Abstract
Microbiome science is revealing that the phenotype and health of animals, including humans, depend on the sustained function of their resident microorganisms. In this essay, I argue for thoughtful choice of model systems for human microbiome science. A greater variety of experimental systems, including wider use of invertebrate models, would benefit biomedical research, while systems ill-suited to experimental and genetic manipulation can be used to address very limited sets of scientific questions. Microbiome science benefits from the coordinated use of multiple systems, which is facilitated by networks of researchers with expertise in different experimental systems.
One of the great biological success stories over the last decade is microbiome science, including a growing understanding of the impact of resident microorganisms on human health. We now have irrefutable evidence that microorganisms are key players that shape many functions of humans and other animals, from metabolism to immunity and behavior [1]. Furthermore, our growing understanding of and capacity to manipulate the resident microorganisms are already translating into real-world benefits. For example, a microbial therapy reliably eliminates life-threatening Clostridium difficile infections in people [2,3], and there are realistic prospects to suppress mosquito transmission of dengue virus by the large-scale release of mosquitoes modified to bear a bacterium Wolbachia that confers vector incompetence [4,5].
Underlying the recent successes and excellent prospects for the discipline of microbiome science, there are, however, differences of opinion, especially concerning the utility of different experimental systems to advance our understanding and application of microbiomes for human health. The appropriate choice of study system is vitally important for everyone from individual researchers and their institutions to national and international funding organizations. The choices facing biomedical applications of microbiome science have many parallels with other disciplines in the life sciences as advancing technologies in molecular biology, cell biology, and microbiology enable us to answer new questions and solve previously intractable problems.
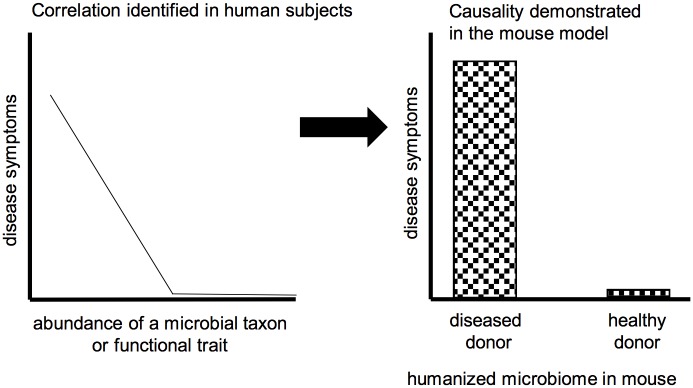
Two linked factors play an important role in the appropriate choice of experimental systems for human microbiome research: history and purpose. Let us start with history. For many, the discipline of microbiome science burst onto the scientific scene some 10 to 15 years ago, made possible by breakthroughs in sequencing technologies [6] that enabled us to identify and study the function of microorganisms in situ, i.e., in their natural communities without isolation into clonal cultures [7,8]. For the first time, it became practicable to study the complex communities associated with humans and other animals. The power of this new science was swiftly recognized by the biomedical community, harnessing microbiome science as a route for improved understanding and treatment of human disease, especially chronic diseases associated with dysfunction of metabolism and immunity, e.g., obesity, inflammatory bowel disease [9–12]. This potent combination of history and purpose identified the human as the ideal biological system for study. For ethical reasons, human studies generally yield correlations between microbial traits (composition and function) and phenotype (including disease state), requiring experimentation on other systems to determine causality and mechanism (Fig 1). To a very large extent, a single traditional model, the laboratory mouse, has fulfilled the important role as experimental model of host–microbe interactions in humans [13].
Fig 1. The laboratory mouse model is widely used to demonstrate causality of correlations between the microbiome and human disease.
(A) Analysis of many humans reveals a negative association between the severity of disease symptoms and an index of the microbiome, e.g., abundance of a specific taxon or functional trait. (B) “Humanized” mice (i.e., mice colonized with microbiome samples from humans) are used to infer a causal role of the microbiome in the human disease: mice display disease symptoms when colonized with microbiome samples from diseased, but not healthy, humans. This figure is a generalized illustration and is not intended to be representative of any specific disease or microbiome dataset.
