Abstract
Natural reservoirs of Yersinia (Y.) enterocolitica comprise different animal species, but little is known about the role of wild animals in the epidemiology of yersiniosis. The aim of the study was to evaluate the prevalence of Y. enterocolitica among game animals in Poland. The bio-serotypes and the pathogenicity markers of the analyzed isolates were determined. The experimental material comprised rectal swabs from 857 free-living animals hunter-harvested over a period of 2 years (2013–2014) in hunting districts across Poland. The isolates from bacteriological studies were confirmed by PCR and bio-serotyped based on the results of biochemical and agglutination tests. In the group of the 218 analyzed isolates of Y. enterocolitica, 133 were derived from wild boars, 70 from red deer, 11 from roe deer and 4 from fallow deer, and they accounted for 61.0%, 32.1%, 5.1% and 1.8% of all isolates, respectively. Bio-serotyping assays revealed that 91.7% of the examined isolates belonged to biotype 1A (200/218). The remaining 18 isolates belonged to bio-serotypes 1B/NI (3/218, 1.4%), 1B/O:8 (1/218, 0.5%), 2/NI (6/218, 2.8%), 2/O:27 (1/218, 0.5%), 2/O:3 (1/218, 0.5%), 2/O:9 (2/218, 0.9%), 3/NI (2/218, 0.9%), 4/O:3 (1/218, 0.5%) and 4/O:9 (1/218, 0.5%). The ail gene, a suggestive virulence gene for Y. enterocolitica, has been found in 30 isolates from 20 wild boars, in 6 isolates from red deer, and in 1 isolate from roe deer. Our study demonstrated that Y. enterocolitica is frequently isolated from game animals in Poland, which poses a risk of spreading these infectious agents to other animal species and humans.
Introduction
Yersinia (Y). enterocolitica, a member of the Enterobacteriaceae family, is the etiological agent of yersiniosis, an important zoonotic disease that causes symptoms ranging from a mild, self-limiting diarrhea to acute mesenteric lymphadenitis, and can sometimes develop into a variety of parenteral forms [1–3]. In Europe, yersiniosis is a notifiable zoonotic disease which must be reported to authorities and presented in the annual report of the European Food Safety Authority (EFSA). For several years, Y. enterocolitica has ranked third among the pathogens that most frequently cause gastrointestinal disorders in Europe, after Campylobacter spp. and Salmonella spp. In 2015, 7202 cases of yersiniosis were recorded in Europe [4].
Y. enterocolitica is relatively resistant to external factors, and as a psychrotolerant pathogens, it can survive in the environment for a long time. Y. enterocolitica has been divided into more than 70 serotypes based on differences in the structure of the somatic antigen, and into 6 biotypes based on its biochemical characteristics. Most of Y. enterocolitica strains that are pathogenic to humans belong to biotypes 1B 2–5, whereas biotype 1A (BT1A) is regarded as nonpathogenic [1,2,5,6]. However, selected strains of this biotype are considered as opportunistic [7,8,9].
The pathogenicity of Y. enterocolitica is related to the presence of chromosomal and plasmid genes which control the production and functions of proteins that facilitate survival in the host body and are responsible for the clinical signs of disease [10–12]. There is a strong relationship between pathogenicity and the presence of chromosomal ail and ystA genes in Y. enterocolitica [10,12]. The BT1A isolates are usually ail and ystA negative, and they may harbor ystB or ystC genes that control the production of heat-stable enterotoxins [8,11]. The presence of ystB has been reported in approximately 80% of Y. enterocolitica BT1A isolates [8].
The epidemiology of yersiniosis is complex and not fully understood. The pig is the main reservoir of human-pathogenic strains, most of which cause asymptomatic infections, colonize tonsils and the digestive tract, and are excreted with feces. Infected animals become long-term carriers [1,13]. Different species of farm, domestic, free-living and game animals can also act as natural reservoirs of Y. enterocolitica. Despite the scant availability of information on the occurrence of Y. enterocolitica in game animals, these hosts seem to be an important reservoir of Y. enterocolitica, and they contribute to the environmental circulation of the bacterium [14–17]. Epidemiological links between free-living animals vs. farm and domestic animals have not been identified to date [17].
Poland has a temperate climate with average annual temperature of 7 0C and relatively cold winters, which enables psychrotolerant bacteria such as Yersinia spp. to survive in the environment [18]. Poland has a long tradition of hunting, and game meat is widely consumed. Game animals of different species make up a substantial part of wildlife in Polish forest ecosystems. Selected species, such as wild boars and free-living ruminants, travel many miles in search of food or for reproductive purposes. Animals that are carriers of human pathogens can contribute to the transmission of pathogens across considerable distances. This study investigates the transmission of Y. enterocolitica by game animals, and it can shed new light on the epidemiology of infections and reduce the prevalence of yersiniosis in humans. This is the first large-scale study into the dissemination of Y. enterocolitica in game animals in Poland.
