Abstract
Plant expansins are proteins involved in cell wall loosening, plant growth, and development, as well as in response to plant diseases and other stresses. In this study, we identified 128 expansin coding sequences from the wheat (Triticum aestivum) genome. These sequences belong to 45 homoeologous copies of TaEXPs, including 26 TaEXPAs, 15 TaEXPBs and four TaEXLAs. No TaEXLB was identified. Gene expression and sub-expression profiles revealed that most of the TaEXPs were expressed either only in root tissues or in multiple organs. Real-time qPCR analysis showed that many TaEXPs were differentially expressed in four different tissues of the two wheat cultivars—the cold-sensitive ‘Chinese Spring (CS)’ and the cold-tolerant ‘Dongnongdongmai 1 (D1)’ cultivars. Our results suggest that the differential expression of TaEXPs could be related to low-temperature tolerance or sensitivity of different wheat cultivars. Our study expands our knowledge on wheat expansins and sheds new light on the functions of expansins in plant development and stress response.
Introduction
Plant expansins are a family of non-enzymatic proteins present in the plant cell wall. They play important roles in cell wall modification, which is critical for cell growth and other biological processes [1]. They bind to glucan-coated cellulose in the cell wall, causing a reversible disruption of hydrogen bonds between cellulose microfibrils and the glucan matrix to loosen the cell wall [2, 3]. Expansins also participate in the processes of seed germination [4] and the creation of yield [5], root growth and development [6–8], stem growth [9] and internode elongation [10]. Expansins are also involved in leaf initiation and expansion [11], flowering and determining flower size [12], pollen germination and fertilization [13], and fruit growth and/or ripening [14, 15]. They were associated with nutrient-uptake efficiency [16] as well as with abiotic and biotic stress tolerance [4, 17]. Most recently, it has been reported that expansins contribute to improving germination, leaf size, fruit growth/ripening, and enhancing plants tolerance to abiotic and biotic stresses [18], suggesting that this may be a promising gene family for improving crop quality and yield.
All expansin proteins have two conserved domains, the DPBB_1 and Pollen_allerg_1 domains. In addition, expansins have a 20–30 amino acid signaling peptide that is important for their expression [2, 19]. Plant expansins can be divided into four subfamilies, expansins A (EXPA), expansins B (EXPB), expansin-like A (EXLA) and expansin-like B (EXLB) [20]. Individual plant species may have all or some of the four subfamilies of expansins. For example, no EXLB has been found in Selaginella, Physcomitrella patens, [21] and maize [22].
Wheat (Triticum aestivum) is an important grain crop worldwide and scientists finds its genome to be one of the most challenging in plants to analyze. Its hexaploid genome features three complete genomes termed A, B and D, that originated from three diploid ancestral species: Triticum urartu (A Genome), Aegilops speltoides (B Genome) and Aegilops tauschii (D Genome) [23]. As a result, most genes in wheat are represented by triplicate homoeologs [24–27]. Currently, only 18 Triticum aestivum EXPs (TaEXPs) have been identified, several of which are homoeologous copies of the same gene [28]. One of the 18 TaEXPs, the TaEXPA1 gene, showed a similar phenotype and function in transgenic Arabidopsis [29].
Genome-wide analysis of expansins has been reported in Arabidopsis and rice [30], tobacco [31], soybean [32], tomato [33], grape [34], and maize [22]. However, no genome-wide expansin screening has been reported in wheat. In this study, we identified 128 expansin sequences from the draft sequences of the hexaploid wheat genome. Cis-acting element analysis revealed that many expansins are regulated by various hormones. They may be directly or indirectly involved in low-temperature stress response. We then compared the expansin expression profiles between a cold-tolerant (Dongnongdongmai 1, or D1) and a cold-sensitive (Chinese Spring, or CS) wheat cultivar. Our results suggest the possible involvement of wheat expansin in the low temperature stress response of wheat. Our study expands our knowledge of wheat expansins and provides new insights into the functions of expansins in plant development and stress response.
Materials and methods
Identification and sequence analysis of the wheat expansin genes
The sequences of the 18 identified previously TaEXP genes [28] were used to identify new TaEXPs using a blastn search against the Triticum aestivum genome sequences from GRAMENE (http://ensembl.gramene.org/) [35]. Sequences of Triticum urartu expansin genes [36] were also used to further identify TaEXPs in Triticum aestivum by BLAST against the IWGSC database. In addition, the expansin protein family Expan_Lol_pI (IPR007118) (https://www.ebi.ac.uk/interpro) was used to further search for expansin genes in the UniProt database (http://www.uniprot.org/). A total of 263 protein sequences were found and sequences that did not originate from the wheat genome were removed. The expansin genes from different databases were integrated and unified using the wheat IWGSC1+popseq.30 genome assembly.
The two essential structures of expansin genes, the DPBB_1 and Pollen_allerg_1 domains, were identified by the Conserved Domains tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). In plants, the p12/PNP/kiwellin proteins had a DPBB_1 domain only, and Grp2 pollen allergens proteins had a Pollen_allerg_1 domain only. Therefore, shorter sequences that had only one of the two domains were removed from further analysis to avoid false positive results [37]. Homoeologous copies were identified by the polyploid theory service in GRAMENE. The homoeologous expansin genes that were not present in all three subgenomes (A, B and D) were considered to be incomplete because of the incomplete genome sequences of wheat (Cheuk and Houde, 2016). The expansin gene library was regarded as the main data source. The TGACv1 database (http://www.gramene.org/) was initially used as supplementary data. Sequences from Triticum urartu (release no. 30) and Aegilops tauschii (release no. 30) were deemed as a second set of supplementary data [38]. The supplemental sequences were identified by the ncbi-blast-2.2.31+ program (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.2.31-last-win32-release/) from the main data by blast against the first and second supplementary data with the following parameters: Identity ≥ 93%, overlap_coverage ≥ 75% [39], overlap_length ≥ 300bp and gene_length ≥ 600 bp [2].
The gene sequences, including genomic sequences, transcript sequences, coding domain sequences (CDS), and promoter sequences, were downloaded from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and GRAMENE. All gene IDs were unified into TGACv1 gene ID. Protein sequences were predicted using the translation tool (http://web.expasy.org/translate/). The molecular weight, isoelectric point (pI), and the grand average of hydrophobicity (GRAVY) of each TaEXPs were calculated by the Protparam tool (http://web.expasy.org/protparam/). The signal peptides were predicted online with SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/).
The TaEXPs proteins were aligned using Clustal X1.83 with the default parameters except the gap opening, which was changed to 4.0 [40]. A phylogenetic tree was constructed using the neighbor-joining method in MEGA 5.0 [41] with default parameters. The bootstrap method was used for the test of phylogeny. The number of bootstrap replications was 500, and the model was equal to the input model. The MEME program (http://meme-suite.org/) was used to detect particular motifs of TaEXP proteins. The optimal motif width was 15 to 200 and maximum number of motifs was 20 [42]. The Web AUGUSTUS program (http://bioinf.uni-greifswald.de/augustus/submission.php) [43] and the GSDS 2.0 program (http://gsds.cbi.pku.edu.cn/) [44] were used to predict the exon–intron structures. Map Inspect software (http://mapinspect.software.informer.com/) was used to analyze the genic physical location on chromosomes. Cis-acting elements analysis in the 2 -kb upstream regions was performed with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [45, 46].
The overall expression and sub-expression of the wheat expansins were analyzed using previously developed databases. The overall expression profile (measured as transcripts per million) was calculated for each tissue by the UniGene EST profile in the US Department of Health National Center for Biotechnology Information website (https://www.ncbi.nlm.nih.gov/unigene/). The sub-expression profile (measured as reads per kilobase of exon model per million mapped reads) was calculated from the E-MTAB-4484 datasets of the Expression Atlas (https://www.ebi.ac.uk/gxa/home) [47]. These datasets were generated using the Triticum aestivum cultivar Chinese Spring. All datasets are publicly available at the above two websites. The profiles and cis-acting elements analysis of TaEXPs were made into heat maps by HemI 1.0 software (http://hemi.biocuckoo.org/) [48].
Plant materials and treatments
Two cultivars of Triticum aestivum, CS and D1 [49], were used as the plant materials to investigate the expression profiles of expansin genes during cold-acclimation. Seeds were soaked into double-distilled water about 24 h at 25°C, and then sprouted on moistened filter paper for 12 h. The seedlings were transplanted into pots (four seedlings per pot) and then grown in a plant culture room at 15 ± 2°C under 16:8 h light:dark cycles using red LED and natural light. For expansin expression profiling at normal growth condition, tissues (root, tillering node, stem, and the third leaf) were collected when plants reached the three-leaf stage (~20 d growth at 15°C) were frozen immediately in liquid nitrogen and stored at −80°C for RNA extraction. Tissues were collected from 4 different plants from one pot and the same tissue was mixed together. Three different pots were used as 3 independent experiments. Results were average of these 3 independent experiments. The plants underwent low-temperature treatment when they reached the three-leaf stage. A total of 30 pots (4 plants per pot) from each cultivar were used, half of which were randomly selected and moved to a 4°C growth chamber for 0, 3, 6, 12 and 24 h. The other half were grown in the original culture room as controls. Four different tissues (root, tillering node, stem, and the third leaf) were collected at the three-leaf stage from both 4°C plants and normal temperature plants. Samples were frozen immediately in liquid nitrogen and stored at −80°C for RNA extraction. At each time point, tissues were collected from 4 different plants in one pot and the same tissue was mixed together. Three different pots were used as 3 independent experiments. Results were average of these three independent experiments. The differential TaEXP expression at 4°C was compared against the expression at normal temperature of the same time point (control).
RNA isolation and real-time quantitative -PCR analysis
Total RNA was isolated from each plant tissue using a TransZol Up Plus RNA Kit (Transgen Biotech, Beijing, China), and treated with DNase I (Transgen). The quality of RNA was examined by agarose gel electrophoresis. The RNA samples were reverse transcribed to cDNAs with TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen). The cDNA was diluted five times before being used in downstream experiments.
Due to the high similarity of the homoeologous copies of each gene, the coding domain sequence (CDS) regions of the 45 TaEXPs were used to design for specific primers using Primer 5.0 program and Primique software (http://cgi-www.daimi.au.dk/cgi-chili/primique/front.py) [50]. Primers were then verified by blasting against genomic sequences in GRAMENE and in EST and NR/NT databases. The β-actin (Traes_1AL_E195290EF.2; Traes_1BL_D0A3F2067.1; Traes_1DL_727F09EF8.1) was used as the reference gene. All primers were listed in Supplemental S1 Table.
The Linegene 9600 fluorescence quantitative PCR detection system (Bioer Technology Co., Inc., Hangzhou, China) was used for real-time PCR experiments with the TransStart Top Green qPCR SuperMix (Transgen) in a total 20 μl reaction system, using SYBR as a reporter dye. Thermal cycling for RT-qPCR was as follows: 95°C for 20 s, 40 cycles of 95°C for 15 s, and 60°C for 20 s with luminescent reading. Melting curves were mapped by increasing the final temperature from 60 to 95°C. All the data were analyzed using the 2 −ΔΔCT method [51].