So where is the problem? It is that many biomedical researchers and their funders are neglecting an important strand of our history and, consequently, may be selling themselves short. The “new” discipline of microbiome science did not enter an empty playing field. Instead, and to its great benefit, microbiome science builds on more than a century of research on microorganisms in healthy animals: the discipline of symbiosis [14]. Eukaryotic organisms—from the first unicellular eukaryotes to complex, multicellular groups (including animals)—have repeatedly entered into alliances with microorganisms, enabling them to exploit otherwise unavailable habitats and, for animals, unsuitable diets [15]. As befits a mature discipline, the goals in symbiosis research are diverse. Some questions are driven by ecological and evolutionary considerations. How do the presence and composition of its microbiota influence the ecological fit and the fitness of an individual host and—at larger scales—the structure of ecological communities and evolutionary trajectory of host lineages [16–20]? Other questions are mechanistic: what is the molecular basis of microbial persistence in their host, including the patterns of host–microbe signal and nutrient exchange, and how are these often coevolved traits influenced by the mode of transmission and evolutionary age of the association [21–23]? Over many decades, the associations used to address these questions have been selected for their microbiological simplicity, ideally a single microbial taxon that is restricted to a specific host organ and is morphologically conspicuous (e.g., the pigmented algal symbionts in corals, the luminescent bacteria in squid). The microbiology of the traditional animal models does not meet these criteria, and symbiosis researchers have been adept at identifying and developing alternative systems to answer their questions. This diversity of system and purpose has led to the opposite mindset from the biomedical microbiome scientist. For the symbiosis researcher, there is an inherent value in diversity of systems. Not only are different biological systems suitable for different questions, but multiple systems used to address the same question are predicted to yield a richer understanding of any one topic.
For the sake of clarity and brevity, the preceding paragraphs portray the assumptions and expectations in our discipline as a simple dichotomy between the “mature” field of symbiosis research and the “new” field of biomedical microbiome science. Although this is an accurate reflection of the view of some colleagues, I believe that the perspective of many practitioners in the discipline is more nuanced. There is a growing sense that the application of microbiome science for human health would be better served by the use of a greater range of experimental models by the biomedical community and, equally, that fundamental discovery will be facilitated by a greater focus on systems that are experimentally tractable and amenable to the latest molecular techniques. How can we achieve these beneficial outcomes?
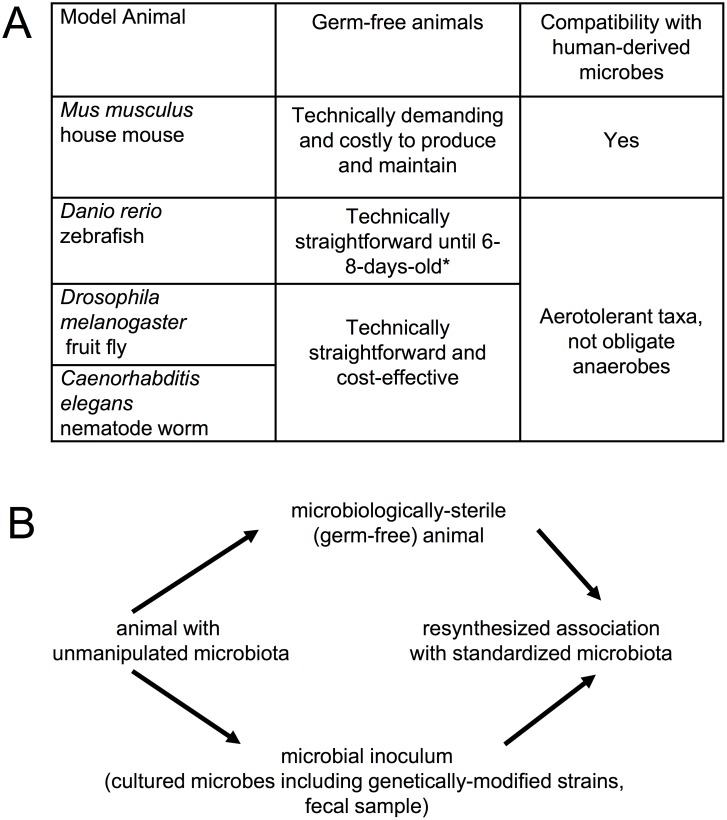
A first-order question for the future of microbiome science is the utility of the traditional animal models that have been the basis for fundamental biological discoveries over many years (Fig 2A). These are the animal systems that are maintained indefinitely under laboratory conditions; are amenable to genetic manipulation; have genetically defined wild-type strains, enabling the phenotype of mutations to be investigated against a common genetic background; and have attracted the critical mass of investigators needed to develop common tools and structures for sharing resources and data, with dissemination via stock centers, databases, etc. These models are also tractable to the key procedures in microbiome science—to generate microbiologically sterile (“germ-free”) animals that can be recolonized with a standardized microbiota (Fig 2B) [24–29]—and their value is being enhanced further by advances in experimental manipulation of the microbial partners. In particular, protocols for enumeration, cultivation, and genetic manipulation of key members of the microbiome are either in place or the subject of intensive method development [30–33]. Nevertheless, some of these traditional models are not currently being used to their full capacity in microbiome science.
Fig 2. The traditional animal models are amenable to microbiome research.
(A) The advantages and limitations of traditional models for microbiome research. *Zebrafish older than 6–8 days require feeding, which is technically demanding under strictly sterile conditions [27]. (B) The key microbiological manipulations required for experimental investigation of microbiome function.
