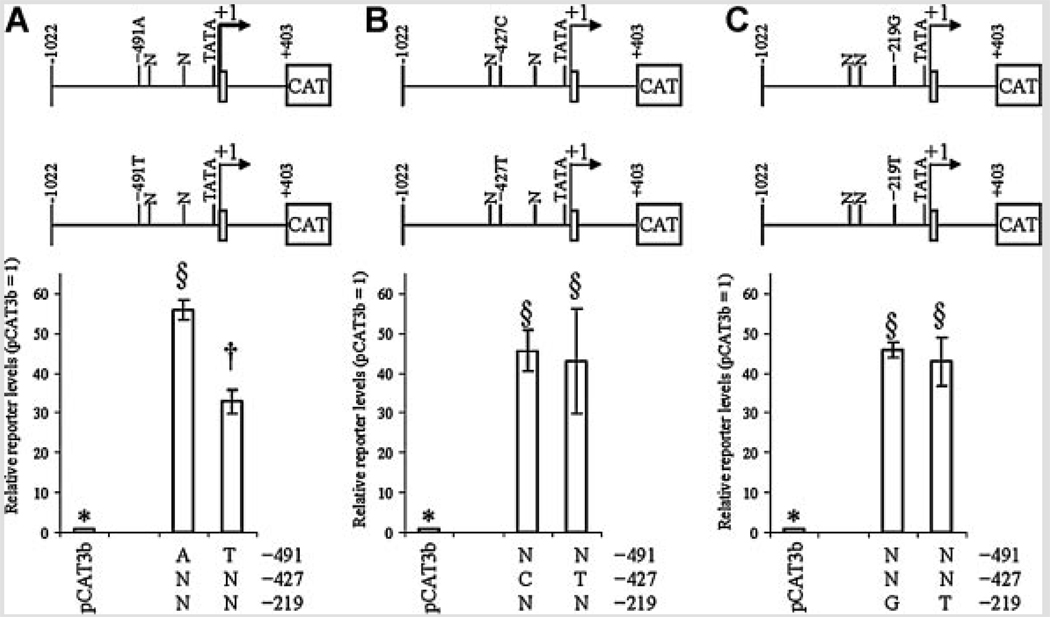
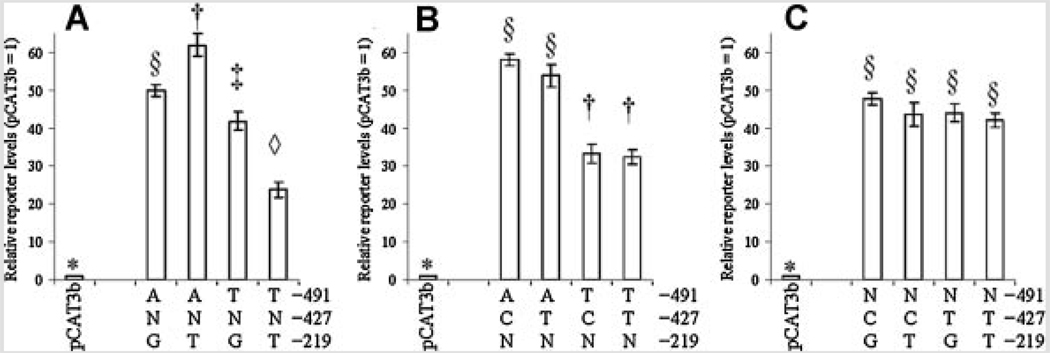
To investigate effects of the three APOE promoter polymorphisms at −491, −427, and −219 on promoter activity, NB and PC12 cell cultures were independently transfected with eight APOE promoter polymorphic clones, as described herein. Cells were also cotransfected with pSVβGAL to monitor transfection efficiency. Cells were collected and extracts used to measure total protein and for ELISA of reporter gene protein. Correlation analysis of raw data revealed that a strong confounding correlation (r = 0.628, P = 0.001) existed between ELISA signal of CAT reporter protein and activity of β-galactosidase (Table IV). Therefore, statistical analysis was continued with CAT reporter protein signal adjusted by total protein. In all cases, APOE promoter-reporter gene fusion clones had significantly higher reporter protein levels than did pCAT3Basic vector backbone. Analysis by each individual polymorphic site revealed that the −491 polymorphism drove significant independent difference in reporter protein levels in both cell lines (Fig. 2A and Fig. 3A). The −427 polymorphism (Fig. 2B and Fig 3B) showed no significant differences in activity between either allele. The −219 polymorphism had a significant difference of reporter levels in NB cells (Fig. 2C) but not in PC12 cells (Fig. 3C).
TABLE IV.
Correlation of Raw CAT Reporter Protein ELISA, β-GAL Activity, and Total Cellular Protein in Transient DNA Transfection Assays
| CAT ELISA | Total protein | β-GAL Activity | ||
|---|---|---|---|---|
| A. | Correlations in NB cells | |||
| CAT ELISA | 1 | |||
| na | ||||
| Total protein | 0.23355 | 1 | ||
| P = 0.2721 | na | |||
| β-GAL activity | 0.54456 | −0.40755 | 1 | |
| P = 0.0059 | P = 0.0481 | na | ||
| B. | Correlations in PC12 cells | |||
| CAT ELISA | 1 | |||
| na | ||||
| Total protein | −0.27462 | 1 | ||
| P = 0.1941 | na | |||
| β-GAL activity | 0.77672 | −0.63253 | 1 | |
| P < .0001 | P = 0.0009 | na |
FIG. 2.
Effects of individual single–nucleotide polymorphism on reporter levels in NB cells. APOε promoter polymorphism-CAT reporter fusion clones were transfected into NB cells as described. Cell lysates were extracted and reporter protein levels measured by ELISA. Data were grouped in three ways, according to the state of each individual polymorphism. All APOε-derived clones drove reporter levels significantly higher than empty pCAT3-basic backbone. “N” refers to either variant at the non-specified polymorphism locus. The top portion of each figure schematically depicts locations of SNPs with respect to the APOε promoter. A: −491 A/T polymorphism. Each state was significantly different from the other. B: −427 T/C polymorphism. Neither state differed significantly from the other. C: −219 G/T polymorphism. Each state was significantly different from the other.
FIG. 3.
Effects of individual single-nucleotide polymorphism on reporter levels in PC12 cells. APOε promoter polymorphism-CAT reporter fusion clones were transfected into PC12 cells as described. Cell lysates were extracted and reporter levels measured by ELISA. Data were grouped in three ways, according to the state of each individual polymorphism. All APOε-derived clones drove reporter protein levels significantly higher than empty pCAT3-basic backbone. “N” refers to either variant at the non-specified polymorphism locus. The top portion of each figure schematically depicts locations of SNPs with respect to the APOε promoter. A: −491 A/T polymorphism. Each state was significantly different from the other. B,C: −427 T/C and −219 G/T polymorphisms. Neither state within either polymorphism differed significantly from the other.