Statistical analysis
Microsoft Excel was used for statistical analysis (Student’s t-tests). Values are presented here as means of three biological replicates; error bars indicate standard derivation (SD). Significant differences are denoted with “*” (P < 0.05) or “**” (P < 0.01).
Results
Identification of expansin sequences in wheat
A total of 128 expansin sequences were identified and these were named according to standard principles for consistency [20]. Sequences of the 128 expansin genes were deposited in the GRAMENE database (http://ensembl.gramene.org/) and can be accessed by the gene IDs listed in Table 1. TaEXPA1-11, TaEXPB1-11 and TaEXPB23 had been named previously [28, 52–54], so the newly identified expansins were named as TaEXPA12 to –29, TaEXPB12 to –24 except for two that were named TaEXPB23 and TaEXLA1 to –4 (Table 1). Fifty-five of the identified expansin genes were confirmed to be located on 16 chromosomes (S1 Fig), but the chromosomal locations of other genes were not determined because some genome sequences were incomplete. Majority of the expansin genes had three copies, one each from chromosomes A, B, or D. TaEXPA3 and TaEXPA22 had two extra copies from B and D. TaEXPA25, TaEXPA26, TaEXPA28, TaEXPA29, TaEXPB20, and TaEXLA4 were not detected in chromosome A; TaEXPA26, TaEXPB17 and TaEXLA4 were not in chromosome B and TaEXPA28 and TaEXPA29 were not in chromosome D.
Table 1. Wheat expansin genes identified in this study.
| Gene ID | Name | Chromo-some | Protien length | Mw(Da) | pI | GRAVY | SP | DPBB_1 domain | Pollen_allerg_1 domain |
|---|---|---|---|---|---|---|---|---|---|
| TRIAE_CS42_3AL_TGACv1_194727_AA0638220 | TaEXPA3-A | 3A | 251 | 26231.3 | 6.78 | -0.004 | 1–24 | 64–149 | 162–233 |
| TRIAE_CS42_3B_TGACv1_223925_AA0789720 | TaEXPA3-B1 | 3B | 251 | 26248.2 | 6.78 | -0.018 | 1–24 | 64–149 | 162–233 |
| TRIAE_CS42_3B_TGACv1_222333_AA0762340 | TaEXPA3-B2 | 3B | 251 | 26232.2 | 6.78 | -0.024 | 1–24 | 64–149 | 162–233 |
| TRIAE_CS42_3DL_TGACv1_250798_AA0873340 | TaEXPA3-D1 | 3D | 251 | 26270.3 | 6.78 | -0.006 | 1–24 | 64–149 | 162–233 |
| TRIAE_CS42_3DL_TGACv1_253727_AA0895440 | TaEXPA3-D2 | 3D | 251 | 26226.3 | 6.78 | 0.006 | 1–24 | 64–149 | 162–233 |
| TRIAE_CS42_3AL_TGACv1_199377_AA0669710 | TaEXPA4-A | 3A | 249 | 27113.4 | 7.48 | -0.122 | 1–22 | 59–146 | 159–230 |
| TRIAE_CS42_3B_TGACv1_223005_AA0774980 | TaEXPA4-B | 3B | 249 | 27035.2 | 6.52 | -0.143 | 1–22 | 59–146 | 159–230 |
| TRIAE_CS42_3DL_TGACv1_250306_AA0865790 | TaEXPA4-D | 3D | 249 | 26914.1 | 6.52 | -0.12 | 1–22 | 59–146 | 159–230 |
| TRIAE_CS42_3AS_TGACv1_211321_AA0688560 | TaEXPA5-A | 3A | 252 | 26769.3 | 9.23 | -0.088 | 1–22 | 60–148 | 161–232 |
| TRIAE_CS42_3B_TGACv1_224402_AA0796400 | TaEXPA5-B | 3B | 257 | 27391.1 | 9.17 | -0.089 | 1–22 | 66–153 | 166–237 |
| F775_32564 | TaEXPA5-D | 277 | 29527.6 | 9.27 | -0.075 | 1–22 | 60–147 | 160–231 | |
| TRIAE_CS42_4AS_TGACv1_306950_AA1015420 | TaEXPA6-A | 4A | 254 | 27799.5 | 7.46 | 0.031 | 1–22 | 63–149 | 162–233 |
| TRIAE_CS42_4BL_TGACv1_320333_AA1035610 | TaEXPA6-B | 4B | 254 | 27718.4 | 7.48 | 0.036 | 1–24 | 63–149 | 162–233 |
| TRIAE_CS42_4DL_TGACv1_344422_AA1147040 | TaEXPA6-D | 4D | 254 | 27746.4 | 7.48 | 0.039 | 1–24 | 63–149 | 162–233 |
| TRIAE_CS42_2AL_TGACv1_095133_AA0307290 | TaEXPA7-A | 2A | 258 | 27670.7 | 8.09 | 0.072 | 1–19 | 68–154 | 167–234 |
| TRIAE_CS42_2BL_TGACv1_129425_AA0383180 | TaEXPA7-B | 2B | 258 | 27670.7 | 8.09 | 0.071 | 1–19 | 68–154 | 167–234 |
| TRIAE_CS42_2DL_TGACv1_161611_AA0559500 | TaEXPA7-D | 2D | 258 | 27684.7 | 8.09 | 0.068 | 1–19 | 68–154 | 167–234 |
| TRIAE_CS42_3AS_TGACv1_212410_AA0700630 | TaEXPA8-A | 3A | 246 | 25486.6 | 7.55 | 0.107 | 1–20 | 60–144 | 157–228 |
| TRIAE_CS42_3B_TGACv1_224406_AA0796470 | TaEXPA8-B | 3B | 246 | 25520.6 | 7.54 | 0.081 | 1–20 | 60–144 | 157–228 |
| TRIAE_CS42_3DS_TGACv1_272528_AA0921790 | TaEXPA8-D | 3D | 246 | 25460.5 | 7.54 | 0.08 | 1–20 | 60–144 | 157–228 |
| TRIAE_CS42_5AL_TGACv1_374454_AA1200520 | TaEXPA9-A | 5A | 268 | 28813.8 | 9.4 | -0.048 | 1–29 | 76–164 | 177–248 |
| TRIAE_CS42_5BL_TGACv1_406965_AA1351930 | TaEXPA9-B | 5B | 266 | 28546.4 | 9.51 | -0.069 | 1–27 | 74–162 | 175–246 |
| TRIAE_CS42_5DL_TGACv1_435145_AA1446640 | TaEXPA9-D | 5D | 266 | 28589.5 | 9.61 | -0.039 | 1–27 | 74–162 | 175–246 |
| TRIAE_CS42_4AS_TGACv1_307970_AA1024860 | TaEXPA12-A | 4A | 259 | 27673.7 | 9.05 | -0.012 | 1–24 | 66–155 | 168–240 |
| TRIAE_CS42_4BL_TGACv1_321137_AA1056060 | TaEXPA12-B | 4B | 282 | 30232.7 | 9.45 | -0.009 | 1–24 | 66–155 | 168–240 |
| TRIAE_CS42_4DL_TGACv1_342474_AA1114540 | TaEXPA12-D | 4D | 259 | 27646.8 | 9.05 | 0.021 | 1–24 | 66–155 | 168–240 |
| TRIAE_CS42_1AL_TGACv1_000407_AA0011360 | TaEXPA13-A | 1A | 256 | 27735.5 | 6.78 | 0.094 | 1–26 | 65–151 | 164–235 |
| TRIAE_CS42_5BL_TGACv1_404293_AA1294240 | TaEXPA13-B | 5B | 256 | 27817.5 | 6.67 | 0.082 | 1–26 | 65–151 | 164–235 |
| TRIAE_CS42_5DL_TGACv1_433714_AA1420290 | TaEXPA13-D | 5D | 256 | 27811.5 | 6.78 | 0.083 | 1–26 | 65–151 | 164–235 |
| TRIAE_CS42_1AL_TGACv1_000407_AA0011380 | TaEXPA14-A | 1A | 254 | 27531.2 | 7.46 | 0.059 | 1–16 | 63–149 | 162–233 |
| TRIAE_CS42_5BL_TGACv1_405270_AA1323630 | TaEXPA14-B | 5B | 254 | 27497.1 | 8.05 | 0.065 | 1–16 | 63–149 | 162–233 |
| TRIAE_CS42_5DL_TGACv1_433714_AA1420300 | TaEXPA14-D | 5D | 255 | 27699.5 | 8.05 | 0.071 | 1–17 | 64–150 | 171–250 |
| TRIAE_CS42_4AS_TGACv1_309318_AA1030760 | TaEXPA15-A | 4A | 258 | 27651.7 | 8.96 | -0.032 | 1–24 | 65–154 | 167–239 |
| TRIAE_CS42_4BL_TGACv1_320437_AA1039060 | TaEXPA15-B | 4B | 259 | 27489.5 | 8.79 | 0.084 | 1–24 | 66–155 | 168–240 |
| F775_12787 | TaEXPA15-D | 259 | 27503.5 | 9.01 | 0.048 | 1–24 | 66–155 | 168–240 | |
| TRIAE_CS42_4AS_TGACv1_308797_AA1029560 | TaEXPA16-A | 4A | 257 | 27081.2 | 9.11 | -0.144 | 1–29 | 66–153 | 166–237 |
| TRIAE_CS42_4BL_TGACv1_321802_AA1065630 | TaEXPA16-B | 4B | 257 | 27217.5 | 8.99 | -0.149 | 1–29 | 66–153 | 166–237 |
| TRIAE_CS42_4DL_TGACv1_343034_AA1128010 | TaEXPA16-D | 4D | 257 | 27108.3 | 9.01 | -0.145 | 1–29 | 66–153 | 166–237 |
| TRIAE_CS42_3AS_TGACv1_210583_AA0675440 | TaEXPA17-A | 3A | 258 | 27696.8 | 8.73 | -0.054 | 1–24 | 67–154 | 166–238 |
| TRIAE_CS42_3B_TGACv1_227374_AA0823100 | TaEXPA17-B | 3B | 261 | 28065.2 | 9.03 | -0.091 | 1–24 | 67–156 | 169–241 |
| TRIAE_CS42_3DS_TGACv1_271478_AA0899830 | TaEXPA17-D | 3D | 261 | 28071.4 | 8.94 | -0.051 | 1–24 | 67–156 | 169–241 |