The purpose of the study was to determine the prevalence of Y. enterocolitica infections among game animals in Poland with the use of traditional culturing and molecular methods. The isolates were divided into different biotype, serotype and virulence marker groups, to explore the significance of game animals as a reservoir and a vector in the spread of Y. enterocolitica infections.
Materials and methods
The investigated material comprised rectal swabs from 857 game animals: wild boars (Sus scrofa) (n = 434), red deer (Cervus elaphus) (n = 291), roe deer (Capreolus capreolus), (n = 117) and fallow deer (Dama dama) (n = 15), hunted in 12 out of 16 Polish regions during the hunting seasons of 2013–2014. The animals were hunted by authorized hunters under a grant from the Polish National Centre for Research and Development. The health status of the animals was not recorded. Samples for analysis were collected in duplicate from each animal before evisceration, they were immediately placed in tubes with the Amies transport medium and delivered to the laboratory within 48 h. A total of 1714 samples were analyzed.
The animals were not hunter-harvested exclusively for the needs of this study. All samples were collected during routine examinations of game meat, therefore, ethical approval for animal experimentation was not required.
Bacteriological analysis
For each duplicate sample, one rectal swab was analyzed by adding 9 ml of ITC warm enrichment broth (irgasan, ticarcillin, potassium chlorate) and incubating at 25 0C for 48 h, and the other sample was placed in 9 ml of cold enrichment PSB broth (peptone, sorbitol, bile salts) and incubated at 4 0C for 3 weeks to determine its ability to grow at a low temperature. Warm and cold enrichments were tested in accordance with PN-EN ISO 10273 [19]. After incubation, 0.5 ml of each enrichment was immersed in 4.5 ml of 0.5% KOH in 0.5% NaCl for 20 s to reduce background microflora. It was then streaked onto the CIN medium (Yersinia Selective Agar, DIFCO) and incubated at 30 0C for 48 h. in an aerobic atmosphere. Isolates with a growth pattern indicative of Y. enterocolitica on CIN (small and smooth, with the a red center and a translucent rim), which were motile at 22 0C, but not at 37 0C were stored on agar slants at 4 0C for biotyping, serotyping and molecular analyses. Additionally, biochemical identification was performed using the API 20E bacterial identification system (bioMerieux) according to the manufacturer’s instructions.
DNA isolation
Bacterial DNA was extracted with the Genomic Mini kit (Biotechnology A&A, Poland) according to the manufacturer’s procedures. The extracted nucleic acid was stored at -20 0C until further analyses.
Multiplex PCR
The identity of Y. enterocolitica isolates was confirmed and their pathogenic characteristics were determined by amplifying ail, yst A and ystB genes as markers of virulence. Primer sequences are specified in S1 Table. The primers were synthesized in the DNA Sequencing Laboratory of the Polish Academy of Sciences, Oligo, Warsaw, Poland. The HotStartTaq Plus DNA Polymerase Kit (Qiagen) and the HotStartTaq Plus Master Mix Kit (Qiagen) were used in the multiplex PCR technique. The reaction was performed according to the following schedule: final concentration of MgCl2−1.5 nM, initial denaturation at 94 0C for 120 s, followed by 30 cycles of DNA amplification: denaturation at 94 0C for 90 s, annealing at 46 0C for 60 s, polymerization at 72 0C for 120 s, and final polymerization at 72 0C for 5 min.
The products were separated by electrophoresis in 2% agarose gel with the Midori Green Advanced DNA Stain (Nippon Genetics Europe GmbH, Germany) in 1x TAE buffer. The products were visualized under UV light. PCR results were analyzed and archived with the use of the GelDoc gel documentation system (Bio-Rad).
The specificity of the reaction was confirmed by sequencing amplicons (Genomed, Poland). The analyzed nucleotide sequences are available in the GenBank database under accession numbers [KJ592623-KJ592627], [KM253258-KM253268] and [KT209961-KT209996].
Biotype identification
Biotypes were identified according to the method proposed by Wauters [5,6] and described in the PN-EN ISO 10273 [19]. PCR-confirmed Y. enterocolitica isolates were examined for their ability to produce salicin acid, trehalose and xylose, and for esculin fermentation. They were also evaluated for the presence of pyrazinamidase and indole as well as the ability to reduce nitrogen compounds. The results of the applied biochemical tests are interpreted in S2 Table.