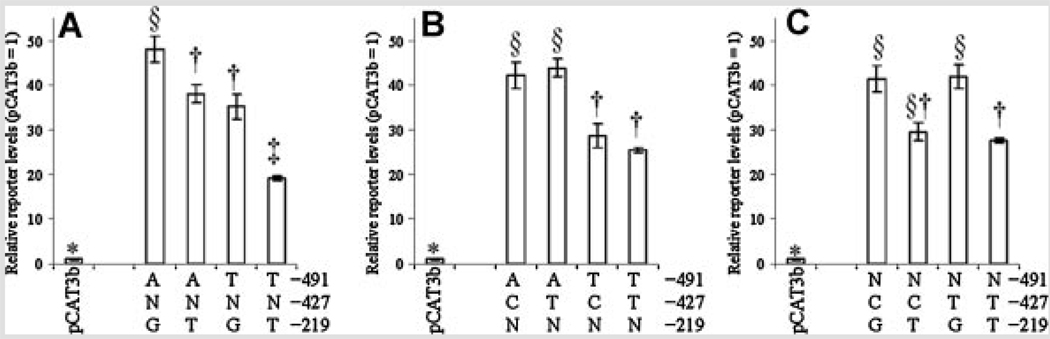
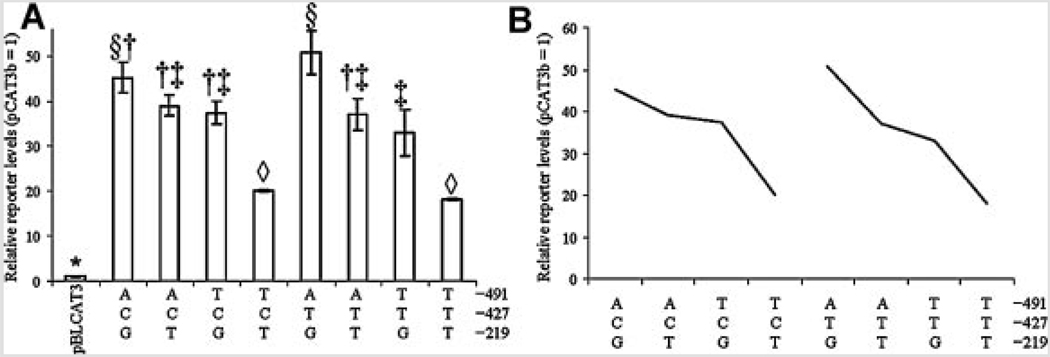
Multiple ANOVA (Table V and Table VI) indicated no significant interaction between any two-polymorphism effects in NB cells (Fig. 4). However, in PC12 cells (Fig. 5), there was a significant interaction between −491 and −219 (P < 0.0001). This interaction remained significant when ranked reporter protein levels were subject to ANOVA (Table VIB). The −219 variants had opposite (significant at kratio = 100) effects depending upon the particular −491 variant each was associated with (Fig. 5B). The −491A/−219G double polymorphism had significantly lower reporter protein levels than did −491A/−219T. On the other hand, −491T/−219G had significantly higher reporter protein levels than did −491T/−219T.
TABLE V.
Multiple ANOVA of SNP–CAT Reporter Fusion Clone Protein Levels in NB Cells
| Source | df | Type III SS | MS | F | P | |
|---|---|---|---|---|---|---|
| A. | Relative reporter levels | |||||
| −491 | 1 | 51.12 | 51.12 | 47.40 | <0.0001 | |
| −427 | 1 | 0.10 | 0.10 | 0.09 | 0.7665 | |
| −219 | 1 | 34.46 | 34.46 | 31.95 | <0.0001 | |
| −491 × −427 | 1 | 1.26 | 1.26 | 1.17 | 0.2960 | |
| −491 × −219 | 1 | 1.87 | 1.87 | 1.73 | 0.2066 | |
| −427 × −219 | 1 | 0.33 | 0.33 | 0.30 | 0.5890 | |
| −491 ×−427 × −219 | 1 | 1.23 | 1.23 | 1.14 | 0.3022 | |
| B. | Ranked relative reporter levels | |||||
| −491 | 1 | 541.5 | 541.5 | 36.71 | <0.0001 | |
| −427 | 1 | 8.17 | 8.17 | 0.55 | 0.4676 | |
| −219 | 1 | 337.5 | 337.5 | 22.88 | 0.0002 | |
| −491 × −427 | 1 | 16.67 | 16.67 | 1.13 | 0.3036 | |
| −491 × −219 | 1 | 6.00 | 6.00 | 0.41 | 0.5326 | |
| −427 × −219 | 1 | 2.67 | 2.67 | 0.18 | 0.6764 | |
| −491 × −427 × −219 | 1 | 1.50 | 1.50 | 0.10 | 0.7539 | |
df, degrees of freedom; SS, sums of squares; MS, mean square.
Bold values indicate a significant result or interaction.
TABLE VI.
Multiple ANOVA of SNP–CAT Reporter Fusion Clone Protein Levels in PC12 Cells
| Source | df | Type III SS | MS | F | P | |
|---|---|---|---|---|---|---|
| A. | Relative reporter levels | |||||
| −491 | 1 | 107.56 | 107.56 | 143.38 | <0.0001 | |
| −427 | 1 | 1.29 | 1.29 | 1.73 | 0.2075 | |
| −219 | 1 | 1.81 | 1.81 | 2.42 | 0.1396 | |
| −491 × −427 | 1 | 0.51 | 0.51 | 0.69 | 0.4198 | |
| −491 × −219 | 1 | 46.34 | 46.34 | 61.77 | <0.0001 | |
| −427 × −219 | 1 | 0.2 | 0.2 | 0.27 | 0.6122 | |
| −491 ×−427× −219 | 1 | 7.91 | 7.91 | 10.55 | 0.0050 | |
| B. | Ranked relative reporter levels | |||||
| −491 | 1 | 726 | 726 | 104.96 | <0.0001 | |
| −427 | 1 | 8.17 | 8.17 | 1.18 | 0.2933 | |
| −219 | 1 | 2.67 | 2.67 | 0.39 | 0.5434 | |
| −491 × −427 | 1 | 0.17 | 0.17 | 0.02 | 0.8786 | |
| −491 × −219 | 1 | 240.67 | 240.67 | 34.8 | <0.0001 | |
| −427 × −219 | 1 | 1.50 | 1.50 | 0.22 | 0.6477 | |
| −491 × −427 × −219 | 1 | 60.17 | 60.17 | 8.70 | 0.0094 | |
df, degrees of freedom; SS, sums of squares; MS, mean square.
Bold values indicate a significant result or interaction.
FIG. 4.
Interactions between any two of three APOε promoter polymorphisms in NB cells. Reporter level activity data from APOε promoter polymorphism-CAT reporter fusion clones transfected in NB cells were combined according to any two polymorphic sites. “N” refers to either variant at the non-specified polymorphism locus. A: Activity of clones arranged according to specific −491/−219 combination. Combinations that share statistical symbols do not significantly differ from each other. B: Activity of clones arranged according to specific −491/−427 combination. Combinations that share statistical symbols do not significantly differ from each other. C: Activity of clones arranged according to specific −427/−219 combination. Combinations that share statistical symbols do not significantly differ from each other.
FIG. 5.
Interactions between any two of three APOε promoter polymorphisms in PC12 cells. Reporter level activity data from APOε promoter polymorphism-CAT reporter fusion clones were combined according to any two polymorphic sites. “N” refers to either variant at the non-specified polymorphism locus. A: Activity of clones arranged according to specific −491/−219 combination. Combinations that share statistical symbols do not significantly differ from each other. B: Activity of clones arranged according to specific −491/−427 combination. Combinations that share statistical symbols do not significantly differ from each other. C: Activity of clones arranged according to specific −427/−219 combination. Combinations that share statistical symbols do not significantly differ from each other.
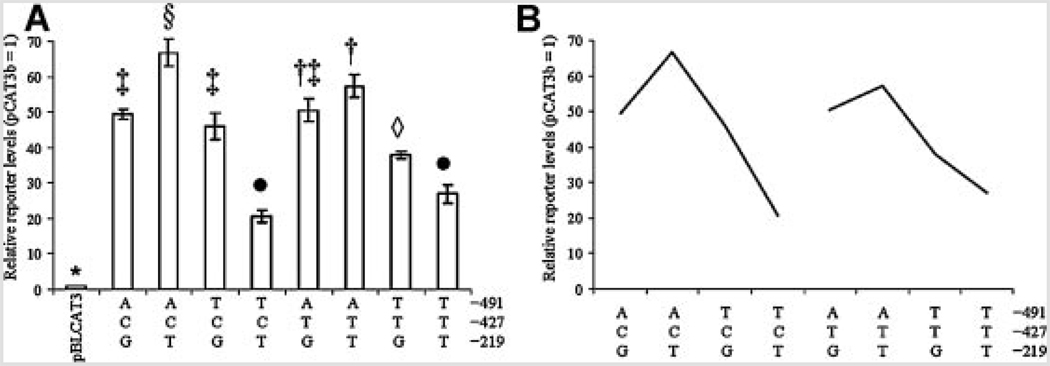
No significant three-way interaction appeared in NB cells (Fig. 6). A significant three-way interaction (P = 0.005) was detected in PC12 cells by multiple ANOVA (Fig. 7). However, this three-way interaction was weak at best (Fig. 7). While some individual polymorphic triads significantly differed from others (Fig. 7A), comparing the three–way interaction “sliced” according to the state of the −427 polymorphism showed little difference that specifically depended upon whether or not −427 was “C” or “T” (Fig. 6B and Fig 7B).