| TRIAE_CS42_1AL_TGACv1_000844_AA0020230 | TaEXPA18-A | 1A | 259 | 27480.3 | 8.91 | 0.069 | 1–24 | 67–153 | 171–238 |
| TRIAE_CS42_1BL_TGACv1_031377_AA0112820 | TaEXPA18-B | 1B | 261 | 27682.6 | 8.91 | 0.081 | 1–29 | 69–155 | 173–240 |
| TRIAE_CS42_1DL_TGACv1_062518_AA0215780 | TaEXPA18-D | 1D | 259 | 27481.4 | 8.91 | 0.096 | 1–24 | 67–153 | 171–238 |
| TRIAE_CS42_4AL_TGACv1_291883_AA0996900 | TaEXPA19-A | 4A | 255 | 27641 | 8.05 | -0.091 | 1–25 | 65–151 | 164–234 |
| TRIAE_CS42_5BL_TGACv1_406879_AA1351000 | TaEXPA19-B | 5B | 255 | 27632.1 | 8.56 | -0.1 | 1–25 | 65–151 | 164–235 |
| TRIAE_CS42_5DL_TGACv1_434954_AA1444020 | TaEXPA19-D | 5D | 255 | 27581.9 | 8.35 | -0.104 | 1–25 | 65–151 | 164–234 |
| TRIAE_CS42_4AS_TGACv1_307135_AA1017450 | TaEXPA20-A | 4A | 253 | 27107.8 | 8.32 | 0.045 | 1–24 | 61–148 | 161–232 |
| TRIAE_CS42_4BL_TGACv1_320333_AA1035620 | TaEXPA20-B | 4B | 253 | 27097.7 | 8.53 | 0.032 | 1–24 | 62–148 | 161–232 |
| TRIAE_CS42_4DL_TGACv1_343034_AA1128030 | TaEXPA20-D | 4D | 250 | 26696.2 | 8.32 | 0.062 | 1–24 | 62–145 | 158–229 |
| TRIAE_CS42_5AL_TGACv1_374723_AA1207410 | TaEXPA21-A | 5A | 282 | 29824.6 | 10.68 | -0.602 | 1–24 | 65–140 | |
| TRIAE_CS42_5BL_TGACv1_407229_AA1354640 | TaEXPA21-B | 5B | 259 | 27737.6 | 8.96 | -0.027 | 1–25 | 66–154 | 167–238 |
| TRIAE_CS42_5DL_TGACv1_433521_AA1415520 | TaEXPA21-D | 5D | 293 | 32414.9 | 10.24 | -0.438 | 45–133 | 146–217 | |
| TRIAE_CS42_4AS_TGACv1_306914_AA1015010 | TaEXPA22-A | 4A | 253 | 27566.2 | 6.24 | 0.091 | 1–24 | 62–148 | 161–232 |
| TRIAE_CS42_4BL_TGACv1_320277_AA1033630 | TaEXPA22-BI | 4B | 253 | 27445.1 | 6.24 | 0.083 | 1–24 | 62–148 | 161–232 |
| TRIAE_CS42_4BL_TGACv1_320277_AA1033640 | TaEXPA22-B2 | 4B | 247 | 27172.6 | 6.38 | -0.031 | 1–20 | 58–144 | 155–226 |
| TRIAE_CS42_4DL_TGACv1_343032_AA1127880 | TaEXPA22-D1 | 4D | 253 | 27545.2 | 5.84 | 0.083 | 1–24 | 62–148 | 161–232 |
| TRIAE_CS42_4DL_TGACv1_345231_AA1152290 | TaEXPA22-D2 | 4D | 253 | 27678.4 | 6.24 | 0.046 | 1–24 | 62–148 | 161–232 |
| TRIAE_CS42_2AS_TGACv1_114363_AA0366370 | TaEXPA23-A | 2A | 261 | 27546 | 8.9 | 0.007 | 1–24 | 69–157 | 170–240 |
| TRIAE_CS42_2BS_TGACv1_146019_AA0453260 | TaEXPA23-B | 2B | 262 | 27524.9 | 8.9 | -0.003 | 1–29 | 70–158 | 171–241 |
| TRIAE_CS42_2DS_TGACv1_177171_AA0567520 | TaEXPA23-D | 2D | 261 | 27547 | 8.9 | 0.035 | 1–28 | 69–157 | 170–240 |
| TRIAE_CS42_3AS_TGACv1_210530_AA0674760 | TaEXPA24-A | 264 | 28433.5 | 8.56 | -0.076 | 1–23 | 71–160 | 173–245 | |
| TRAES3BF143800010CFD | TaEXPA24-B | 3B | 264 | 28442.4 | 8.06 | -0.058 | 1–23 | 71–160 | 173–245 |
| F775_19619 | TaEXPA24-D | 264 | 28386.3 | 8.61 | -0.089 | 1–23 | 71–160 | 173–245 | |
| TRIAE_CS42_4BL_TGACv1_320333_AA1035630 | TaEXPA25-B | 4B | 259 | 27424 | 6.87 | 0.043 | 1–22 | 66–154 | 167–237 |
| TRIAE_CS42_4DL_TGACv1_343034_AA1128020 | TaEXPA25-D | 4D | 258 | 27123.7 | 7.55 | 0.153 | 1–25 | 65–153 | 166–236 |
| Traes_7DL_5812FED4E | TaEXPA26-D | 7D | 268 | 28389.3 | 8.98 | -0.075 | 1–22 | 61–174 | 184–264 |
| TRIAE_CS42_7AL_TGACv1_556942_AA1773860 | TaEXPA27-A | 7A | 260 | 27811.6 | 7.48 | -0.017 | 1–20 | 69–156 | 169–240 |
| TRIAE_CS42_7BL_TGACv1_577039_AA1863550 | TaEXPA27-B | 7B | 262 | 28059.9 | 7.48 | -0.021 | 1–22 | 71–158 | 171–242 |
| TRIAE_CS42_7DL_TGACv1_604808_AA2001830 | TaEXPA27-D | 7D | 261 | 27972.8 | 7.46 | -0.001 | 1–21 | 70–157 | 170–241 |
| TRIAE_CS42_4BL_TGACv1_320437_AA1039040 | TaEXPA28-B | 4B | 259 | 27607.8 | 8.7 | 0.116 | 1–24 | 49–165 | 175–255 |
| TRIAE_CS42_4BL_TGACv1_320277_AA1033650 | TaEXPA29-B | 4B | 253 | 27737.4 | 6.78 | 0.041 | 1–24 | 44–158 | 168–247 |
| TRIAE_CS42_1AL_TGACv1_002200_AA0039680 | TaEXPB1-A | 1A | 265 | 28714.1 | 4.98 | -0.264 | 1–24 | 78–156 | 170–245 |
| TRIAE_CS42_1BL_TGACv1_034214_AA0143800 | TaEXPB1-B | 1B | 270 | 29277.7 | 4.98 | -0.236 | 1–24 | 83–161 | 175–250 |
| TRIAE_CS42_1DL_TGACv1_062107_AA0208850 | TaEXPB1-D | 1D | 264 | 28687.1 | 4.98 | -0.248 | 1–24 | 77–155 | 169–244 |
| TRIAE_CS42_1AL_TGACv1_000456_AA0012510 | TaEXPB8-A | 6A | 273 | 29962.9 | 9.39 | -0.274 | 1–26 | 80–160 | 174–253 |
| TRIAE_CS42_1BL_TGACv1_031113_AA0108080 | TaEXPB8-B | 6B | 273 | 29736.7 | 9.06 | -0.194 | 1–26 | 80–160 | 174–253 |
| TRIAE_CS42_1DL_TGACv1_063167_AA0224540 | TaEXPB8-D | 6D | 273 | 29890.8 | 9.48 | -0.259 | 1–26 | 80–160 | 174–253 |
| TRIAE_CS42_6AS_TGACv1_485310_AA1542890 | TaEXPB7-A | 1A | 270 | 29137.9 | 6.5 | -0.181 | 1–26 | 83–161 | 175–250 |
| TRIAE_CS42_6BS_TGACv1_513750_AA1648570 | TaEXPB7-B | 1B | 270 | 29219.1 | 6.5 | -0.127 | 1–26 | 83–161 | 175–250 |
| TRIAE_CS42_6DS_TGACv1_542511_AA1723160 | TaEXPB7-D | 1D | 270 | 29203 | 6.93 | -0.174 | 1–26 | 83–161 | 175–250 |
| TRIAE_CS42_1AL_TGACv1_003247_AA0048900 | TaEXPB10-A | 1A | 271 | 29853.7 | 8.81 | -0.256 | 1–24 | 78–158 | 172–251 |
| TRIAE_CS42_1BL_TGACv1_032979_AA0135990 | TaEXPB10-B | 1B | 271 | 29976.9 | 8.81 | -0.269 | 1–24 | 78–158 | 172–251 |
| TRIAE_CS42_1DL_TGACv1_062861_AA0221130 | TaEXPB10-D | 1D | 271 | 30037 | 8.81 | -0.259 | 1–24 | 78–158 | 172–251 |
| TRIAE_CS42_2AL_TGACv1_093830_AA0287660 | TaEXPB12-A | 2A | 281 | 29498.3 | 9.17 | -0.125 | 1–25 | 80–160 | 174–260 |
| TRIAE_CS42_2BL_TGACv1_129731_AA0394090 | TaEXPB12-B | 2B | 281 | 29494.3 | 9.16 | -0.104 | 1–25 | 80–160 | 174–260 |
| TRIAE_CS42_2DL_TGACv1_158079_AA0508190 | TaEXPB12-D | 2D | 281 | 29496.3 | 9.16 | -0.105 | 1–25 | 80–160 | 174–260 |
| TRIAE_CS42_6AL_TGACv1_471607_AA1511690 | TaEXPB13-A | 6A | 289 | 30322.7 | 9.45 | -0.136 | 1–25 | 99–180 | 194–269 |
| TRIAE_CS42_6BL_TGACv1_500765_AA1609400 | TaEXPB13-B | 3B | 289 | 30349.1 | 9.49 | 0.002 | 1–25 | 99–180 | 194–269 |
| TRIAE_CS42_6DL_TGACv1_527377_AA1703040 | TaEXPB13-D | 6D | 289 | 30308.7 | 9.45 | -0.139 | 1–25 | 99–180 | 194–269 |
| TRIAE_CS42_5AS_TGACv1_393897_AA1277020 | TaEXPB14-A | 5A | 323 | 35331.1 | 6.64 | -0.215 | 1–26 | 108–186 | 200–281 |
| TRIAE_CS42_5BS_TGACv1_423497_AA1378190 | TaEXPB14-B | 5B | 341 | 37470.6 | 7.51 | -0.094 | 1–27 | 107–185 | 199–280 |
| TRIAE_CS42_5DS_TGACv1_457565_AA1487840 | TaEXPB14-D | 5D | 347 | 38014.2 | 7.95 | -0.135 | 1–28 | 113–191 | 205–286 |
| TRIAE_CS42_2AL_TGACv1_094343_AA0296260 | TaEXPB15-A | 2A | 265 | 28794.1 | 9.07 | -0.163 | 1–22 | 78–156 | 171–245 |
| TRIAE_CS42_2BL_TGACv1_131181_AA0424020 | TaEXPB15-B | 2B | 265 | 28791.1 | 9.22 | -0.174 | 1–22 | 78–156 | 171–245 |
| TRIAE_CS42_2DL_TGACv1_159779_AA0542610 | TaEXPB15-D | 2D | 265 | 28779 | 9.24 | -0.217 | 1–23 | 78–156 | 171–245 |