Serotype identification
The serotype of Y. enterocolitica isolates was determined by the slide agglutination test with the use of commercial sera for antigens O:3, O:5, O:8, O:9 (ITEST, Hradec Kralove, Czech Republic) and O:27 (SEFIN, Berlin, Germany). Live bacterial cells cultured for 24 hours on blood agar (Grasco) were used as the antigen. Bacterial cells were suspended in a drop of 0.85% NaCl, placed on slides and combined with a drop of each of the sera. Agglutination after 1 min of shaking in an orbital shaker was regarded as a positive result. In the absence of agglutination with any of the available diagnostic sera, the isolate was marked as NI (non-identified).
Statistical analysis
The results were processed statistically with the use of a descriptive statistics module, correlation analysis and a non-parametric statistics module. Calculations were performed in STATISTICA PL v.12 software. The Chi-squared non parametric test with Yates’ correction [20] was used. The calculations were performed at a significance level α = 0.05.
Results
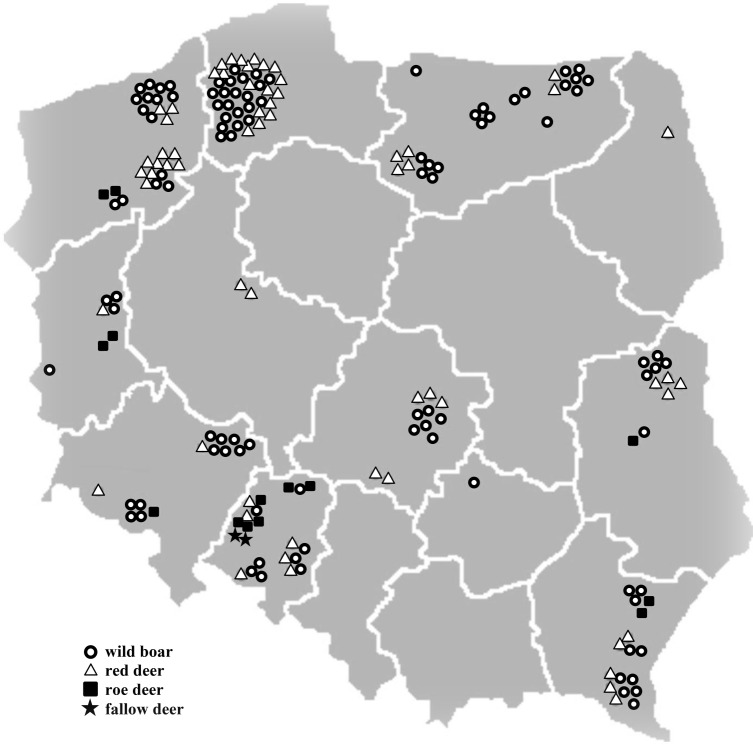
Y. enterocolitica isolates were detected in the rectal swabs of the 21.7% (186/857) of the tested animals. The results of culture tests and molecular analyses are shown in Table 1, and the geographical origin of the positive samples is presented in Fig 1.
Table 1. Results of the bacteriological study, including the number of Yersinia spp. confirmed by PCR.
| Species | Examined animals | Y. enterocolitica-positive animals (%) | Y. enterocolitica isolates confirmed by PCR | ||
|---|---|---|---|---|---|
| ITC | PSB | total no. of isolates | |||
| Wild boar | 434 | 110 (25.3%) | 49 | 84 | 133 |
| Red deer | 291 | 63 (21.6%) | 27 | 43 | 70 |
| Roe deer | 117 | 11 (9.4%) | 3 | 8 | 11 |
| Fallow deer | 15 | 2 (13,3%) | 2 | 2 | 4 |
| Total | 857 | 186 (21.7%) | 81 | 137 | 218 |
ITC–Yersinia isolates from warm enrichment; PSB—Yersinia isolates from cold enrichment
Fig 1. Geographical distribution of Yersinia enterocolitica positive samples in the Polish regions.
The prevalence of Y. enterocolitica infections (110/434) was highest in wild boars, where 25.3% of the examined animals were infected. In comparison, only 21.6% red deer (63/291), 9.4% roe deer (11/117) and 13.3% fallow deer (2/15) were infected. Highly significant (p<0.01) differences were confirmed in the frequency of the isolation of Y. enterocolitica isolates from wild boar and red deer versus roe deer and fallow deer.