FIG. 6.
Reporter protein levels of APOε promoter polymorphism triplet-CAT fusion clones in NB cells. A: Reporter level activity data from APOε promoter polymorphism-CAT reporter fusion clones were combined according to all three polymorphic sites. Data were analyzed by Waller–Duncan multiple range test as described in the text. Individual combination of each of two states for three polymorphic sites (eight total combinations) is shown. Combinations that share statistical symbols do not significantly differ from each other. B: “Slice” of reporter protein levels according to state of −427 polymorphic site. Similarity of response “curves” between states indicates that effect of the −427 polymorphism on the overall system is very low.
FIG. 7.
Reporter protein levels of APOε promoter polymorphism triplet-CAT reporter fusion clones in PC12 cells. A: Reporter level activity data from APOε promoter polymorphism-CAT reporter fusion clones were combined according to all three polymorphic sites. Data were analyzed by Waller–Duncan multiple range test as described. Each individual combination of states for three polymorphic sites (eight total combinations) is shown. Combinations that share statistical symbols do not significantly differ from each other. B: “Slice” of reporter protein levels according to state of −427 polymorphic site. Similarity of response “curves” between states indicates that effect of the −427 polymorphism on the overall system is very low, despite a “significant” three-way interaction ANOVA result.
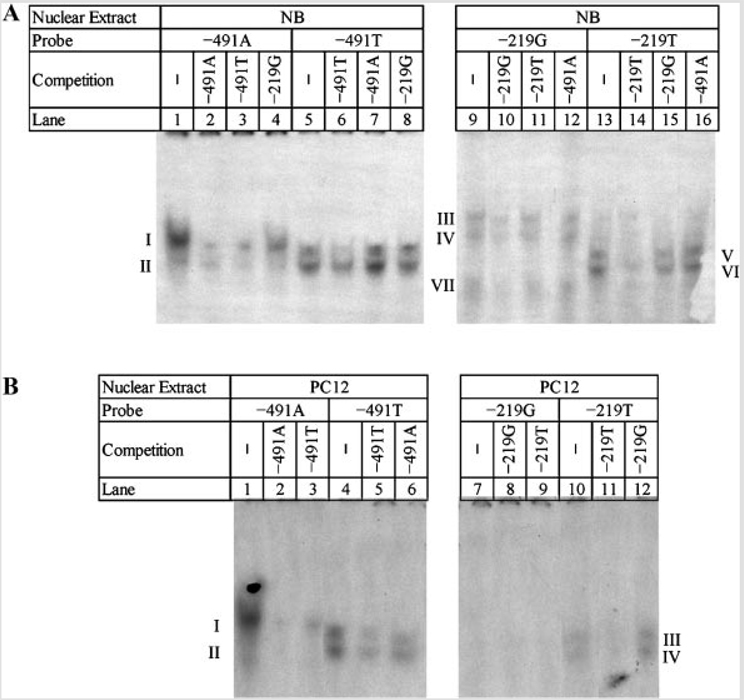
Electrophoretic Mobility Shift Assays (EMSA) of APOε Polymorphisms
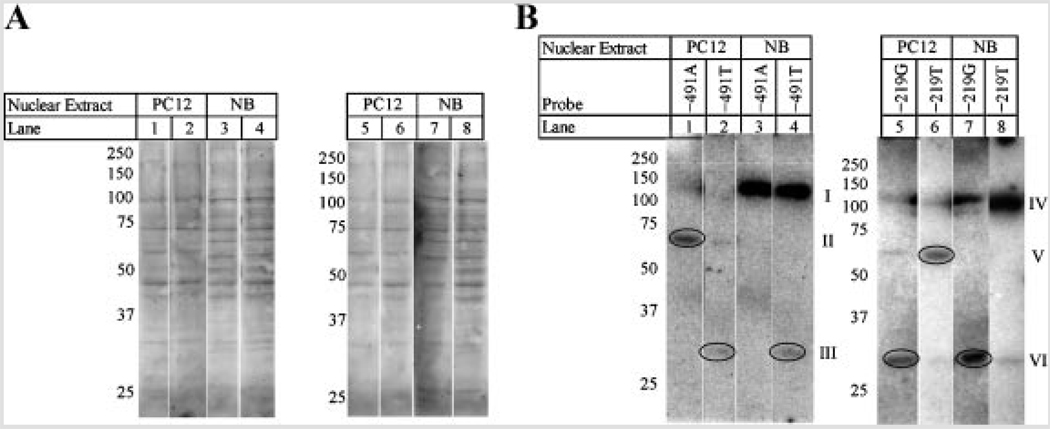
Given that the −427 T/C polymorphism produced no significant results in our constitutive expression assay, it was excluded from EMSA analysis. Double-stranded oligomers corresponding to the −491 A/T and −219 G/T polymorphisms were obtained from two different sources and used to perform EMSA and competitive EMSA twice, as described herein. Reactions were performed with nuclear extracts from NB and PC12 cells. Competition was against unlabeled corresponding polymorphic ds-oligomer and against alternate polymorphic ds-oligomer for the same site in both NB and PC12 extracts. Additionally, in NB extracts, the −491A and T probes were competed against unlabeled −219G ds-oligomer, and the −219G and T probes were competed against unabeled −491A ds-oligomer, both as “negative controls” for the competition. It is likely that differences between DNA–protein interactions at the −491 polymorphic locus may be more quantitative in nature (altered affinity for the same transcription factors) than qualitative in NB nuclear extracts.
When NB nuclear extracts were probed (Fig. 8A), differences appeared between the −491A and −491T polymorphisms (lanes 1–8) and between the −219G and −219T polymorphisms (lanes 9–16). Both of the −491 variants had two DNA interaction bands (I, II) with NB nuclear extracts. However, interaction at band “I” was much stronger than at band “II” for −491A, while interaction was approximately equal between the two bands for −491T. Competition with 150× molar excess unlabeled ds-oligomers indicated that the −491A and T variants equally competed against radiolabeled −491A, while unlabeled −491T competed more efficiently against labeled −491T than did unlabeled −491A. There was also some limited competition of unlabeled −219G with the −491A probe. EMSA of the −219 polymorphisms also showed differences between the two variants. When probing with −219G, three interactions (III, IV, VII) appeared. When probing with −219T, two different interactions (V, VI) were present. Competition of labeled −219G with unlabeled −219G caused reduction in signal for all bands, but competition was incomplete. Competition with unlabeled −219T did not reduce any signal. Competition of labeled −219T with unlabeled −219T reduced the signal at “V” and “VI”. There was little to no signal reduction when competing with unlabeled −219G. Competition with unlabeled −491A had no effect on either labeled probe. Differences in DNA–protein interactions at the −291 locus are qualitative in NB nuclear extracts, indicating a change in specific transcription factor binding.
FIG. 8.