| TRIAE_CS42_6AL_TGACv1_471087_AA1502540 | TaEXPB16-A | 6A | 287 | 29956.1 | 8.74 | -0.114 | 1–26 | 94–176 | 190–267 |
| TRIAE_CS42_6BL_TGACv1_503053_AA1627120 | TaEXPB16-B | 6B | 287 | 30016.3 | 8.85 | -0.083 | 1–26 | 94–176 | 190–267 |
| TRIAE_CS42_6DL_TGACv1_526554_AA1687000 | TaEXPB16-D | 6D | 287 | 30107.2 | 8.74 | -0.135 | 1–26 | 94–176 | 190–267 |
| TRIAE_CS42_6AL_TGACv1_471607_AA1511700 | TaEXPB17-A | 6A | 265 | 27780.1 | 5.56 | -0.099 | 1–27 | 74–155 | 169–245 |
| TRIAE_CS42_6DL_TGACv1_526693_AA1690080 | TaEXPB17-D | 6D | 266 | 27750.3 | 6.27 | -0.042 | 1–27 | 74–156 | 170–246 |
| TRIAE_CS42_2AL_TGACv1_093830_AA0287650 | TaEXPB18-A | 2A | 261 | 27787.7 | 8.69 | -0.269 | 1–27 | 65–145 | 159–240 |
| TRIAE_CS42_2BL_TGACv1_129731_AA0394080 | TaEXPB18-B | 2B | 261 | 27912.8 | 8.8 | -0.271 | 1–27 | 65–145 | 159–240 |
| TRIAE_CS42_2DL_TGACv1_158079_AA0508200 | TaEXPB18-D | 2D | 261 | 27859.7 | 8.57 | -0.281 | 1–27 | 65–145 | 159–240 |
| TRIAE_CS42_1AL_TGACv1_003486_AA0050020 | TaEXPB19-A | 1A | 274 | 30222.4 | 9.44 | -0.362 | 1–25 | 83–161 | 175–254 |
| TRIAE_CS42_1BL_TGACv1_030851_AA0102320 | TaEXPB19-B | 1B | 274 | 30186.5 | 9.36 | -0.303 | 1–25 | 83–161 | 175–254 |
| TRIAE_CS42_1DL_TGACv1_062107_AA0208840 | TaEXPB19-D | 1D | 274 | 30189.4 | 9.44 | -0.339 | 1–25 | 83–161 | 175–254 |
| TRIAE_CS42_3B_TGACv1_222229_AA0760180 | TaEXPB20-B | 3B | 300 | 31366.2 | 5.17 | -0.205 | 1–25 | 108–188 | 202–279 |
| TRIAE_CS42_3DL_TGACv1_250163_AA0863360 | TaEXPB20-D | 3D | 314 | 32644.6 | 5.15 | -0.239 | 1–27 | 122–202 | 216–293 |
| TRIAE_CS42_5AL_TGACv1_375143_AA1216890 | TaEXPB21-A | 5A | 308 | 32331.6 | 5.13 | 0.122 | 1–26 | 73–164 | 178–254 |
| TRIAE_CS42_5BL_TGACv1_404585_AA1305050 | TaEXPB21-B | 5B | 309 | 32456.8 | 5.48 | 0.09 | 1–26 | 73–164 | 178–255 |
| TRIAE_CS42_5DL_TGACv1_433716_AA1420380 | TaEXPB21-D | 5D | 308 | 32213.5 | 5.43 | 0.119 | 1–26 | 73–164 | 178–254 |
| TRIAE_CS42_2AS_TGACv1_115226_AA0371610 | TaEXPB22-A | 2A | 253 | 26463 | 8.31 | -0.02 | 1–27 | 65–146 | 160–233 |
| TRIAE_CS42_2BS_TGACv1_146193_AA0458020 | TaEXPB22-B | 2B | 252 | 26420.9 | 8.31 | 0 | 1–26 | 64–145 | 159–232 |
| TRIAE_CS42_2DS_TGACv1_179985_AA0609770 | TaEXPB22-D | 2D | 254 | 26741.4 | 7.5 | 0.045 | 1–27 | 65–146 | 160–233 |
| TRIAE_CS42_2AL_TGACv1_093830_AA0287670 | TaEXPB24-A | 2A | 262 | 26924.6 | 7.52 | 0.188 | 1–26 | 73–153 | 167–242 |
| TRIAE_CS42_2BL_TGACv1_129731_AA0394100 | TaEXPB24-B | 2B | 262 | 26981.6 | 8.05 | 0.121 | 1–26 | 73–153 | 167–242 |
| TRIAE_CS42_2DL_TGACv1_158079_AA0508180 | TaEXPB24-D | 2D | 262 | 26873.5 | 7.52 | 0.2 | 1–26 | 73–153 | 167–242 |
| TRIAE_CS42_5AL_TGACv1_374686_AA1206430 | TaEXLA1-A | 5A | 275 | 29628 | 9.5 | -0.062 | 1–30 | 69–132 | 161–238 |
| TRIAE_CS42_4BL_TGACv1_320494_AA1041290 | TaEXLA1-B | 4B | 275 | 29527 | 9.39 | -0.013 | 1–30 | 69–132 | 161–238 |
| Traes_4DL_74A03B3BE | TaEXLA1-D | 4D | 277 | 29663.2 | 9.35 | -0.01 | 1–32 | 71–134 | 163–240 |
| TRIAE_CS42_5AL_TGACv1_377017_AA1243240 | TaEXLA2-A | 5A | 273 | 28875 | 8.78 | 0.025 | 1–27 | 66–144 | 159–236 |
| TRIAE_CS42_4BL_TGACv1_322143_AA1068920 | TaEXLA2-B | 4B | 268 | 28615.6 | 8.31 | -0.045 | 1–22 | 61–139 | 154–231 |
| TRIAE_CS42_4DL_TGACv1_344616_AA1148420 | TaEXLA2-D | 4D | 273 | 28934.9 | 7.98 | -0.004 | 1–27 | 66–144 | 159–236 |
| TRIUR3_10907 | TaEXLA3-A | 270 | 28368.8 | 5.61 | -0.083 | 63–141 | 155–232 | ||
| TRIAE_CS42_2BL_TGACv1_129773_AA0395490 | TaEXLA3-B | 2B | 271 | 28685.5 | 5.75 | -0.023 | 1–25 | 64–142 | 157–225 |
| Traes_4DL_7453A191F | TaEXLA3-D | 4D | 271 | 28883.9 | 6.71 | -0.023 | 1–25 | 64–142 | 156–233 |
| Traes_4DL_C95A997A8 | TaEXLA4-D | 4D | 270 | 28670.5 | 5.39 | 0.048 | 1–25 | 68–142 | 156–232 |
New genes were named following the known genes. Protein length, SP, and two domains were shown beside amino acids numbers. MW, molecular weight; pI, isoelectric point.
Sequence comparison in the GRAMENE database revealed that the 128 expansin sequences belong to 45 homoeologous TaEXPs, including 26 members of TaEXPAs, 15 members of TaEXPBs and four members of TaEXLAs. No EXLB sequence was identified. The 45 homoeologous TaEXPs were further confirmed by local BLAST analysis with the following parameters: identity ≥ 93%, overlap_coverage ≥ 75% [39], overlap_length ≥ 300 bp and gene_length ≥ 600 bp [2].
Only 11 of the 45 TaEXPs have been previously reported [28] and the remaining 34 were newly identified expansins in wheat. All the identified expansin sequences had two conserved essential domains, DPBB_1 and Pollen_allerg_1. The predicted proteins ranged from 246 to 347 amino acids with an average of 266.64 amino acids. Their predicted molecular weights ranged from 25.46 to 38.14 kDa. The pI values ranged from 4.98 to 10.68. The GRAVY values ranged from −0.602 to 0.2. The number of predicted DPBB_1 domains were 85–114 amino acids in TaEXPAs, 78–82 in TaEXPBs and 63–78 in TaEXLAs. The number of predicted Pollen_allerg_1 domains were 67–79 amino acids in TaEXPAs, 71–81 in TaEXPBs and 77–78 in TaEXLAs (Table 1). The homoeologous TaEXPs were similar in their protein properties, but different TaEXPs were significantly different in their protein properties.
Phylogenetic and structural analysis of the identified wheat expansins
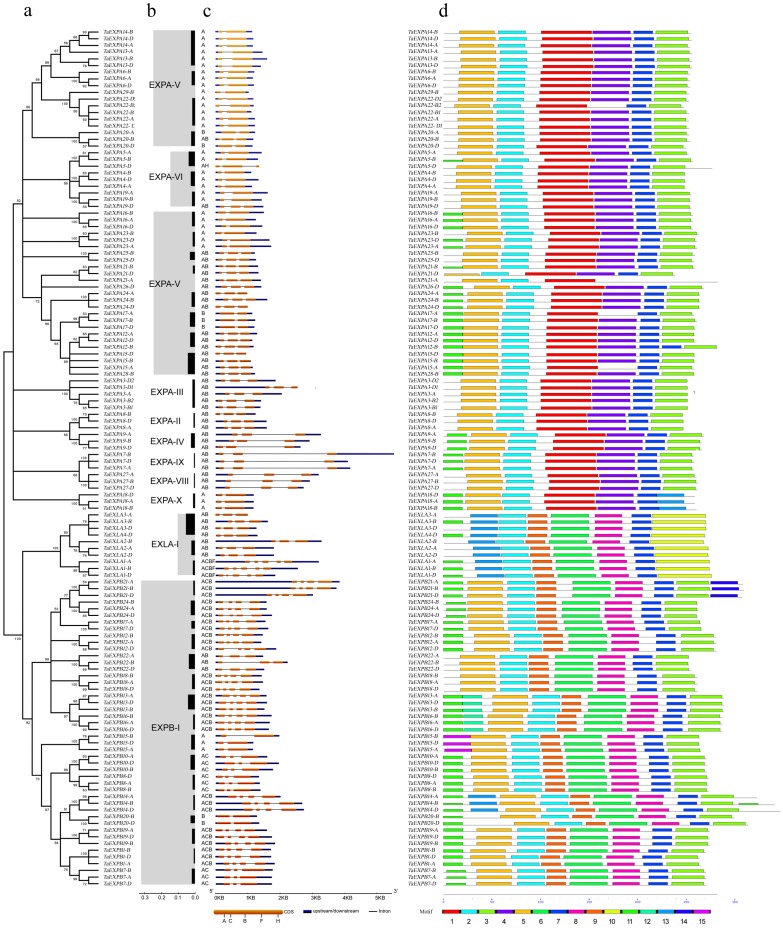
To better understand the relationships among the different expansins, the 128 predicted expansin proteins were used to establish a phylogenetic tree. This phylogenetic tree had three major branches that separate TaEXPAs, TaEXPBs and TaEXLAs (Fig 1a). Nine evolutionary branches were found, based on the grouping method of the Arabidopsis and rice expansin gene families [2] with bootstrap support values > 50% (Fig 1b, longitudinal). TaEXPAs had seven branches (EXPA-II, EXPA-III, EXPA-IV, EXPA-V, EXPA-VI, EXPA-VIII, and EXPA-IX), while only one branch was found in TaEXPBs (EXPB-I) and TaEXLAs (EXLA-I). The EXPA-V clade was the largest clade with 15 TaEXPA members. TaEXPBs only had the EXPB-I branch and TaEXLAs belonged to the EXLA-I branch. This showed that TaEXPAs were intricate in the phylogeny process. Two abundant EXPB-I sub-branches in wheat have been previously verified in a study related to the gramineous EXPB subfamily [3]. However, the EXPA-V branch had not been previously identified in wheat and the EXPA-IV branch was the largest clade in tobacco [31].