The biotype and serotype characteristics of Y. enterocolitica isolates are shown in Table 2. Bioserotyping tests demonstrated that 91.7% of the isolates belonged to BT1A (200/218), serotypes O:27 (n = 4), O:3 (n = 7), O:5 (n = 10), O:8 (n = 10) and O:9 (n = 9). Most isolates of BT1A (160/200) did not agglutinate with the applied diagnostic sera, and they were determined as NI (non-identified). The remaining 18 isolates belonged to the bio-serotypes 1B/NI (3/218, 1.4%), 1B/O:8 (1/218, 0.5%), 2/NI (6/218, 2.8%), 2/O:27 (1/218, 0.5%), 2/O:3 (1/218, 0.5%), 2/O:9 (2/218, 0.9%), 3/NI (2/218, 0.9%), 4/O:3 (1/218, 0.5%) and 4/O:9 (1/218, 0.5%).
Table 2. Biotype and serotype characteristics of Yersinia enterocolitica isolates.
|
Biotype/serotype |
Number of Yersinia enterocolitica isolates number | |||
|---|---|---|---|---|
| Wild boar | Red deer | Roe deer | Fallow deer | |
| 1A/NI | 107 | 46 | 5 | 2 |
| 1A/O:27 | 3 | 1 | ||
| 1A/O:3 | 1 | 6 | ||
| 1A/O:5 | 2 | 4 | 3 | 1 |
| 1A/O:8 | 7 | 3 | ||
| 1A/O:9 | 3 | 3 | 2 | 1 |
| 1B/NI | 3 | |||
| 1B/O:8 | 1 | |||
| 2/NI | 2 | 3 | 1 | |
| 2/O:27 | 1 | |||
| 2/O:3 | 1 | |||
| 2/O:9 | 2 | |||
| 3/NI | 1 | 1 | ||
| 4/O:3 | 1 | |||
| 4/O:9 | 1 | |||
| TOTAL | 133 | 70 | 11 | 4 |
NI–non-identified serotypes
Most of the 218 analyzed Y. enterocolitica isolates (n = 137, 62.8%) were obtained from cold enrichment. In 36 cases (wild boar n = 24, red deer n = 10, fallow deer n = 2), isolates were obtained from both warm and cold enrichments, and in this group, biotype/serotype differences were noted in 14 cases. In 8 wild boars isolates were obtained simultaneously from warm and cold enrichments and they were classified into the following biotypes/serotypes: 2/NI and 1A/NI, 1A/O:27 and 1A/NI, 1B/NI and 1A/NI, 2/NI and 1A/NI, 1A/NI and 1A/O:27, 1B/O:8 and 2/O:9, 1A/NI and 1A/O:8, 1A/NI and 2/NI. The isolates from 4 red deer were classified as: 1A/O:8 and 1A/NI, 1A/NI and 2/NI, 1A/O:5 and 1A/O:3, 1A/NI and BT2/NI. The isolates from 2 fallow deer were classified as: 1A/O:9 and 1A/NI, 1A/O:5 and 1A/NI.
The prevalence of ail, ystA and ystB genes in Y. enterocolitica isolates is presented in Tables 3 and 4. The ail gene, which is highly suggestive for pathogenic Y. enterocolitica isolates, was found in 30 isolates from 20 wild boars and it appeared simultaneously with the ystA gene in only 2 cases. In the remaining 28 isolates, the ail gene occurred simultaneously with the ystB gene, and 21 of these isolates belonged to BT1A.
Table 3. The prevalence of ail, ystA and ystB genes in Yersinia enterocolitica isolates from game animals.
| Animal species | Prevalence of genes | ||
|---|---|---|---|
| ail | ystA | ystB | |
| Wild boar | 30 | 2 | 131 |
| Red deer | 6 | 1 | 70 |
| Roe deer | 1 | 1 | 10 |
| Fallow deer | 0 | 0 | 4 |
| Total | 37 | 4 | 215 |
Table 4. The prevalence of ail, ystA and ystB genes in Yersinia enterocolitica biotype 1B, 2–4 isolates from game animals.
| NO | ANIMAL SPECIES | BIOTYPE | SEROTYPE | GENES |
|---|---|---|---|---|
| 1 | Wild boar | 2 | O:9 | ail, ystB |
| 2 | 2 | NI | ail, ystB | |
| 3 | 2 | O:3 | ail, ystA | |
| 4 | 1B | NI | ystB | |
| 5 | 1B | NI | ystB | |
| 6 | 1B | NI | ystB | |
| 7 | 3 | NI | ystB | |
| 8 | 2 | NI | ystB | |
| 9 | 1B | O:8 | ystB | |
| 10 | 2 | O:9 | ystB | |
| 11 | Red deer | 4 | O:3 | ail, ystA |
| 12 | 3 | NI | ystB | |
| 13 | 2 | O:27 | ystB | |
| 14 | 4 | O:9 | ail, ystA | |
| 15 | 2 | NI | ystB | |
| 16 | 2 | NI | ystB | |
| 17 | 2 | NI | ystB | |
| 18 | Roe deer | 2 | NI | ystB |
NI- non-identified serotypes
The ail gene was found in 6/70 Y. enterocolitica isolates from red deer. Only in one case, the ail gene occurred together with the ystA gene. In the remaining 5 cases, the ail gene was present with the ystB gene. Most of Y. enterocolitica isolates from red deer harbored only the ystB gene. In roe deer, the ail gene was found in 1 out of 11 isolates, and it was accompanied by the ystB gene. Single ystB genes were found in 11 isolates.