Electrophoretic mobility shift assay (EMSA) of APOε promoter polymorphic ds-oligomers (A) EMSA in NB nuclear extracts. Polymorphic ds-oligomers were synthesized and labeled with [γ32P]-ATP as described in the text. ds-Oligomers were incubated with nuclear extracts from NB cells as described in the text. Reactions were run on native 5% TGE (Tris/Glycine/EDTA)–PAG (polyacrylamide gel) electrophoresis, gel was dried, and subject to radiofluorography. Bands corresponding to DNA–protein interactions are labeled according to blotted protein migration rate; unbound probe ran at bottom of gel (not shown). B: EMSA in PC12 nuclear extracts. Polymorphic ds-oligomers were synthesized and labeled with [γ32P]-ATP. ds-Oligomers were incubated with nuclear extracts from PC12 cells as described in the text. Reactions were run on native 5% TGE–PAG, gel was dried, and subject to radiofluorography. EMSA bands are indicated.
EMSA assay with the −491 probes and PC12 nuclear extracts (Fig. 8B, lanes 1–6) resembled the EMSA pattern found when NB nuclear extracts were probed with the same ds-oligomers (Fig. 8A). Two bands (I, II) appeared in uncompeted reactions (Fig. 8B, lanes 1 and 4), although “II” was either very weak or absent with the −491A probe. Both −491 unlabeled ds-oligomers strongly competed against labeled −491A (lanes 2 and 3), while competition was not as complete when unlabeled ds-oligomers were competed against labeled −491T (lanes 5 and 6). No specific signal was apparent when the −219G ds-oligomer was used to probe PC12 nuclear extracts, but when −219T was used to probe PC12 extracts (lanes 10–12), interactions appeared at “III” and “IV.” This interaction was blocked by unlabeled −219T but not by unlabeled −219G ds-oligomer. This indicates that a DNA–protein interaction existed with PC12 nuclear extract to the −219T ds-oligomer, and it is specific to the “T” state of the polymorphism but is not present with the polymorphism’s “G” state in a rodent nuclear extract. Migration rates may be similar enough to consider “I” and “II” to be the same bands as “III” and “IV”, respectively.
Southwestern Blotting of APOε Polymorphisms
Nuclear extracts (10 µg) from PC12 and NB cells were subject to 10% SDS–PAGE and blotted to nitrocellulose. Uniformity of transfer was assessed by temporary staining with Ponceau S (Fig. 9A). The double–stranded oligomers that were used for EMSA were also used to probe the nitrocellulose membranes for Southwestern blotting. Assays were done in duplicate and figures are representative of results. Southwestern blotting with the −491A, −491T, −219G, and −219T probes produced at least one band with each probe in both PC12 and NB nuclear extracts (Fig. 9B). Oligomers for the −491 A/T polymorphism had different binding patterns in PC12 and NB extracts, and these patterns differed between “A” and “T” variants (Fig. 9B). In PC12 extracts, the “A” variant bound a protein at approximately 60–70 kDa, while the “T” variant lacked this binding. In contrast to the “A” SNP, “T” bound a protein that ran at approximately 30 kDa. In NB extracts, both −491 variants strongly interacted with a protein at approximately 125 kDa, and the “T” variant had an additional interaction at approximately 30 kDa.
FIG. 9.
Southwestern blots of APOε promoter polymorphic ds-oligomers in NB and PC12 cell nuclear extracts. Nuclear proteins from NB (lanes 3, 4, 7, and 8) and PC12 (lanes 1, 2, 5, and 6) cells were separated via denaturing 10% SDS–PAGE and blotted to nitrocellulose. A: Blots were stained with Ponceau S and photographed. B: Blots were probed with [γ32P]-ATP labeled oligomers for APOε promoter polymorphisms −491A (lanes 1 and 3), −491T (lanes 2 and 4), −219G (lanes 5 and 7), and −219T (lanes 6 and 8) under renaturing conditions as described in the text. Blots were exposed to X-ray film for fluorography.
The −219 G/T polymorphic ds-oligomers also had different binding patterns between PC12 and NB nuclear extracts, and differences existed between “G” and “T” variants (Fig. 9B). Specifically, in PC12 extracts, the “G” variant had a distinct interaction at approximately 30 kDa with a possible much weaker interaction at approximately 60–70 kDa, while the “T” variant interacted at 60–70 kDa. In NB nuclear extracts, both “G” and “T” variants interacted with a protein at approximately 125 kDa, while the “G” variant had an additional interaction at approximately 30 kDa.
Alterations of Predicted TF Sites Around the APOε Polymorphisms
When sequences flanking each polymorphism were used to further probe the TransFac database, substituting minority polymorphic variant alleles at −491 (A → T), −427 (T → C), and −219 (G → T), several interesting predictions came to light. The 20 bp region between −481/−501 was predicted to gain likely binding sites for GATA-1, -2, and -3, while losing potential binding sites for p300, RXR-α, SP1, and SRY with the A → T polymorphism. The −417/−437 region was predicted to lose no putative binding sites while gaining AP-1, GC box, SP1, and T-Antigen binding sites with the T → C polymorphism. The −209/−229 region was predicted to lose Bcd, E47/Th1, GATA, LBP-1, and Prd sites while gaining a Cdx-1 site with the G → T polymorphism.
DISCUSSION
The ε4 APOE genotype is the strongest known genetic risk factor for AD. This genotype instills gain of function in the ApoE protein that corresponds to increased accumulation of Aβ peptide. Our underlying hypothesis is that increased expression of a lower risk factor genotype (e.g., ε3) can give analogous results to gain of function, upon the presumption that the function “gained” actually exists in the lower-risk phenotype, albeit operating at significantly lower efficiency. Association studies between ApoE levels and Aβ accumulation support this hypothesis [Lambert et al., 2005]. Selected APOE promoter SNPs, specifically occurring at −491 (A/T), −427 (T/C), and −219 (G/T) have been variously shown to potentially associate, independently or in tandem with each other, with incidence of AD [Belbin et al., 2007]. Meta-analysis has revealed associations between each of these sites and AD risk [Bertram et al., 2007]. Specific mechanisms of these SNPs have been previously investigated in cell culture and EMSA studies [Bullido et al., 1998; Artiga et al., 1998b]. These studies were done in hepatic cell lines and determined that the −491A polymorphic variant drove higher reporter expression than did −491T, but only in a single clone pair, not a full battery of possible variants. The previous studies also determined that the −219G variant drove greater reporter expression than did −219T, again only with a single clone pair.
In the population tested herein of 735 individuals, 310 of whom were diagnosed with sporadic AD, we determined that homozygosity for the −491A allele and for the −219T allele associated with significant risk (OR 1.55 and OR 1.64, respectively) of sporadic AD. After stratification for the presence of the ε4 allele, the −491AA genotype remained a risk factor for AD among non-ε4 subjects (OR 1.58).