Fig 1. Phylogenetic analysis of wheat expansins.
a. Phylogenetic tree of the 128 expansin sequences; b. Evolutionary branches of wheat expansins; c. Analysis of gene structure, coding domain sequences (CDSs), 5’/3’ untranslated region and exson/introns; d. Motif analysis of the 128 expansin sequences.
The phylogenetic tree showed the linearized distance that exists among TaEXPs and demonstrated the nature of all of their evolutionary branches (Fig 1b, horizontal). The linearity of all expansin genes was shorter than 0.1. The EXPB-I genes were longer than 3.0, the EXPA-V were 2.0 to 3.0, the EXLA-I and EXPA-VI were 1.0 to 2.0, and other genes were shorter than 0.05.
Gene structure analysis revealed the relationship among the CDS region, 5’/3’ untranslated region and exon/intron structure using GSDS software (Fig 1c). It showed that the gene region and structure of each TaEXPs homoeologous copy were similar. The pattern of intron insertion was classified into five categories of ACBFH [2, 30, 34]. TaEXPAs had one or two introns (A, B, AB or AH); TaEXPBs had one to three introns (A, B, AB, AC or ACB); TaEXLAs had two or four introns (AB and ACBF). By comparing among each of the TaEXPs homoeologous copies, the insertions of H, B and A were identified from TaEXPA5-D, TaEXPA19-D and TaEXPA20-B. Extension of A and B introns, which were longer than exons, were identified from TaEXPA7, TaEXPA9, TaEXPA27, TaEXPB12-D, TaEXPB15-B, and TaEXPB22-B. The proportion of intron extended genes was about 9.4% in wheat genome and 14.8% in maize [22], but it was 86.5% in tobacco [31], 76.3% in tomato [33] and 81.3% in soybean [32].
Analysis of TaEXP protein domains and motifs
The TaEXP proteins contain three major domains: the signal peptide (SP), the DPBB_1 domain, and the Pollen_allerg_1 domain (S2 Fig). Similar to a previous study [33], the α and β insertions [55] were identified in our newly identified TaEXPs. The α-insertions were present in the TaEXPAs with seven conserved amino acids, GGWCNPP; the β-insertions were present in TaEXPBs and TaEXLAs, with only one conserved amino acid G.
A total of 15 motifs were identified (S3 Fig). Motifs 2 and 7 were present in most TaEXPs (Fig 1d). Motif 2 contained five specific amino acids (GCGCC) and was present in the conserved DPBB_1 domain (pfam03330) before α and β insertions. Motif 7 also contained five specific amino acids [WM (Met) WGW] and this motif was mapped in the 3’ part of the Pollen_allerg_1 domain (pfam01357). In TaEXPB13 and TaEXPB16, Motif 7 was present in the signal peptide region. Motifs 3 and 5 were found in both TaEXPAs and TaEXPBs, while Motifs 10 and 13 were identified in TaEXLAs. Motifs 1 and 4 were found in TaEXPAs, while Motifs 6, 8 and 9 were localized in TaEXPBs and TaEXLAs. Most motifs of TaEXPA were conserved except TaEXPA21-A.
Analysis of overall expression and subexpression of the wheat expansins
To better understand the functions of the expansins, the overall expression and subexpression of expansins were investigated using previously developed databases as described in the Materials and Methods section.
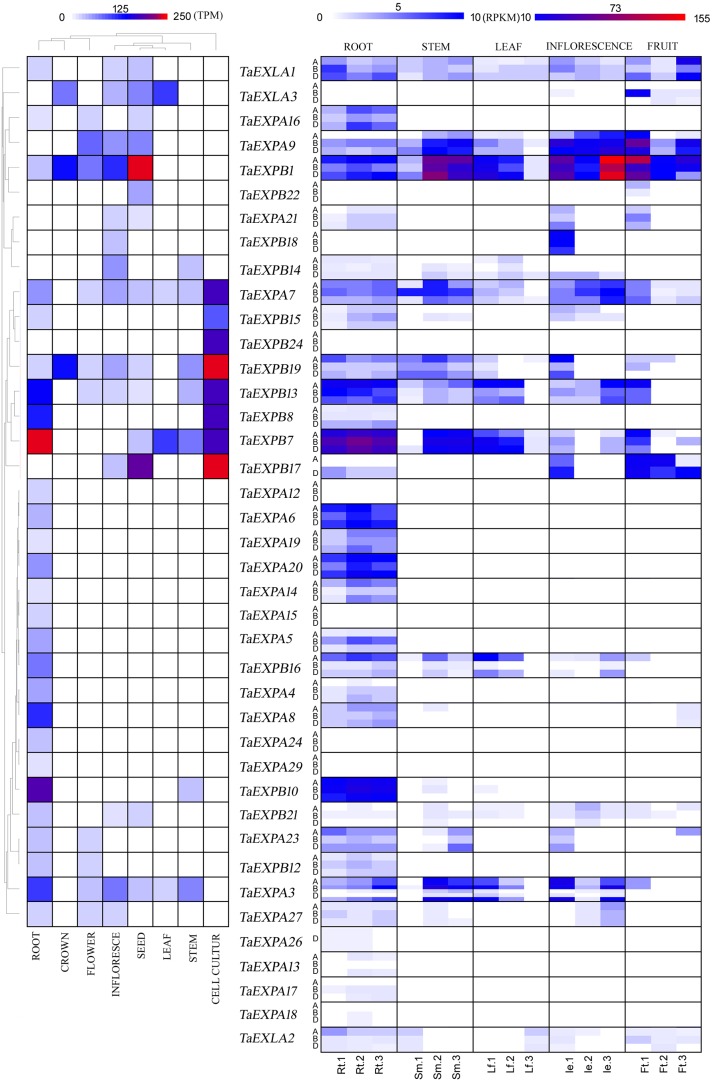
The overall expression measures the expression of organs mixed from different time points. Our results showed that 12 genes were expressed only in root tissue. TaEXPA3, TaEXPA8, TaEXPB7, TaEXPB8, TaEXPB10, and TaEXPB13, were expressed at a higher level in root than in other organs. Eighteen TaEXPs were expressed in some of the reproductive organs, including crown, flower, inflorescence, and seed. TaEXPB1 and TaEXPB19 were expressed in all these four organs. Only four genes, specifically TaEXPA3, TaEXPA7, TaEXPB7, and TaEXLA3, were expressed in leaves. Two TaEXPAs and five TaEXPBs were expressed in stems. TaEXPA7 and seven TaEXPBs showed prominent expression in cell culture. TaEXPA3, TaEXPA7, TaEXPB7, TaEXPB13, and TaEXPB19 were expressed in six of the eight tissues and/or organs (Fig 2).
Fig 2. Expansin gene expression and subexpression profiles in different tissues. Expression profiles were expressed as transcripts per million.
The subexpression profiles were expressed as reads per kilobase of exon model per Million mapped reads (FPKM). A, B and D represent subgenomes A, B, and D. Columns are labeled to show expansion expression as follows: Rt 1: in root during the cotyledon emergence stage, Rt 2: in root during the three leaf stage, Rt 3: in root during the maximum stem length stage; Sm 1: in stem during the half of flower open stage, Sm 2: in stem at the beginning of stem elongation, Sm 3: in stem during two nodes or internodes visible stages; Lf 1: in leaf during the main shoot and axillary shoots visible at three nodes stage, Lf 2: in stem during cotyledon emergence, Lf 3: in leaf during fruit formation; Ie 1: in inflorescence during the half of flower open stage, Ie 2: in inflorescence during the two nodes or internodes visible stage; Ie 3: in inflorescence at maximum stem length; Ft 1: in fruit during fruit formation (30 to 50% fruit formation), Ft 2: in fruit during fruit formation (70% to final size), Ft 3: in fruit during fruit ripening.
For the subexpression profile, which measures the expression in different organs under the same growth conditions, our results showed that most of the homoeologous copies had different expressions, but the differences were not statistically significant. No preferred expression was observed in the three subgenomes (A, B, or D), while different expressions were observed in the same organ but different developmental stages. Overall, expansin genes were expressed broadly in roots. TaEXPB1 had the highest expression among all the expansin genes. The expression reached its highest level at the late stage of inflorescence (le. 3 in Fig 2) and early fruit formation (ft. 1 in Fig 2). Its expression was also relatively high during early stem elongation stage (sm. 2 in Fig 2). The expressions of TaEXPA3, TaEXPA7, TaEXPA9, TaEXPB7, TaEXPB16, and TaEXPB19 were significantly higher during the early stem elongation stage (sm. 2 in Fig 2), but decreased during the second half of the flower opening stage, suggesting that these genes were involved in stem elongation.
Analysis of wheat expansin cis-acting elements
To further understand the expression and regulation of the 128 wheat expansins identified in the present study, the cis-acting elements, which play important roles in gene expression, were analyzed. Cis-acting elements are regions of non-coding DNA and are located in the promoters region. Seven cis-acting elements were identified (Fig 3 and S2 Table) including ABRE (cis-acting element involved in the abscisic acid response), ERE (ethylene-responsive element), GARE-motif (gibberellin-responsive element), TGA-element (auxin-responsive element), TCA (cis-acting element involved in salicylic acid response), CGTCA/TGACG-motif (cis-acting regulatory element involved in the MeJA-response) and LTR (cis-acting element involved in low-temperature response). The CGTCA/TGACG-motifs were the most abundant cis-acting elements, with an average of 4.9 elements per gene. The CGTCA/TGACG-motifs activate the defense mechanisms in plants in response to environmental stress, such as low temperature or drought. The second most abundant cis-acting element was ABRE, which is involved in the response to abscisic acid. Under severe environmental changes, such as drought, low temperature, or high salt, plants produce abscisic acid, which in turn, activates genes that respond to these stressors via the ABRE cis-acting element. In addition, LTR, a cis-acting element that is responsible for gene expression under low temperature stress, was identified in 48 of the 128 expansin genes examined (S2 Table). These results strongly suggest that some of the expansin genes might be responsible for the low-temperature stress response in wheat.
Fig 3. Heat map of cis-acting elements of expansin genes.