The ail gene was not detected in samples from fallow deer, and all 4 Y. enterocolitica isolates from this animal species contained the ystB gene. In our study, 37 out 218 (17.0%) isolates contained the ail gene, but 21 of BT1A isolates with the ail gene also harbored ystB. Detailed molecular characteristics and the results of a phylogenetic analysis of isolates harboring ail and ystB were described in a previous paper [21].
Y. enterocolitica isolates were detected in rectal swabs from all examined animal species, and they were most prevalent in wild boars. Most isolates belonged to the BT1A/NI group and harbored the ystB gene.
Discussion
The present study makes the first ever attempt to identify the prevalence of Y. enterocolitica in wild animals based on samples collected from animals that were hunter-harvested in all controlled hunting zones in Poland. The pathogen was isolated from 21.7% of 857 examined animals, which clearly indicates that this bacterium is widespread among game animals in Poland. Most controlled hunting zones in Poland are situated in large forests which occupy vast parts of the country, except central Poland. Y. enterocolitica was isolated in each hunting district where test samples were collected (Fig 1).
Free-living animals seem to play an important role in the epidemiology of yersiniosis. It should be noted that the consumption of game meat increases the risk of foodborne diseases [14,17,22]. There is a general scarcity of published data on the prevalence of foodborne infections caused by the consumption of game meat contaminated with Y. enterocolitica. However, the pathogen can contaminate the carcasses of infected game animals when the intestines are damaged by shot pellets or during evisceration. The contamination of game meat with Y. enterocolitica also remains insufficiently investigated. Bucher et al. [23] detected pathogenic Y. enterocolitica on the surface of 38.8% raw game meat samples in Bavaria, Germany. Avagnina et al. [24] isolated Y. enterocolitica from 2.4% examined carcasses of large game animals in Italy. In Poland, Bancerz-Kisiel et al. [25] detected Y. enterocolitica isolates in cold stored carcasses in 60% of the examined roe deer, 43.8% of red deer and 55% of wild boars.
Relatively little is known about the occurrence of Y. enterocolitica infections in wild animals in the world. According to some studies, wild boars are a significant reservoir of strains pathogenic to humans [14,22,26–28]. Serological investigations in northeastern Germany and Switzerland demonstrated the presence of antibodies against Yersinia spp. in the sera of more than 60% of the examined wild boars [22,29]. Bacteriological analyses conducted by von Altrock [28] revealed the presence of Y. enterocolitica in the tonsils of 17.1% of the examined wild boars in Lower Saxony, Germany.
Our study demonstrated that other species of game animals, including red deer, roe deer and fallow deer, can also be a source of infections of Y. enterocolitica. The highest number of positive samples was found in wild boars (25.3%), which is consistent with the results of the cited studies. This result could be partly attributed to the high genetic affinity of wild boars and pigs. Pigs are the natural hosts and reservoirs of the discussed pathogen. However, significant percentages of Y. enterocolitica isolates also obtained from red deer, roe deer and fallow deer (21.7%, 9.4% and 13.3%, respectively) in our study (Table 1). These findings provide evidence that wild boars are not the only species of game animals that play a role in dissemination of Y. enterocolitica.
Most isolates were classified as BT1A (91.7%). Similar results were observed by other researchers [17,27,28] and in our previous study into Y. enterocolitica in wild animals [14,26,30–32]. However, we also detected 18 isolates (0.8%) belonging to biotypes 2, 3, 4 and 1B (Tables 2 and 4). In Europe, the most common pathogenic bio-serotypes isolated in clinical cases of yersiniosis are serotypes O:9 and O:5.27 of BT2, serotypes O:1, 2, 3, O:5.27 of BT3, serotype O:3 of BT4 and serotypes O:2, 3 of BT5 [1]. In our study, only 6 out of 18 isolates of BT2 (O:9, O:27, O:3), BT4 (O:3, O:9) and 1B (O:8) could be considered as virulent on the basis of bio-serotype typing (Table 2). The pathogenic characteristics of 3 isolates were confirmed by molecular analysis which revealed the presence of ail and ystA genes (Table 4).