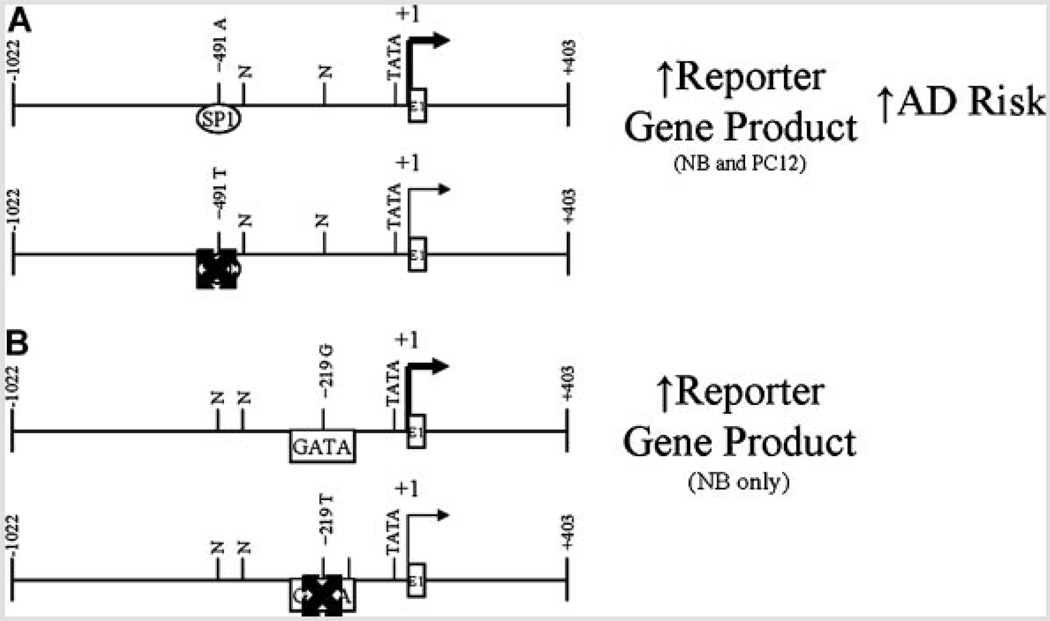
We further investigated these SNPs’ responses in human neuroblastoma (NB) and rat neuronal (PC12) cells. In addition, we considered potential interactions between and among each polymorphic state. We have determined that the −491A variant independently produced greater levels of reporter protein than did the −491T variant in both NB and PC12 cell cultures. This corresponds to other studies that have linked the −491A variant to greater levels of ApoE in vivo [Laws et al., 2002] and in vitro [Artiga et al., 1998a] and to greater risk of AD [Casadei et al., 1999; Lambert et al., 2002, 2004]. These studies, along with our own work, have led us to propose the model outlined in Figure 10. Briefly, our reporter assays determined that the −491A variant resulted in higher levels of reporter protein while −491T corresponded to lower reporter levels. In our own data, −491AA genotype corresponded to greater risk of AD when our sample was stratified for APOEε4 status. On the other hand, while we did observe a correspondence between −219 G/T CAT reporter fusion clone construction and reporter gene product levels, we did not observe a corresponding APOEε4-stratified AD risk (Fig. 11C–D). However, it should be noted that the −491AA genotype has also been associated with reduced levels of ApoE in vivo [Roks et al., 2002]. In addition, other studies have failed to find linkage between the −491 A/T SNP and effects on AD frequency [Roks et al., 1998; Toji et al., 1999].
FIG. 10.
Schematic representation of APOε promoter polymorphism activity. Diagrams represent relative activities of each of two APOε promoter polymorphisms (−491 A/T and −219 G/T) found to be active in our reporter gene assay. A: Activity of −491 A/T polymorphism in NB and PC12 cells. Taken together, the −491A clones showed significantly higher expression of reporter gene product than did −491T clones. This corresponds with our observation that −491AA genotype correlates with increased risk of AD. B: Activity of −219 G/T polymorphism in NB cells. Taken together, the −219G clones showed significantly higher expression of reporter gene product than did −219T clones. However, associated risk for AD was not independent at the −219 G/T polymorphism when stratified for APOεε4 status.
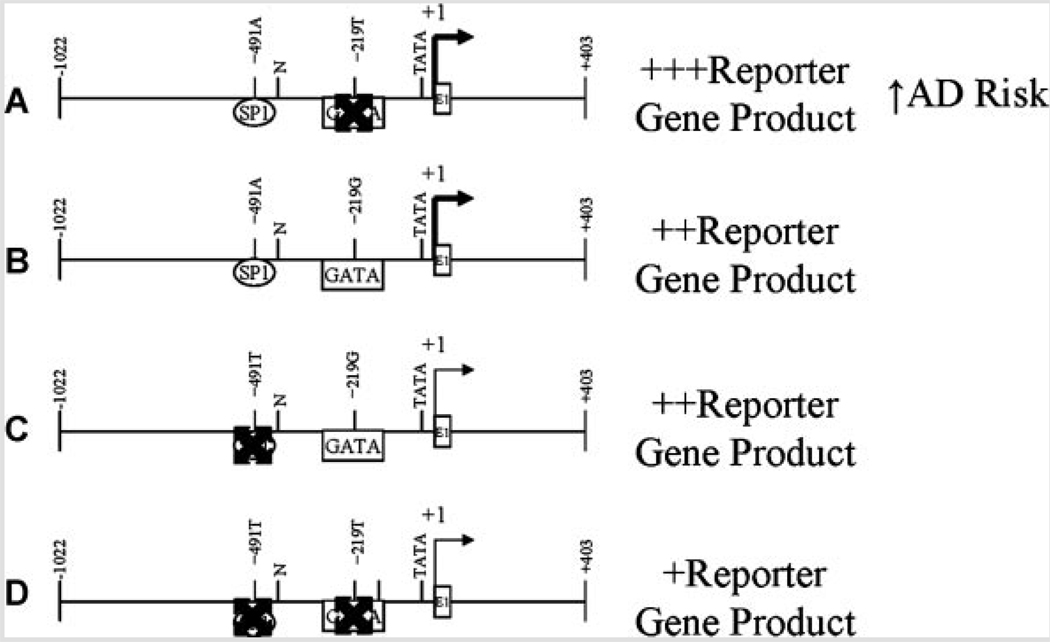
FIG. 11.
Interaction of −491 A/T and −219 G/T polymorphisms. The −219 G/T polymorphism did not bring about significant changes in reporter gene product levels in PC12 cells. However, multiple ANOVA revealed interaction between −491 A/T and −219 G/T. Examination of the data revealed that the −219T variant appears to “intensify” the effects of −491A versus T while −219G “mutes” this effect. When compared to putative loss or gain of transcription factor binding sites, the (A) presence of an SP1 site and absence of a GATA site corresponded to greatest reporter levels. This corresponded to increased AD risk in “haplogroup” analysis performed by other laboratories [Parra-Bonilla et al., 2003]. The presence of a GATA site, regardless of whether an SP1 site was (B) present or (C) absent corresponded to intermediate levels of CAT reporter product. D: When both sites were absent, CAT reporter levels were lowest in our study.
In addition to effects in cell culture due to −491 A/T, our work herein has shown that the −219G variant independently produced greater levels of reporter protein in NB cells, but not in PC12 cells. The −219G polymorphism has been shown to associate with increased [Beyer et al., 2002] risk of AD in some populations, but other studies have shown that the T allele is associated with increased AD risk [Lambert et al., 2002, 2004]. In addition, multiple ANOVA of our data indicated interactive effects between the −491 A/T and −219 G/T alleles. A previous study by other workers has determined that the −491A/−219T “haplogroup” may confer AD risk [Lambert et al., 2004] and that these SNPs may work in vivo as a “haplogroup” more strongly than they do as independent alleles [Parra-Bonilla et al., 2003]. Such behavior would be similar to risk conferred by two promoter polymorphisms we have previously studied in the amyloid β precursor protein (APP) promoter [Lahiri, 2004b; Lahiri et al., 2005]. Combination of our reporter assay results with our analysis of potential changes in transcription factor binding sites at these two polymorphic sites suggests a model that may explain both our results and those noted by Lambert’s group (Fig. 11). Briefly, the −419A variant is predicted to have greater affinity for SP1 than does −491T, while −219G is predicted to have greater affinity for GATA family factors than does −219T. If the combination is −491A/−219T, SP1 binding drives greater expression of the APOE gene, resulting in greater risk of AD. On the other hand, our reporter assay determined nearly equal levels of reporter protein for both −491A/−219G and −419T/−219G. In this case, GATA factor(s) binding to the −219 site “override” the more distal site in regulation of the APOE gene promoter. When both sites are low affinity, as in −491T/−219T, then the APOE promoter would lack either additional stimulus, predicting lower levels of ApoE and reduced risk of AD. This model does not explain the behavior of −219 oligomers in EMSA with PC12 nuclear extracts. We have elsewhere investigated potentially important differences in human APOE promoter activity in human versus rodent cell cultures and extracts [Maloney et al., 2007]. As AD is a human disorder, we have given preference to results found with NB extracts in our model.
The SP1 transcription factor has been determined to be present in SK–N–SH cells (NB cells used herein) [Carrillo et al., 1999], and the GATA2, GATA3, GATA4, and GATA6 factors [Aoyama et al., 2005] are highly expressed in several neuroblastoma cell lines. Likewise, SP1 [Atkins et al., 2003; Nguyen et al., 2005] and GATA factors have been determined to be active in PC12 cells [Jia and Takimoto 2003; Lange-Dohna et al., 2003].