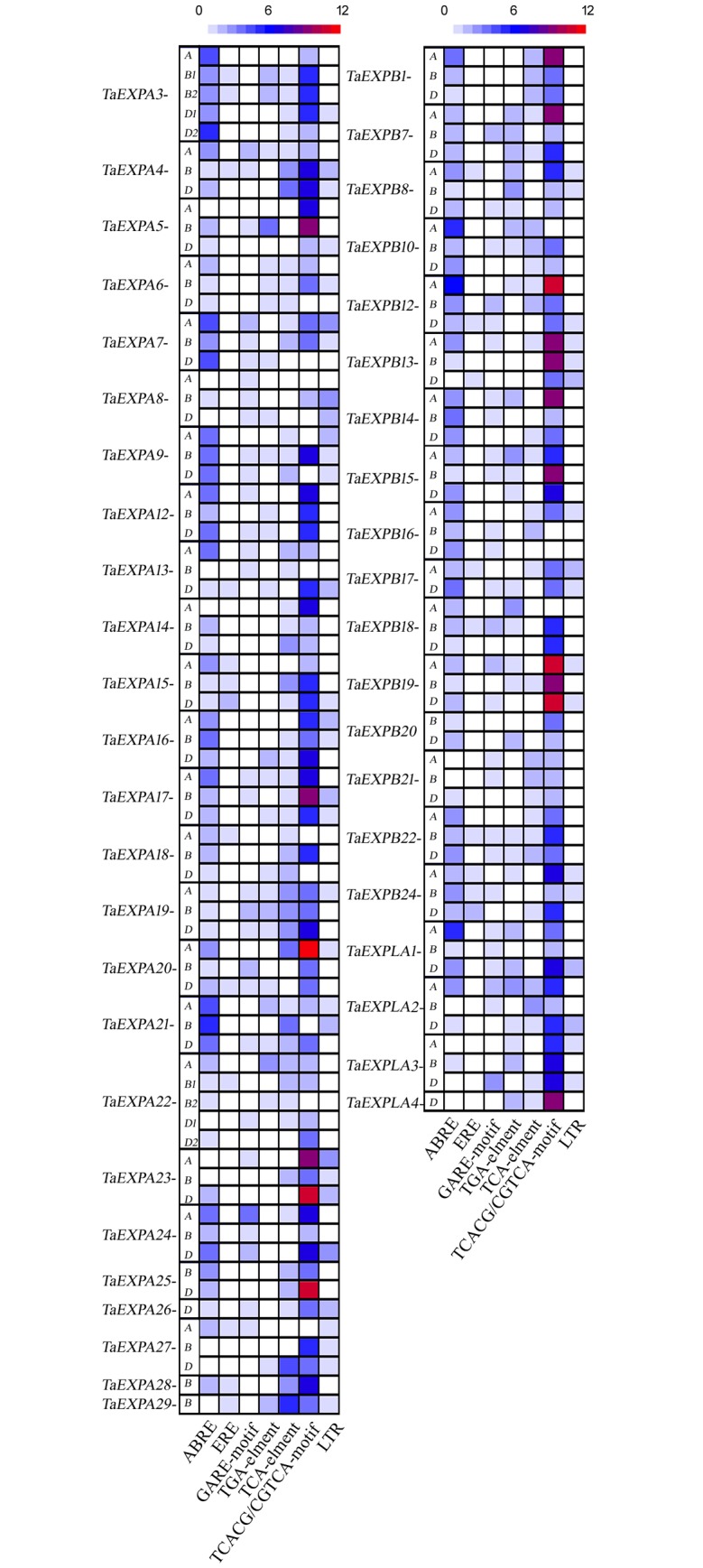
Color indicates the number of cis-acting elements (0–12) in each expansin gene.
Comparison of expansin expressions between cold-tolerant and cold-sensitive wheat cultivars
To investigate if expansins were involved in the low-temperature response of wheat, two wheat (Triticum aestivum) cultivars, the cold-tolerant D1 and the cold-sensitive CS cultivars, were used to compare the expression of expansin under normal and low temperatures at the three-leaf stage.
Phenotypical differences between CS and D1 cultivars
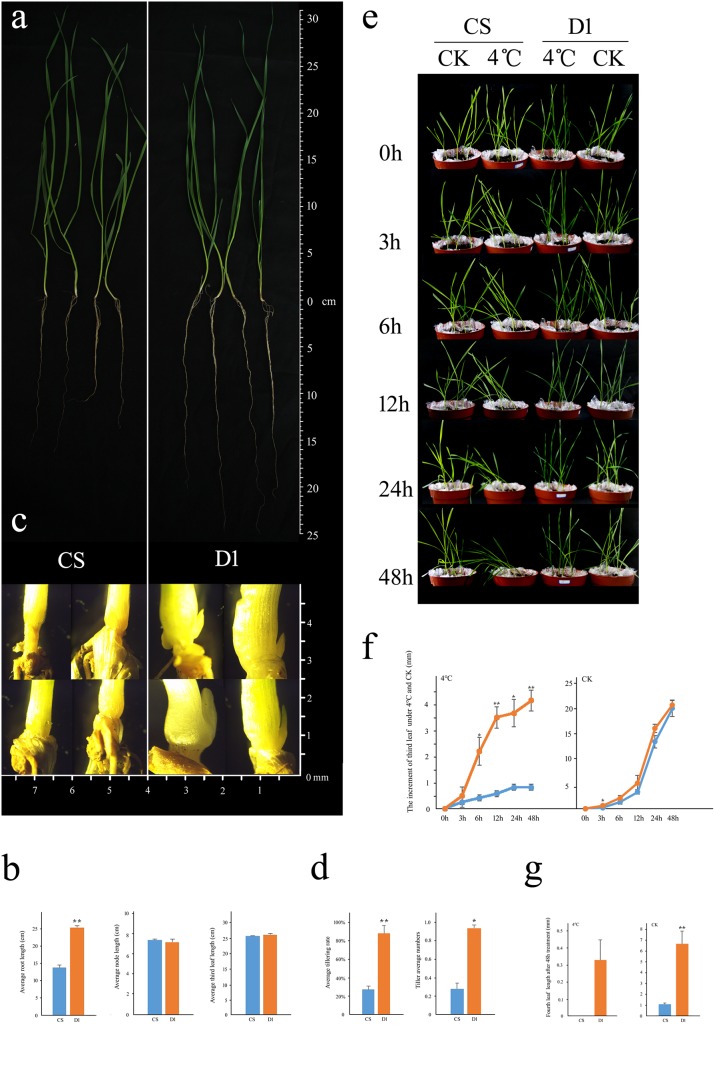
Under normal growth condition (15°C), the aboveground parts of the plants were similar, but the roots of D1 were significantly longer than roots of CS plants (Fig 4a and 4b). The tillering capacity was also much higher in D1 than in CS plants (Fig 4c). Both the tillering rate and number of tillers produced in D1 plants were significantly higher than that of CS plants (Fig 4d). Under low temperature treatment (4°C), the leaves of the CS cultivar gradually collapsed while the control (CS at 15°C) and the D1 leaves grew normally (Fig 4e). The growth of the third leaf was slower for both CS and D1 plants, but the growth of D1 was significantly faster than that of the CS third leaves (P < 0.05; Fig 4f). After 48 h low temperature treatment, the fourth leaf emerged in D1, but was not observed in CS plants (Fig 4g).
Fig 4. Phenotypic analysis of the wheat cultivars Chinese Spring (CS) and Dongnongdongmai 1 (D1).
a. Morphology of CS and D1 plants (including roots); b. Average root length, node length and third leaf length of CS and D1, unit: cm; c. Morphology of tilling nodes of CS and D1 at the same magnification (unit: mm); d. Average tilling rate and tiller numbers of CS and D1. e. Morphology of CS and D1 plants under 4°C treatment; CK: control (growing at 15°C); f. growth of the third leaf under 4°C treatment.
TaEXP expression of CS and D1 cultivars under normal temperature
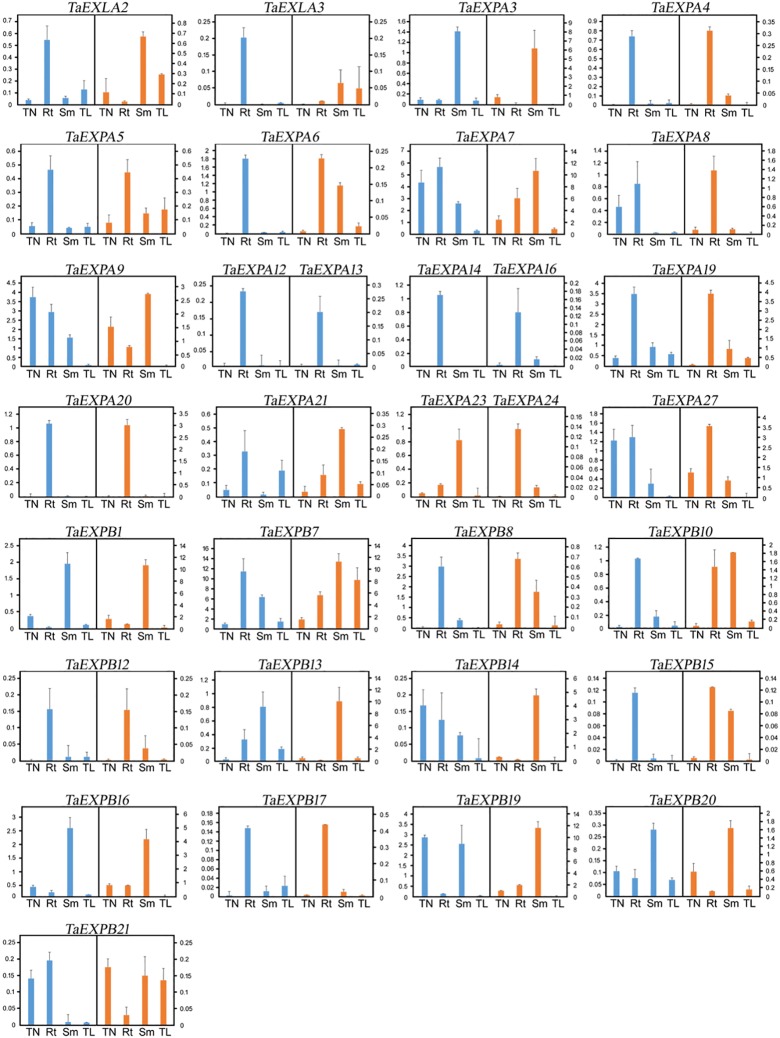
Our preliminary data showed that some of the TaEXP genes had a very low (< 0.1) or undetectable level of expression. After excluding these genes, we selected 32 genes to examine the expression in CS and D1 plants under normal growth conditions to receive a baseline expression profile (Fig 5). Our results showed that some of the TaEXP genes, such as TaEXLA2, TaEXPA3, TaEXPA4, TaEXPA19, TaEXPB1, and TaEXPB12 had similar expression patterns in these two cultivars. Many other TaEXP genes showed either a different expression in a particular tissue (such as TaEXLA 2 and TaEXLA3 in roots), or different expression in all four tested tissues (such as TaEXB21). TaEXPA23 and TaEXPA24 were only expressed in D1, while TaEXP12, TaEXPA13, TaEXP14, and TaEXPA16 were only expressed in CS, suggesting that these genes might be responsible for the phenotypic differences (Fig 4) between these two cultivars.