In 2014, 244 cases of yersiniosis were recorded in Poland. The incidence rate was 0.63/100,000 inhabitants, and most infections were caused by Y. enterocolitica bio-serotypes 4/O:3 (88%), 1B/O:8 (6.9%), and 2/O:9 (5.2%) [33]. The clinical significance of Y. enterocolitica BT1A is difficult to determine, mainly due to the lack of pYV plasmids which are the most important markers of the pathogenic Y. enterocolitica. Until recently, BT1A strains were defined as non-pathogenic. In recent years, they have been increasingly isolated from clinical cases of yersiniosis, although in some cases, the symptoms of yersiniosis were not specific and could be caused by another pathogen [7–9]. However, there is clinical evidence that selected BT1A strains of Y. enterocolitica can cause gastrointestinal symptoms [8]. Little is known about the pathogenic mechanisms of disease caused by Y. enterocolitica BT1A. This study revealed that the YST-b enterotoxin encoded by the ystB gene in most BT1A isolates was the major contributor to diarrhea caused by Y. enterocolitica of this biotype [7,8,11]
The identification of one 1B/O:8 Y. enterocolitica isolate in our study is noteworthy. This bio-serotype is rare, and it is isolated mainly in North America. The first highly pathogenic 1B/O:8 strain of Y. enterocolitica was isolated in Europe in 2003. In Poland, the prevalence of yersiniosis has increased gradually since 2004, and in 2008 O:8 strains of Y. enterocolitica outnumbered O:3 and O:9 serotypes which has been predominant in previous years [34]. The reservoir of Y. enterocolitica 1B/O:8 in Poland has not yet been identified [34]. However, our isolate harbored only the ystB gene (Table 4).
The EFSA has recommended biotyping as the simplest method of evaluating the pathogenicity of Y. enterocolitica [4]. Our experience suggests that determination of the pathogenicity by biotyping/serotyping is a time-consuming and laborious process. In some cases, the results were ambiguous and required replication. Virulence markers were quickly and reliably identified by PCR, but the results were difficult to interpret when ail and ystB were detected simultaneously. Therefore, both methods should be used to Y. enterocolitica in wildlife specimens.
The isolation and characterization of Y. enterocolitica require some technical effort because the growth of this microorganism could be inhibited by the antagonistic effects of background flora [35]. The method applied in the present study is useful for isolating Y. enterocolitica from environmental samples [32]. The high frequency (186/857, 21.7%) of Y. enterocolitica detections in game animals can be attributed to the combined application of both methods, and most of the isolates in the present study originated from cold enrichment (137/218, 62.8%).
In this study, virulence markers ail, ystA and ystB were amplified in a molecular analysis to determine the pathogenicity of isolates. More importantly, this procedure was applied to confirm and verify the results of the bacteriological methods. The amplification of the ail gene is particularly useful for determining the virulence properties of Y. enterocolitica [2,10,36,37].
In conclusion, most Y. enterocolitica isolates from game animals belonged to the BT1A, but enteropathogenic Y. enterocolitica bio-serotypes which are common in European pigs were also identified in hunted game animals. Our results confirm the high prevalence of Y. enterocolitica in Poland and indicate that game animals, especially wild boars, are important vectors of infections. Red deer appear to be more susceptible to Y. enterocolitica infections than other wild ruminants. The prevalence of the identified virulence factors and the bio-serotype affiliation of the isolates from game animals inhabiting different Polish regions clearly indicate that Y. enterocolitica is widely disseminated in Poland.
Supporting information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Centre for Research and Development (NCBiR, grant no. NR12-0126-10) and Baitursynov Kostanay State University, Kazakhstan (contract no. 89). This publication was supported by KNOW (Leading National Research Centre) Scientific Consortium "Healthy Animal - Safe Food", decision of Ministry of Science and Higher Education no. 05-1/KNOW2/2015.