Based on the functional and Southwestern blotting results, we suggest that loss of function in −491T may be due to loss of SP1 binding or altered affinity to an alternatively spliced form of SP1, which may be found in PC12 cells. This explanation, alone, would not suffice to explain activity changes for −219T due to loss of binding for another important TF of 30–32 kDa (most likely GATA2 or 4). However, it has been previously shown that GATA1 acted in apparent cooperation with SP1 at the pyruvate kinase promoter and Tal-1 gene [Gregory et al., 1996] and that GATA4 and GATA6 interact directly with SP1 in modulation of tissue-specific transcription of the cytochrome P450c17 gene [Fluck and Miller, 2004].
In PC12 nuclear extracts, the −491 “A” variant bound a protein at approximately 60–70 kDa, while the −491 “T” variant lacked this binding. This band may correspond to an alternatively spliced form of SP1 transcription factor, though SP1 is typically found to be a higher molecular weight protein (>110 kDa) [Thomas et al., 2007]. In contrast to the “A” variant, the “T” variant bound a protein that migrated at approximately 30 kDa, which may correspond to the predicted gain of a GATA binding site. In NB nuclear extracts, both −491 variants strongly interacted with a protein at approximately 125 kDa, and the “T” variant had an additional interaction at approximately 30 kDa, again corresponding to a predicted gain of a GATA binding site. However, these binding experiments were not repeated with unlabeled −491 or −219 oligomers, leaving the specificity of these interactions unanswered.
We are, therefore, cognizant that our case in this paper for SP1 as the critical transcription factor in the activity of these APOE promoter polymorphisms is important but primarily circumstantial, based upon gel migration rates and predicted binding site affinity. The SP1 transcription factor may be further implicated if evaluated in the broader context of AD-related protein expression. We have previously determined that SP1 and APP co–localize in both mouse and monkey brain regions [Brock et al., 2008]. In addition, lifespan studies of SP1 and APP expression show that both genes’ mRNA levels tightly mirror each other in both mice and monkeys [Dosunmu et al., 2009].
The APOEε4 polymorphism is a gain-of-function variant, with chaperone activity that brings about plaque formation from oligomers of Aβ [Ma et al., 1994], however, gain-of-function may be effectively mimicked by increase of “non-functional” variant levels if the “non-functional” variant actually has some small level of the function in question. We propose an explanation of the independent activity of the −491AA genotype in increasing AD risk in non-APOEε4 individuals. Specifically, the −491A allele is preferentially activated by the SP1 transcription factor over the −491T variant. This activation leads to increased levels of APOE gene transcription and of ApoE protein. When an individual’s APOE genotype lacks the ε4 allele, a “double dose” of increased APOE expression, via SP1 activation of −491AA genotype, would still result in sufficiently greater amounts of APOE gene expression to partially “make up for” deficiency of APOEε4 chaperone activity. However, this “de facto gain-of-function” would still not be equivalent to the true gain-of-function found in ApoE ε4 protein, which accounts for the lower, albeit still significant, increase in risk for AD found in −491AA individuals. There is increasing evidence that cholesterol plays a role in AD pathology, perhaps through its effects on amyloid deposition [Sparks et al., 1994, 2002; Pappolla et al., 2003; Sambamurti et al., 2004]. Furthermore, elevated low–density lipoprotein (LDL) levels correspond to greater brain amyloid β peptide deposition [Kuo et al., 1998]. Likewise, study of an African population determined an association between higher levels of cholesterol and LDL and AD risk in individuals who lack the APOEε4 genotype [Hall et al., 2006]. In essence, we suggest that total levels of ApoE, and by extension, levels of LDL, and levels of circulating lipids in general, have an important influence on lifetime risk of developing AD for the majority of cases of the disease, specifically sporadic AD in individuals lacking the APOEε4 genotype.
This hypothesis would lead to parallel consideration of gene–environment interactions potentially influencing the effect of a promoter polymorphism. On the one hand, gene expression could be acutely perturbed due to inflammation, nutritional fluctuation, or stress. However, environment can also alter gene expression in a long-term fashion. One manner in which this can occur is by induction of a somatic epitype, persistent, non-heritable alterations in DNA methylation and/or oxidation in response to extrinsic factors, such as exposure to lead (Pb) [Lahiri and Maloney, 2006]. Pb exposure has further been determined to alter levels of SP1 in a latent early-life regulation (LEARn) fashion [Lahiri et al., 2007]. It should be noted that Pb exposure and APOE genotype have been found to interact in development of central nervous system toxicity [Stewart et al., 2002], and Pb was the specific agent determined to influence SP1 and APP levels in a LEARn fashion in two recently studied species, mice and monkeys [Basha et al., 2005; Wu et al., 2008]. This suggests a further possibility that variation in an SP1 site, such as found at the APOE−491 A/T polymorphism, could be a factor in individual response to stressors such as Pb exposure potentially altering APOE levels, resulting in a similar effect to the Pb/APOE genotype interaction. This would present another avenue whereby variations in APOE gene promoter sequences would be influenced by environment, explaining the “incomplete” affect that the APOE promoter polymorphisms have been observed to have on incidence of AD.
Since the cell culture in the present work is carried out with PC12 and SK–N–SH cells, we briefly argue for their utility as CNS/neuronal cellular models. Our recent work [Ge et al., 2004; Maloney et al., 2007] has shown distinct similarities of DNA–nuclear protein interactions for portions of the APP and APOE promoters, respectively, in PC12 and SK–N–SH cell nuclear extracts versus both mouse and, importantly, post-mortem human brain cell nuclear extracts. In addition, several workers have shown that these cell lines express APP, synaptic proteins, and the secretases. However, even if these cells are accepted as suitable stand-ins for CNS neuronal cells, the valid issue can be raised that neurons do not typically express ApoE unless injured. We contend that pre-AD conditions are a type of stress or injury to neurons, which could cascade into full-blown AD, in part through altered expression of the APOE gene, thereby validating the use of neuronal cultures to study APOE expression in the context of AD etiology.
In addition, there is currently no complete model of AD available. Even most transgenic animal models do not use the “native” promoters of AD-associated genes. Instead, high-throughput promoters, such as the promoter of the human prion protein PrP, are used to ensure certain and rapid development of AD-like symptoms in the animal.
Our study is also unique in that it does not solely measure effects of individual alleles on expression, but also potential interactions between and among the three polymorphic sites, some of which interactions we have found to be significant by reporter expression assay. Likewise, most of the studies of these polymorphic sites have used a European population, while ours used a USA population, extending the size of the genetic pool from which all studies of these polymorphisms are drawn. It also examines allele effects at the multiple levels of population, gene expression, and DNA–protein interaction levels.
ACKNOWLEDGMENTS
This work was supported by the Alzheimer’s Association [to D.K.L.]; National Institutes of Health [grant numbers AG18379, AG18884 to D.K.L.]; and the Ministerio de Educación y Ciencia [grant number SAF2006-00724 to J.P.-T.]. We gratefully acknowledge the advice and help from Dennis Dickson and Clare Ellis, Mayo Clinic, Jacksonville, FL.