Fig 5. RT-qPCR of wheat expansin expressions in CS (blue) and D1 (orange) during the three leaf stage under normal growth condition.
Note that certain genes (such as TaEXPA12 and TaEXPA13) were only shown their expression in CS (blue) and certain other genes (such as TaEXPA23 and TaEXPA24) were only shown their expression in D1 (orange). It indicates that the expression of these genes was only detected in cultivar, but not the other.
Differential TaEXP expression between CS and D1 cultivars under low-temperature treatment
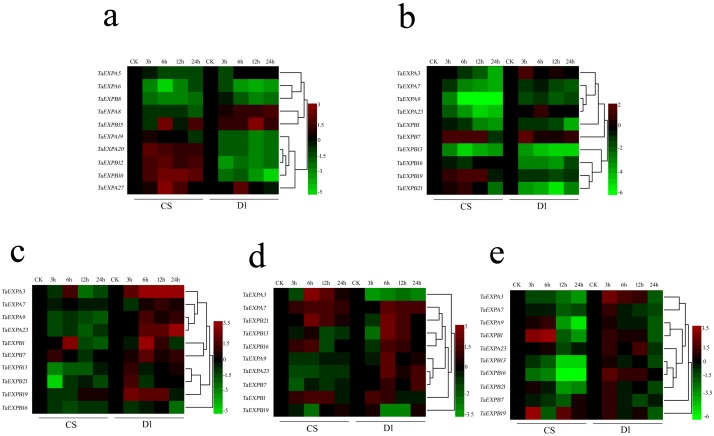
We further selected 10–20 TaEXP genes to compare their expressions under low-temperature (4°C) treatment at different time points (from 0 to 24 h). Our results from each independent experiment had a high consistency. So the following results (heat maps) were shown as the average of three independent experiments. Many of the TaEXP genes showed significant differences in expression between CS and D1, especially after 24 h of low temperature treatment (Fig 6a–6e). In root (Fig 6a and 6b), the expression levels of most of the TaEXP genes decreased as the time at 4°C increased from 3 to 24 h. Many of the TaEXP genes were found differentially expressed. The expressions of TaEXPA20, TaEXPB10, and TaEXPB12 increased in CS but decreased in D1 at 4°C, while the expression of TaEXPA8 decreased in CS but increased in D1. The expressions of TaEXPA5, TaEXPA6, and TaEXPB8 decreased in both CS and D1.
Fig 6. Differential expansin expression in CS and D1 during the three leaf stage at 4°C, compared against the expression at normal growth condition at the same time point.
a–b. roots in CS and D1; c. tillering nodes in CS and D1; d. stem in CS and D1; e. leaves in CS and D1. Results were shown as average of three independent experiments.
In tillering nodes (Fig 6c), overall, the expression of TaEXP decreased in CS, but increased in D1. Considering the phenotype of CS at 4°C, the decrease of TaEXP expression may be related to the collapse of the CS plants. In D1, the expressions of five genes increased during 3–6 h of 4°C treatment. A similar increase of expression of TaEXPA9, TaEXPA23, and TaEXPB7 was also observed in both CS and D1 during the tillering stage, suggesting that these three genes may play important roles in tillering node development. In stem (Fig 6d), the expression of most of genes in CS decreased, while they increased in D1, suggesting that the decrease in CS may be involved in the collapse of the plants under low-temperature stress. Fig 6e shows the TaEXP expression changes in leaves. The expressions of most of the genes decreased in both CS and D1, but the decrease, which occurred mostly during 12–24 h of low-temperature treatment, was more significant in CS than in D1. The slower decrease of TaEXP expression in D1 may facilitate the low-temperature tolerance by giving the plants more time to adjust this type of environment change.
Overall, the differential expression was more prominent between CS and D1 in root-specific TaEXP genes. This could be one of the causes of the significant difference of root development between the two cultivars. The expression differences in leaves and nodes were less significant between CS and D1, suggesting the TaEXP genes may have similar functions in the development of leaves and nodes.
Discussion
Expansins are a large family of proteins that play broad roles in plant development, from the emergence of root hairs to fruit softening. Recent studies have shown documented the presence of 36 expansin genes in Arabidopsis, 75 in soybean, 52 in tobacco, 38 in tomato, 39 in potato, 58 in rice, and 88 in maize [31]. Because of the complexity of the wheat genome, only 18 expansins have been identified so far [28]. In this study, we identified 128 expansin sequences in wheat using a genome-wide approach. These sequences belong to 45 different wheat expansins. Further study of these expansins will significantly enhance our understanding the physiological functions of wheat expansins.
It has been reported that the number of expansins in each of the four subfamilies of expansins differs significantly [31]. In most cases, the number of EXPAs are larger than that of EXPBs. Consistent with this report, we identified 26 TaEXPAs and 15 TaEXPBs. One interesting phenomenon is that the number of TaEXPBs is higher in monocots than in dicots [31]. One possible explanation for this is that EXPAs are more specific to xyloglucans, which are more abundant in the primary walls of non-graminaceous plants [56]. However, EXPBs are more specific to xylan [2], which is a minor component of the primary walls of dicots but abundant in the walls of grasses of the Poaceae [57]. Another difference of expansin genes in monocots and dicots was the proportion of introns extended genes, which was less than 15% in wheat (Fig 1c) and maize (monocotyledon), but more than 75% in dicots [31].
Cis-acting elements are short DNA sequences on the promoter regions. These elements are recognized and bonded by trans-acting factors that control gene expression. Having the same or similar cis-acting elements on different genes often suggests that these genes are being transcribed in a coordinated way. In this study, we found that stress-related cis-acting elements are abundant and common in many EXPs. For example, 10 to 14 TGACG/CGTCA-motifs were found in 17 EXPs, including ten EXPBs, six EXPAs and one EXLA (S2 Table). TGACG/CGTCA-motifs are involved in jasmonic acid methyl ester (MeJA) responses and play important roles in the response of plants to drought and low temperature stress. MeJA, a volatile organic compound, is used in plants for stress response and in many diverse developmental pathways such as root growth or flowering. It could accelerate the expression of EgEXPA2 and EgEXPA3 in petals [58]. Aside from cis-acting elements, gene expression of TaEXPs is also regulated by DNA methylation in plants [59]. A higher level of cytosine methylation was detected in the TaEXPA1-B promoter, making TaEXPA1-B silent and activating TaEXPA1-A and D [27]. However, these three homoeologous TaEXPA1 genes showed a similar phenotype and function in transgenic Arabidopsis [29].
The development of wheat roots is essential to overwintering and regeneration; studies have shown that expansins play important roles in root development. In Brachypodium root, auxin influenced the up-regulated expression of 23 expansins [60]. In barley (Hordeum vulgare), expression of the HvEXPB1 was found to be root specific. A lack of HvEXPB1 expression led to the spontaneous generation of a root-hairless mutant barley [61]. TaEXPB12 shares 97% similarity with HvEXPB1 and may have similar functions. Our current results showed that TaEXPB12 was expressed in root at a low level in wheat. The rice (Oryza sativa) expansin OsEXPA17 was exclusively expressed in root hair cells and mutants of OsEXPA17 had shorter root hairs [8]. Limited AtEXPA7 expression also led to shorter root hair development in Arabidopsis [7]. This suggests that root hair development requires a synergistic action between EXPAs and EXPBs [37]. Moreover, AtEXPA17 and AtEXPA18 were shown to be involved in the formation of primordia in lateral root [62, 63]. In our current study, we showed that the cold-tolerant D1 wheat cultivar had longer and stronger roots, compared to the cold-sensitive CS cultivar.
Studies have shown that the major function of expansins is to change the strength and elasticity of cell walls by modifying the polysaccharide bonds in those walls. The activation of expansins loosens the cell wall, while the inhibition of expansin expression reduces the flexibility of cell walls (Cosgrove 2000a). Expansin is the only type of protein discovered to date that can change the elasticity of cell walls but not damage its viability, suggesting that expansin only breaks the hydrogen bonds of the cell wall polysaccharides and that this process is reversible [2, 3].
The differential expressions of many expansin genes between the CS and D1 cultivars suggest that expansins in D1 could help enhance its low-temperature tolerance by modifying the cell wall elasticity. Further studies will be required to understand which expansin genes are involved in this process.
Wheat and other plants have evolved multiple mechanisms to respond to environmental stress, such drought, high levels of salt in soil, and high/low temperatures. Understanding the molecular mechanisms may help us select stress-tolerant cultivars and improve crop yields. Our real-time qPCR analysis showed that many TaEXPs were differently expressed in the CS and D1 wheat cultivars. These differential expressions may cause differential development of different organs. For example, we found that the expression of TaEXPB10 and TaEXPB12 was much higher in CS than in D1 (Fig 6a). The differential expressions in root at 4°C (Fig 6a) might have partially caused of the collapse of CS under the low-temperature treatment. TaEXPB12 has a 97% similarity to HvEXPB1, which has been shown to be directly related to the formation of root hair in barley [58]. We speculate that the D1 strain may enhance its low-temperature resistance by limiting its root hair development and therefore reducing the contact with soil. One limitation of our current study is the lack of comprehensive comparison of gene expressions of the same variant (CS or D1) between normal growth condition and cold treatment, due to the complexity of expansin expression at different time points under low temperature treatment. As a result, the differences of expansin gene expression between CS and D1 under cold treatment could potentially be contributed in part by their genetic background. Studies are underway in our laboratory to comprehensively compare the expansin gene expression under different growth conditions and in different tissues.
In conclusion, our genome-wide screening of wheat expansins identified more expansin genes than had been previously reported. Their differential expressions in different tissues suggest wheat expansins have complex functions. Our comparative expression study between cold-tolerant and cold-sensitive wheat cultivars revealed that expansins in wheat may be involved in a low-temperature response. Our study sheds new light on the functions of expansins in plant development and stress response.
Supporting information
Genes from IWGSC database were showed in physical map of wheat chromosomes.
(DOC)
DPBB_1 domains and Pollen_allerg_1 domains were contained in TaEXP proteins.
(DOC)
There were 15 motifs in TaEXP genes.
(DOC)
These primers were used in qPCR.
(DOC)
The number of ABRE, ERE, GARE-motif, TGA-element, TCA-element, TGACG/CGTCA-motif and LTR was showed respectively.