References
- 1.Bottone EJ. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997; 10: 257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tirziu E, Cumpanasoiu C, Gros RV, Seres M. Yersinia enterocolitica monographic study. J Anim Sci Biotechnol. 2011; 44: 144–149. [Google Scholar]
- 3.Wren BW. The Yersiniae–a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol. 2003; 1: 55–64. doi: 10.1038/nrmicro730 [DOI] [PubMed] [Google Scholar]
- 4.EFSA. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Foodborne Outbreaks in 2015. The EFSA J. 2016; 14: 4634 doi: 10.2903/j.efsa.2016.4634 [Google Scholar]
- 5.Wauters G, Kandolo K, Janssens M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987; 9: 14–21. [PubMed] [Google Scholar]
- 6.Wauters G. Antigens of Yersinia enterocolitica 1987. In: Yersinia enterocolitica. Bottone E.J. (ed), CRC Press, Inc., Boca Raton, Fla: 1981, pp. 41–53. [Google Scholar]
- 7.Huovinen E, Sihvonen LM, Virtanen MJ, Haukka K, Siitonen A, Kuusi M. Symptoms and sources of Yersinia enterocolitica-infection: a case-control study. BMC Infect Dis. 2010; 10: 122 doi: 10.1186/1471-2334-10-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tennant SM, Grant TH, Robins-Browne RM. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol Med Microbiol. 2003; 38: 127–137. [DOI] [PubMed] [Google Scholar]
- 9.Sihvonen LM, Hallanvuo S, Haukka KK, Skurnik M, Siitonen A. The ail gene is present in some Yersinia biotype 1A strains. Foodborne Pathog Dis. 2011; 8: 456–457. [DOI] [PubMed] [Google Scholar]
- 10.Harnett N, Lin YP, Krishan C. Detection of pathogenic Yersinia enterocolitica using the multiplex polymerase chain reaction. Epidemiol Infect. 1996; 117: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramamurthy T, Yoshino K, Huang X, Balakrish NG, Carniel E, Maruyama T, et al. The novel heat-stable enterotoxin subtype gene (ystB) of Yersinia enterocolitica: nucleotide sequence and distribution of the yst genes. Microb Pathogenesis. 1997; 23: 189–200. [DOI] [PubMed] [Google Scholar]
- 12.Fredriksson-Ahomaa M, Stolle A, Korkeala H. Molecular epidemiology of Yersinia enterocolitica infections. FEMS Immunol Med Microbiol. 2006; 47: 315–329. doi: 10.1111/j.1574-695X.2006.00095.x [DOI] [PubMed] [Google Scholar]
- 13.Gurtler M, Alter T, Kasimir S, Fehlhaber K. Prevalence of Yersinia enterocolitica in fattening pigs. J Food Prot. 2005; 68: 850–854. [DOI] [PubMed] [Google Scholar]
- 14.Bancerz-Kisiel A, Platt-Samoraj A, Szczerba-Turek A, Syczyło K, Szweda W. The first pathogenic Yersinia enterocolitica bioserotype 4/0:3 strain isolated from a hunter wild boar (Sus scrofa) in Poland. Epidemiol Infect. 2015; 143: 2758–2765. doi: 10.1017/S0950268814003872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolova S, Tzvetkov Y, Najdenski H, Vesselinova A. Isolation of pathogenic Yersiniae from wild animals in Bulgaria. J Vet Med B. 2001; 48: 203–209. [DOI] [PubMed] [Google Scholar]
- 16.Hacking MA, Sileo L. Yersinia enterocolitica and Yersinia pseudotuberculosis from wildlife in Ontario. J Wildl Dis. 1974; 10: 452–457. [DOI] [PubMed] [Google Scholar]
- 17.Fredriksson-Ahomaa M, Wacheck S, Bonke R, Stephan R. Different enteropathogenic Yersinia strains found in in wild boars and domestic pigs. Foodborne Pathog Dis. 2011; 8: 733–737. doi: 10.1089/fpd.2010.0711 [DOI] [PubMed] [Google Scholar]
- 18.Przybylak R. Poland’s Climate in the Last Millennium. Past Climates, Regional and Local Climates. 2016. doi: 10.1093/acrefore/9780190228620.013.2 [Google Scholar]
- 19.PN-EN ISO 10273: Microbiology of food and animal feed. Horizontal method for the detection of Microbiology of food and animal feed. Horizontal method for the detection of presumably pathogenic Yersinia enterocolitica. 2005. Polish Committee for Standardization: Polish Norm–European Norm (with appendix PN-EN ISO 10273:2005/Ap1, 2005; PN-EN ISO 10273:2005/Ap2, 2006).
- 20.Stanisz A. Przystępny kurs statystyki w oparciu o program STATISTICA PL na przykładach z medycyny vol. 1. Wydawnictwo StatSoft Polska Sp. z o. o.; 2007.