REFERENCES
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65(11):4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Frank A, Sastre I, Recuero M, Garcia MA, Lendon CL, Han SW, Morris JC, Vazquez J, et al. Risk for Alzheimer’s disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet. 1998a;7(12):1887–1892. doi: 10.1093/hmg/7.12.1887. [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Garcia MA, Aldudo J, Vazquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998b;421(2):105–108. doi: 10.1016/s0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- Atkins DS, Basha MR, Zawia NH. Intracellular signaling pathways involved in mediating the effects of lead on the transcription factor Sp1. Int J Dev Neurosci. 2003;21(5):235–244. doi: 10.1016/s0736-5748(03)00067-4. [DOI] [PubMed] [Google Scholar]
- Basha MR, Wei W, Bakheet SA, Benitez N, Siddiqi HK, Ge YW, Lahiri DK, Zawia NH. The fetal basis of amyloidogenesis: Exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J Neurosci. 2005;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belbin O, Dunn JL, Ling Y, Morgan L, Chappell S, Beaumont H, Warden D, Smith DA, Kalsheker N, Morgan K. Regulatory region single nucleotide polymorphisms of the apolipoprotein E gene and the rate of cognitive decline in Alzheimer’s disease. Hum Mol Genet. 2007;16(18):2199–2208. doi: 10.1093/hmg/ddm171. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Beyer K, Lao JI, Gomez M, Riutort N, Latorre P, Mate JL, Ariza A. The Th1/E47cs-G apolipoprotein E (APOE) promoter allele is a risk factor for Alzheimer disease of very later onset. Neurosci Lett. 2002;326(3):187–190. doi: 10.1016/s0304-3940(02)00355-5. [DOI] [PubMed] [Google Scholar]
- Brock B, Basha MR, DiPalma K, Anderson A, Harry GJ, Rice DC, Maloney B, Lahiri DK, Zawia NH. Co-localization and Distribution of Cerebral APP and SP1 and its Relationship to Amyloidogenesis. J Alzheimers Dis. 2008;13(1):71–80. doi: 10.3233/jad-2008-13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, Lendon C, Han SW, Morris JC, Frank A, et al. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer’s dementia. Nat Genet. 1998;18(1):69–71. doi: 10.1038/ng0198-69. [DOI] [PubMed] [Google Scholar]
- Carrillo C, Cisneros B, Montanez C. Sp1 and AP2 transcription factors are required for the human fragile mental retardation promoter activity in SK-N-SH neuronal cells. Neurosci Lett. 1999;276(3):149–152. doi: 10.1016/s0304-3940(99)00798-3. [DOI] [PubMed] [Google Scholar]
- Casadei VM, Ferri C, Veglia F, Gavazzi A, Salani G, Cattaneo M, Sorbi S, Annoni G, Licastro F, Mariani C, et al. APOE-491 promoter polymorphism is a risk factor for late-onset Alzheimer’s disease. Neurology. 1999;53(8):1888–1889. doi: 10.1212/wnl.53.8.1888. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Crook R, Hardy J, Duff K. Single-day apolipoprotein E genotyping. J Neurosci Methods. 1994;53(2):125–127. doi: 10.1016/0165-0270(94)90168-6. [DOI] [PubMed] [Google Scholar]
- Dosunmu R, Wu J, Adwan L, Maloney B, Basha MdR, McPherson CA, Harry GJ, Rice DC, Zawia NH, Lahiri DK. Lifespan profiles of Alzheimer’s disease-associated genes and their products in monkeys and mice. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1138. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chen X, Wei X, Bales KR, Berg DT, Paul SM, Farlow MR, Maloney B, Ge Y-W, Lahiri DK. NF-kappaB mediates amyloid beta peptide-stimulated activity of the human apolipoprotein E gene promoter in human astroglial cells. Brain Res Mol Brain Res. 2005;136(1–3):177–188. doi: 10.1016/j.molbrainres.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Fluck CE, Miller WL. GATA-4 and GATA-6 modulate tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1. Mol Endocrinol. 2004;18(5):1144–1157. doi: 10.1210/me.2003-0342. [DOI] [PubMed] [Google Scholar]
- Ge Y-W, Ghosh M, Song W, Maloney B, Lahiri D. Mechanism of promoter activity of the beta-amyloid precursor protein gene in different cell types. Identification of a specific 30 bp fragment in the proximal promoter region. J Neurochem. 2004;90(6):1432–1444. doi: 10.1111/j.1471-4159.2004.02608.x. [DOI] [PubMed] [Google Scholar]
- Ghosh C, Song W, Lahiri DK. Efficient DNA transfection in neuronal and astrocytic cell lines. Mol Biol Rep. 2000;27(2):113–121. doi: 10.1023/a:1007173906990. [DOI] [PubMed] [Google Scholar]
- Gregory RC, Taxman DJ, Seshasayee D, Kensinger MH, Bieker JJ, Wojchowski DM. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996;87(5):1793–1801. [PubMed] [Google Scholar]
- Hall K, Murrell J, Ogunniyi A, Deeg M, Baiyewu O, Gao S, Gureje O, Dickens J, Evans R, Smith-Gamble V, et al. Cholesterol, APOE genotype, and Alzheimer disease: An epidemiologic study of Nigerian Yoruba. Neurology. 2006;66(2):223–227. doi: 10.1212/01.wnl.0000194507.39504.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: Prevalence estimates using the 2000 census. Arch Neurol. 2003;60(8):1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Jia Y, Takimoto K. GATA and FOG2 transcription factors differentially regulate the promoter for Kv4.2 K(+) channel gene in cardiac myocytes and PC12 cells. Cardiovasc Res. 2003;60(2):278–287. doi: 10.1016/s0008-6363(03)00528-5. [DOI] [PubMed] [Google Scholar]
- Kuo YM, Emmerling MR, Bisgaier CL, Essenburg AD, Lampert HC, Drumm D, Roher AE. Elevated low-density lipoprotein in Alzheimer’s disease correlates with brain abeta1–4 levels. Biochem Biophys Res Commun. 1998;252(3):711–715. doi: 10.1006/bbrc.1998.9652. [DOI] [PubMed] [Google Scholar]
- Lahiri DK. A region upstream of the gene promoter for the beta-amyloid precursor protein interacts with proteins from nuclear extracts of the human brain and PC12 cells. Brain Res Mol Brain Res. 1998;58(1–2):112–122. doi: 10.1016/s0169-328x(98)00115-6. [DOI] [PubMed] [Google Scholar]
- Lahiri DK. Apolipoprotein e as a target for developing new therapeutics for Alzheimer’s disease based on studies from protein, RNA, and regulatory region of the gene. J Mol Neurosci. 2004a;23(3):225–234. doi: 10.1385/JMN:23:3:225. [DOI] [PubMed] [Google Scholar]
- Lahiri DK. Functional characterization of APP regulatory elements: Rationale for the identification of genetic polymorphism. Ann NY Acad Sci. 2004b;1030(1):282–288. doi: 10.1196/annals.1329.035. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B. Genes are not our destiny: The somatic epitype bridges between the genotype and the phenotype. Nat Rev Neurosci. 2006;7 10.1038/nrn2022-c1. [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19(19):5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Lewis S, Farlow MR. Tacrine alters the secretion of the beta-amyloid precursor protein in cell lines. J Neurosci Res. 1994;37(6):777–7787. doi: 10.1002/jnr.490370612. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Alley GM, Ge YW, Du Y. Functional characterization of the 5′-regulatory region of the murine apolipoprotein gene. Ann NY Acad Sci. 2002;973:340–344. doi: 10.1111/j.1749-6632.2002.tb04662.x. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Wavrant De-Vrieze F, Ge Y-W, Maloney B, Hardy J. Characterization of two APP gene promoter polymorphisms that appear to influence risk of late-onset Alzheimer’s disease. Neurobiol Aging. 