(DOC)
Acknowledgments
This study was supported by the National Natural Science Foundation Fostering Talents Project of China (No. J1210069).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation Fostering Talents Project of China (No. J1210069). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000; 407: 321–326. doi: 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- 2.Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005; 6: 242 doi: 10.1186/gb-2005-6-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampedro J, Guttman M, Li LC, Cosgrove DJ. Evolutionary divergence of beta-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits. Plant J. 2015; 81: 108–120. doi: 10.1111/tpj.12715 [DOI] [PubMed] [Google Scholar]
- 4.Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS One. 2014; 9: e85208 doi: 10.1371/journal.pone.0085208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae JM, Kwak MS, Noh SA, Oh MJ, Kim YS, Shin JS. Overexpression of sweetpotato expansin cDNA (IbEXP1) increases seed yield in Arabidopsis. Transgenic Res. 2014; 23: 657–667. doi: 10.1007/s11248-014-9804-1 [DOI] [PubMed] [Google Scholar]
- 6.Guo W, Zhao J, Li X, Qin L, Yan X, Liao H. A soybean beta-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011; 66: 541–552. doi: 10.1111/j.1365-313X.2011.04511.x [DOI] [PubMed] [Google Scholar]
- 7.Lin C, Choi HS, Cho HT. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol Cells. 2011; 31: 393–397. doi: 10.1007/s10059-011-0046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ZhiMing Y, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, et al. Root hair-specific expansins modulate root hair elongation in rice. Plant J. 2011; 66: 725–734. doi: 10.1111/j.1365-313X.2011.04533.x [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Gao Y, Wang J, Yang L, Song R, Li X, et al. Overexpression of two cambium-abundant Chinese fir (Cunninghamia lanceolata) alpha-expansin genes ClEXPA1 and ClEXPA2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol J. 2011; 9: 486–502. doi: 10.1111/j.1467-7652.2010.00569.x [DOI] [PubMed] [Google Scholar]
- 10.Kuluev BR, Kniazev AV, Nikonorov Iu M, Chemeris AV. Role of the expansin genes NtEXPA1 and NtEXPA4 in the regulation of cell extension during tobacco leaf growth. Genetika. 2014; 50: 560–569. [PubMed] [Google Scholar]
- 11.Lu P, Kang M, Jiang X, Dai F, Gao J, Zhang C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta. 2013; 237: 1547–1559. doi: 10.1007/s00425-013-1867-3 [DOI] [PubMed] [Google Scholar]
- 12.Kuluev BR, Kniazev AV, Il’iasova AA, Chemeris AV. Ectopic expression of the PnANTL1 and PnANTL2 black poplar genes in transgenic tobacco plants. Genetika. 2012; 48: 1162–1170. [PubMed] [Google Scholar]
- 13.Cosgrove DJ. New genes and new biological roles for expansins. Curr Opin Plant Biol. 2000; 3: 73–78. [DOI] [PubMed] [Google Scholar]
- 14.Rose JK, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci U S A. 1997; 94: 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell. 1999; 11: 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Xie J, Liao H, Wang X. Overexpression of beta-expansin gene GmEXPB2 improves phosphorus efficiency in soybean. Physiol Plant. 2014; 150: 194–204. doi: 10.1111/ppl.12077 [DOI] [PubMed] [Google Scholar]
- 17.Li F, Xing S, Guo Q, Zhao M, Zhang J, Gao Q, et al. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J Plant Physiol. 2011; 168: 960–966. doi: 10.1016/j.jplph.2010.11.023 [DOI] [PubMed] [Google Scholar]
- 18.Marowa P, Ding A, Kong Y. Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016; 35: 949–965. doi: 10.1007/s00299-016-1948-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Jones L, McQueen-Mason S. Expansins and cell growth. Curr Opin Plant Biol. 2003; 6: 603–610. [DOI] [PubMed] [Google Scholar]
- 20.Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol. 2004; 55: 311–314. doi: 10.1007/s11103-004-0158-6 [DOI] [PubMed] [Google Scholar]
- 21.Carey RE, Hepler NK, Cosgrove DJ. Selaginella moellendorffii has a reduced and highly conserved expansin superfamily with genes more closely related to angiosperms than to bryophytes. BMC Plant Biol. 2013; 13: 4 doi: 10.1186/1471-2229-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Yan H, Chen W, Liu J, Jiang C, Jiang H, et al. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics. 2014; 289: 1061–1074. doi: 10.1007/s00438-014-0867-8 [DOI] [PubMed] [Google Scholar]
- 23.Feldman M, Levy AA. Allopolyploidy—a shaping force in the evolution of wheat genomes. Cytogenetic and Genome Research. 2005; 109: 250–258. doi: 10.1159/000082407 [DOI] [PubMed] [Google Scholar]
- 24.Bottley A, Xia GM, Koebner RM. Homoeologous gene silencing in hexaploid wheat. Plant J. 2006; 47: 897–906. doi: 10.1111/j.1365-313X.2006.02841.x [DOI] [PubMed] [Google Scholar]
- 25.Sharma R, Lodhi S, DeRoode D, Sahota P, Thakkar M. Epigenetic Modifications in the Wake-Promoting Basal Forebrain May Contribute to Insomnia during Ethanol Withdrawal. Alcoholism-Clinical and Experimental Research. 2012; 36: 183a–183a. [Google Scholar]
- 26.Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, et al. The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst. 2007; 82: 167–170. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Han Z, Song N, Chai L, Yao Y, Peng H, et al. Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 2013; 197: 1344–1352. doi: 10.1111/nph.12131 [DOI] [PubMed] [Google Scholar]
- 28.Lin Z, Ni Z, Zhang Y, Yao Y, Wu H, Sun Q. Isolation and characterization of 18 genes encoding alpha- and beta-expansins in wheat (Triticum aestivum L.). Mol Genet Genomics. 2005; 274: 548–556. doi: 10.1007/s00438-005-0029-0 [DOI] [PubMed] [Google Scholar]
- 29.Hu Z, Song N, Xing J, Chen Y, Han Z, Yao Y, et al. Overexpression of three TaEXPA1 homoeologous genes with distinct expression divergence in hexaploid wheat exhibit functional retention in Arabidopsis. PLoS One. 2013; 8: e63667 doi: 10.1371/journal.pone.0063667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001; 4: 527–532. [DOI] [PubMed] [Google Scholar]
- 31.Ding A, Marowa P, Kong Y. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol Genet Genomics. 2016; 291: 1891–1907. doi: 10.1007/s00438-016-1226-8 [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Wu N, Song W, Yin G, Qin Y, Yan Y, et al. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014; 14: 93 doi: 10.1186/1471-2229-14-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Liu L, Wang X, Han Z, Ouyang B, Zhang J, et al. Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol Genet Genomics. 2016; 291: 597–608. doi: 10.1007/s00438-015-1133-4 [DOI] [PubMed] [Google Scholar]
- 34.Dal Santo S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, et al. Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS One. 2013; 8: e62206 doi: 10.1371/journal.pone.0062206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tello-Ruiz MK, Stein J, Wei S, Youens-Clark K, Jaiswal P, Ware D. Gramene: A Resource for Comparative Analysis of Plants Genomes and Pathways. Methods Mol Biol. 2016; 1374: 141–163. doi: 10.1007/978-1-4939-3167-5_7 [DOI] [PubMed] [Google Scholar]
- 36.Ling HQ, Zhao SC, Liu DC, Wang JY, Sun H, Zhang C, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013; 496: 87–90. doi: 10.1038/nature11997 [DOI] [PubMed] [Google Scholar]
- 37.Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015; 25: 162–172. doi: 10.1016/j.pbi.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheuk A, Houde M. Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol Genet Genomics. 2016; 291: 873–890. doi: 10.1007/s00438-015-1152-1 [DOI] [PubMed] [Google Scholar]
- 39.Schreiber AW, Hayden MJ, Forrest KL, Kong SL, Langridge P, Baumann U. Transcriptome-scale homoeolog-specific transcript assemblies of bread wheat. BMC Genomics. 2012; 13: 492 doi: 10.1186/1471-2164-13-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23: 2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37: W202–208. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoff KJ, Stanke M. WebAUGUSTUS—a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013; 41: W123–128. doi: 10.1093/nar/gkt418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015; 31: 1296–1297. doi: 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999; 27: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002; 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pingault L, Choulet F, Alberti A, Glover N, Wincker P, Feuillet C, et al. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015; 16: 29 doi: 10.1186/s13059-015-0601-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014; 9: e111988 doi: 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, Zhang L, Cang J, Hao ZB, Yang Y, Li ZF. Comparison of low temperature- induced proteins in tillering node of winter wheat cultivars with different cold resistance. Ying Yong Sheng Tai Xue Bao. 2009; 20: 1092–1098. [PubMed] [Google Scholar]
- 50.Fredslund J, Lange M. Primique: automatic design of specific PCR primers for each sequence in a family. BMC Bioinformatics. 2007; 8: 369 doi: 10.1186/1471-2105-8-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 52.Breen J, Li D, Dunn DS, Bekes F, Kong X, Zhang J, et al. Wheat beta-expansin (EXPB11) genes: Identification of the expressed gene on chromosome 3BS carrying a pollen allergen domain. BMC Plant Biol. 2010; 10: 99 doi: 10.1186/1471-2229-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Q, Zhao M, Li F, Guo Q, Xing S, Wang W. Expansins and coleoptile elongation in wheat. Protoplasma. 2008; 233: 73–81. doi: 10.1007/s00709-008-0303-1 [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Liu D, Zhang H, Gao H, Guo X, Wang D, et al. The alpha- and beta-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: molecular cloning, gene expression, and EST data mining. Genomics. 2007; 90: 516–529. doi: 10.1016/j.ygeno.2007.06.012 [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, et al. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002; 128: 854–864. doi: 10.1104/pp.010658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Neill Y. The composition and structure of plant primary walls. Plant Cell Wall. 2003: 1–54. [Google Scholar]
- 57.Ebringerová A, Heinze T. Xylan and xylan derivatives—biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun. 2015; 21: 542–556. [Google Scholar]
- 58.Igarashi M, Hippo Y, Ochiai M, Fukuda H, Nakagama H. AKT is critically involved in cooperation between obesity and the dietary carcinogen amino-1-methyl-6-phenylimidazo [4,5-b] (PhIP) toward colon carcinogenesis in rats. Biochem Biophys Res Commun. 2014; 443: 852–857. doi: 10.1016/j.bbrc.2013.12.059 [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006; 126: 1189–1201. doi: 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 60.Pacheco-Villalobos D, Diaz-Moreno SM, van der Schuren A, Tamaki T, Kang YH, Gujas B, et al. The Effects of High Steady State Auxin Levels on Root Cell Elongation in Brachypodium. Plant Cell. 2016; 28: 1009–1024. doi: 10.1105/tpc.15.01057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwasniewski M, Szarejko I. Molecular cloning and characterization of beta-expansin gene related to root hair formation in barley. Plant Physiol. 2006; 141: 1149–1158. doi: 10.1104/pp.106.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee HW, Kim J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013; 54: 1600–1611. doi: 10.1093/pcp/pct105 [DOI] [PubMed] [Google Scholar]
- 63.Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013; 73: 212–224. doi: 10.1111/tpj.12013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes from IWGSC database were showed in physical map of wheat chromosomes.
(DOC)
DPBB_1 domains and Pollen_allerg_1 domains were contained in TaEXP proteins.
(DOC)
There were 15 motifs in TaEXP genes.
(DOC)
These primers were used in qPCR.
(DOC)
The number of ABRE, ERE, GARE-motif, TGA-element, TCA-element, TGACG/CGTCA-motif and LTR was showed respectively.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.