- 21.Platt-Samoraj A, Syczyło K, Szczerba-Turek A, Bancerz-Kisiel A, Jabłoński A, Łabuć S, et al. Presence of ail and ystB genes in Yersinia enterocolitica biotype 1A isolates from game animals in Poland. Vet J. 2017; 221: 11–13. doi: 10.1016/j.tvjl.2017.01.013 [DOI] [PubMed] [Google Scholar]
- 22.Fredriksson-Ahomaa M, Wacheck S, Koenig M, Stolle A, Stephan R. Prevalence of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in Switzerland. Int J Food Microbiol. 2009; 135: 199–202. doi: 10.1016/j.ijfoodmicro.2009.08.019 [DOI] [PubMed] [Google Scholar]
- 23.Bucher M, Meyer C, Grötzbach B, Stolle A, Fredriksson-Ahomaa M. Epidemiological data on pathogenic Yersinia enterocolitica in Southern Germany during 2000–2006. Foodborne Pathog Dis. 2008; 5: 273–280. doi: 10.1089/fpd.2007.0076 [DOI] [PubMed] [Google Scholar]
- 24.Avagnina A, Nucera D, Grassi MA, Ferroglio E, Dalmasso A, Civera T. The microbiological conditions of carcases from large game animals in Italy. Meat Sci. 2012; 91: 266–271. doi: 10.1016/j.meatsci.2012.01.025 [DOI] [PubMed] [Google Scholar]
- 25.Bancerz-Kisiel A, Socha P, Szweda W. Detection and characterisation of Yersinia enterocolitica strains in cold-stored carcasses of large game animals in Poland. Vet J. 2016; 208: 102–103. doi: 10.1016/j.tvjl.2015.08.010 [DOI] [PubMed] [Google Scholar]
- 26.Jakubczak A, Platt-Samoraj A, Siemionek J, Terech-Majewska E. The presence of Yersinia enterocolitica in breeding animals, domestic animals, wild boars and fish the Olsztyn District. Med Weter. 1993; 49: 301–303. [Google Scholar]
- 27.Arrausi-Subiza M, Gerrikagoita X, Alvarez V, Ibabe JC, Barral M. Prevalence of Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in the Basque Country, northern Spain. Acta Vet Scand. 2016; 58: 4 doi: 10.1186/s13028-016-0184-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Von Altrock A, Seinige D, Kehrenberg C. Yersinia enterocolitica isolates from wild boars hunted in Lower Saxony, Germany. Appl Environ Microb. 2015; 81: 4835–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al Dahouk S, Nockler K, Tomaso H, Splettstoesser WD, Jungersen G, Riber U, et al. Seroprevalence of brucellosis, tularemia and yersiniosis in wild boars (Sus scrofa) from north-eastern Germany. J Vet Med. 2005; 52: 444–455. doi: 10.1111/j.1439-0450.2005.00898.x [DOI] [PubMed] [Google Scholar]
- 30.Bancerz-Kisiel A, Szczerba-Turek A, Platt-Samoraj A, Socha P, Szweda W. Bioserotypes and virulence markers of Yersinia enterocolitica strains isolated from roe deer (Capreolus capreolus) and red deer (Cervus elaphus). Pol J Vet Sci. 2014; 17: 315–319. [DOI] [PubMed] [Google Scholar]
- 31.Platt-Samoraj A, Syczyło K, Bancerz-Kisiel A, Szczerba-Turek A, Giżejewska A, Szweda W. Yersinia enterocolitica strains isolated from beavers (Castor fiber). Pol J Vet Sci. 2015; 18: 449–451. doi: 10.1515/pjvs-2015-0058 [DOI] [PubMed] [Google Scholar]
- 32.Syczyło K, Platt-Samoraj A, Bancerz–Kisiel A, Szczerba-Turek A, Lipczyńska K, Jabłoński A, et al. Monitoring of Yersinia enterocolitica strains from free-living animals using different methods. Pol J Vet Sci. 2016; 19: 211–213. [DOI] [PubMed] [Google Scholar]
- 33.Kamińska S, Sadkowska-Todys M. Yersiniosis in Poland in 2014. Przegl Epidemiol. 2016; 70: 367–374. [PubMed] [Google Scholar]
- 34.Rastawicki W, Szych J, Rokosz N, Zacharczuk K, Gierczyński R. Seasonality of Yersinia enterocolitica bioserotype 1B/O:8 infections in Poland. Epidemiol Infect. 2013; 141: 2039–2042. doi: 10.1017/S0950268812002786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fredriksson-Ahomaa, Korkeala H. Low Occurrence of Pathogenic Yersinia enterocolitica in Clinical, Food, and Environmental Samples: a Methodological Problem. Clin Microbiol. 2003; 16: 223–229. doi: 10.1128/CMR.16.2.220–229.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bancerz-Kisiel A., Szczerba-Turek A., Platt-Samoraj A, Szweda W. Distribution of the ymoA and ystA genes and enterotoxins Yst production by Yersinia enterocolitica strains isolated from humans and pigs. Pol J Vet Sci. 2012; 15: 609–614. [DOI] [PubMed] [Google Scholar]
- 37.Platt-Samoraj A, Ugorski M, Szweda W, Szczerba-Turek A, Wojciech Ł, Procajło Z. Analysis of the presence of ail, ystA and ystB genes in Yersinia enterocolitica strains isolated from aborting sows and aborted fetuses. J Vet Med B. 2006; 53: 341–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.