2005;26(10):1329–1341. doi: 10.1016/j.neurobiolaging.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Basha MR, Ge YW, Zawia NH. How and when environmental agents and dietary factors affect the course of Alzheimer’s disease: The “LEARn” model (Latent Early Associated Regulation) may explain the triggering of AD. Curr Alzheimer Res. 2007;4(2):219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Pasquier F, Delacourte A, Frigard B, Cottel D, Perez-Tur J, Mouroux V, Mohr M, Cecyre D, et al. Pronounced impact of Th1/E47cs mutation compared with −491 AT mutation on neural APOE gene expression and risk of developing Alzheimer’s disease. Hum Mol Genet. 1998;7(9):1511–1516. doi: 10.1093/hmg/7.9.1511. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Araria-Goumidi L, Myllykangas L, Ellis C, Wang JC, Bullido MJ, Harris JM, Artiga MJ, Hernandez D, Kwon JM, et al. Contribution of APOE promoter polymorphisms to Alzheimer’s disease risk. Neurology. 2002;59(1):59–66. doi: 10.1212/wnl.59.1.59. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Cottel D, Amouyel P, Helbecque N. APOE promoter polymorphisms and dementia in the elderly. Neurosci Lett. 2004;365(2):116–119. doi: 10.1016/j.neulet.2004.04.063. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Mann D, Richard F, Tian J, Shi J, Thaker U, Merrot S, Harris J, Frigard B, Iwatsubo T, et al. Is there a relation between APOE expression and brain amyloid load in Alzheimer’s disease? J Neurol Neurosurg Psychiatry. 2005;76(7):928–933. doi: 10.1136/jnnp.2004.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange-Dohna C, Zeitschel U, Gaunitz F, Perez-Polo JR, Bigl V, Rossner S. Cloning and expression of the rat BACE1 promoter. J Neurosci Res. 2003;73(1):73–80. doi: 10.1002/jnr.10639. [DOI] [PubMed] [Google Scholar]
- Laws SM, Clarnette RM, Taddei K, Martins G, Paton A, Hallmayer J, Almeida OP, Groth DM, Gandy SE, Forstl H, et al. APOE-epsilon4 and APOE-491A polymorphisms in individuals with subjective memory loss. Mol Psychiatry. 2002;7(7):768–775. doi: 10.1038/sj.mp.4001083. [DOI] [PubMed] [Google Scholar]
- Ma J, Yee A, Brewer HB, Jr, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372(6501):92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- Maloney B, Ge Y-W, Alley GM, Lahiri DK. Important differences between human and mouse APOE gene promoters with implications for Alzheimer’s disease. J Neurochem. 2007;103(3):1237–1257. doi: 10.1111/j.1471-4159.2007.04831.x. [DOI] [PubMed] [Google Scholar]
- Nguyen HN, Lee SY, Hwang DY, Kim YK, Yuk DY, Lee JS, Hong JT. Decrease in NF-kappaB, AP-1 and SP-1 activities in neuronal cells expressing presenilin 2. Neuroreport. 2005;16(7):731–735. doi: 10.1097/00001756-200505120-00015. [DOI] [PubMed] [Google Scholar]
- Pappolla MA, Bryant-Thomas TK, Herbert D, Pacheco J, Fabra Garcia M, Manjon M, Girones X, Henry TL, Matsubara E, Zambon D, et al. Mild hypercholesterolemia is an early risk factor for the development of Alzheimer amyloid pathology. Neurology. 2003;61(2):199–205. doi: 10.1212/01.wnl.0000070182.02537.84. [DOI] [PubMed] [Google Scholar]
- Parra-Bonilla G, Arboleda G, Yunis J, Solano E, Pardo R, Arango G, Hedmont D, Arboleda H. Haplogroup analysis of the risk associated with APOE promoter polymorphisms (−219T/G, −491A/T and −427T/C) in Colombian Alzheimer’s disease patients. Neurosci Lett. 2003;349(3):159–162. doi: 10.1016/s0304-3940(03)00816-4. [DOI] [PubMed] [Google Scholar]
- Ramos MC, Matias S, Artiga MJ, Pozueta J, Sastre I, Valdivieso F, Bullido MJ. Neuronal specific regulatory elements in apolipoprotein E gene proximal promoter. Neuroreport. 2005;16(9):1027–1030. doi: 10.1097/00001756-200506210-00029. [DOI] [PubMed] [Google Scholar]
- Roks G, Cruts M, Bullido MJ, Backhovens H, Artiga MJ, Hofman A, Valdivieso F, Van Broeckhoven C, Van Duijn CM. The −491 A/T polymorphism in the regulatory region of the apolipoprotein E gene and early-onset Alzheimer’s disease. Neurosci Lett. 1998;258(2):65–68. doi: 10.1016/s0304-3940(98)00857-x. [DOI] [PubMed] [Google Scholar]
- Roks G, Cruts M, Houwing-Duistermaat JJ, Dermaut B, Serneels S, Havekes LM, Hofman A, Breteler MM, Van Broeckhoven C, van Duijn CM. Effect of the APOE-491A/T promoter polymorphism on apolipoprotein E levels and risk of Alzheimer disease: The Rotterdam Study. Am J Med Genet. 2002;114(5):570–573. doi: 10.1002/ajmg.10407. [DOI] [PubMed] [Google Scholar]
- Sambamurti K, Granholm A-C, Kindy M, Greig N, Lahiri DK, Mintzer J. Cholesterol and Alzheimer’s disease: Clinical and experimental models suggest interactions of different genetic, dietary and environmental risk factors. Curr Drug Targets. 2004;5(6):517–528. doi: 10.2174/1389450043345335. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Sparks DL, Scheff SW, Hunsaker JC, III, Liu H, Landers T, Gross DR. Induction of Alzheimer-like beta-amyloid immunoreactivity in the brains of rabbits with dietary cholesterol. Exp Neurol. 1994;126(1):88–94. doi: 10.1006/exnr.1994.1044. [DOI] [PubMed] [Google Scholar]
- Sparks DL, Lochhead J, Horstman D, Wagoner T, Martin T. Water quality has a pronounced effect on cholesterol-induced accumulation of Alzheimer amyloid beta (Abeta) in rabbit brain. J Alzheimers Dis. 2002;4(6):523–529. doi: 10.3233/jad-2002-4609. [DOI] [PubMed] [Google Scholar]
- Stewart WF, Schwartz BS, Simon D, Kelsey K, Todd AC. ApoE genotype, past adult lead exposure, and neurobehavioral function. Environ Health Perspect. 2002;110(5):501–505. doi: 10.1289/ehp.02110501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K, Wu J, Sung DY, Thompson W, Powell M, McCarrey J, Gibbs R, Walker W. SP1 transcription factors in male germ cell development and differentiation. Mol Cell Endocrinol. 2007;270(1–2):1–7. doi: 10.1016/j.mce.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Thome J, Gewirtz JC, Sakai N, Zachariou V, Retz-Junginger P, Retz W, Duman RS, Rosler M. Polymorphisms of the human apolipoprotein E promoter and bleomycin hydrolase gene: Risk factors for Alzheimer’s dementia? Neurosci Lett. 1999;274(1):37–40. doi: 10.1016/s0304-3940(99)00662-x. [DOI] [PubMed] [Google Scholar]
- Toji H, Maruyama H, Sasaki K, Nakamura S, Kawakami H. Apolipoprotein E promoter polymorphism and sporadic Alzheimer’s disease in a Japanese population. Neurosci Lett. 1999;259(1):56–58. doi: 10.1016/s0304-3940(98)00855-6. [DOI] [PubMed] [Google Scholar]
- Tycko B, Lee JH, Ciappa A, Saxena A, Li CM, Feng L, Arriaga A, Stern Y, Lantigua R, Shachter N, et al. APOE and APOC1 promoter polymorphisms and the risk of Alzheimer disease in African American and Caribbean Hispanic individuals. Arch Neurol. 2004;61(9):1434–1439. doi: 10.1001/archneur.61.9.1434. [DOI] [PubMed] [Google Scholar]
- Wu J, Basha MR, Brock B, Maloney B, Cox D, Harry J, Cardozo-Paleaz F, Rice DC, Lahiri DK, Zawia NH. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): Evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurutuza L, Verpillat P, Raux G, Hannequin D, Puel M, Belliard S, Michon A, Pothin Y, Camuzat A, Penet C, et al. APOE promoter polymorphisms do not confer independent risk for Alzheimer’s disease in a French population. Eur J Hum Genet. 2000;8(9):713–716. doi: 10.1038/sj.ejhg.5200513. [DOI] [PubMed] [Google Scholar]