Many microbiome scientists focused on human health make extensive use of the experimental protocol illustrated in Fig 1 to identify a causal role of the microbiome in human disease. This approach requires that the animal model be colonized by human-derived microbes and respond to them in the same way as humans. To a certain extent, the laboratory mouse meets these expectations. Nevertheless, great caution is needed to extrapolate mouse data back to the human. There are major anatomical differences between the gastrointestinal (GI) tract of the mouse and human, and the laboratory mouse is highly inbred and maintained under pathogen-free conditions that bear no resemblance to the human condition. The use of mice bearing human-derived microbes is particularly problematic because an appreciable proportion of human-associated gut microorganisms fail to colonize the mouse gut, and those microbial taxa that do colonize fail to induce some responses elicited by the native mouse microbiota [34–36]. Two responses to these limitations of the mouse model are appropriate. The first is to investigate microbial interactions with human cells grown in vitro as sheets of gut epithelium or as GI-tract organoids, and we are witnessing rapid progress with these models [13,37,38]. The second response builds on insights from symbiosis research, specifically that the interactions between the human and its microbiota are the product of a long evolutionary history of interactions between animals and benign or beneficial microorganisms. The eukaryotic forbears of animals lived in a world that had been inhabited by bacteria for billions of years, with the expectation that—as for modern unicellular eukaryotes and basal animals—the ancestral animals were colonized by a diversity of microorganisms, with which they interacted [39]. Consequently, we should expect (and indeed, are finding) conserved genetic and cellular mechanisms underlying interactions between various animals and their resident microbiota [1]. The implication is that fundamental discoveries in microbiome science can be made using the most tractable animal systems, including nonmammal vertebrates such as the zebrafish and invertebrates such as Drosophila and Caenorhabditis elegans (Fig 2A). The human may, in a perfect world, be the best model for the human—but arguably, we will obtain a better understanding of the human–microbiome interactions faster and more cost-effectively by more extensive integration of “simple” animal models into the collective endeavor.
Other practitioners in the discipline are wary of the traditional animal models because of the perception that the microbiota of these models is unnatural as a result of laboratory cultivation. There is some truth in these concerns. The gut microbiota of the laboratory mouse is different from wild conspecifics [40], taxonomically indistinguishable bacteria in wild and laboratory Drosophila differ in functional gene content [41], and the oft-repeated claim that C. elegans has no microbiota arises from a standard laboratory practice that maintains this species in a microbiologically sterile condition apart from a single Escherichia coli strain used as food [25]. These constraints can, however, be overcome by thoughtful experimental design. We are already witnessing evidence-based discussion about the wisdom of maintaining laboratory mouse cultures under intensely hygienic conditions [42,43], and research on C. elegans reunited with its natural microbiome is yielding new insights into the immune function of this model [25].
What about the systems that are being used to study animal–microbe interactions but lack the in-depth infrastructure available to the traditional models? A number of systems used by relatively small communities of researchers have yielded major insights of general significance. I cite a few among many examples here: hydra for the role of the microbiota in orchestrating peristalsis of the gut epithelium [44], squid for integration of microbial function into the circadian rhythm of the host [45], the aphid for coevolved metabolite exchange [46], and the killifish for microbial determinants of lifespan [47]. For these systems, the future is bright because of the progressive democratization of technologies. Tools and resources that once demanded an army of researchers with funds to match can now be developed by small but well-integrated teams. For your favorite organism, you can sequence its genome or modify its genes and their expression by a growing list of genetic technologies, including RNA interference (RNAi), genome editing, and the use (or set up) of publicly available online databases to share information and resources. An early-career researcher entering the discipline is well-advised to consider whether their preferred system has, or has good prospects of, these resources for the host or microbial partners.
My conclusion is somewhat akin to Goldilocks’ porridge: neither too few nor too many. Let us not invest our resources in a narrow set of systems, especially when other systems (often less familiar to the biomedical community) would yield results faster and more economically. And let us be cautious of systems that are ill-suited to experimental and genetic manipulation. The middle ground does, however, create a substantial constraint for the individual research group. The use of any biological system has large front-end costs, including the financial cost of setting up facilities and the opportunity cost of developing the expertise. These costs reduce our flexibility. It is difficult for researchers to use multiple systems in one study, for example, to first investigate a problem in a “simple” system and then to test for its relevance to mammals (but see [48]); and it is difficult for a single researcher to switch between different systems as their research questions change over time. I believe that it is important for the future of our discipline to mitigate this constraint. One powerful solution is already starting to happen: networks of researchers who can share access to multiple different systems (traditional models, nontraditional models, in vitro systems, and human research) and the associated expertise. These networks can be (partly or entirely) colocated as centers at one (or several) institution(s). Combined with a greater awareness of the opportunities available from different biological systems, these institutional innovations will make it easier for us all to choose the best biological model, and to use it well.
Acknowledgments
I am indebted to many colleagues, both established scientists and especially graduate students and other early career scientists, for all those countless conversations that inspired me to write this article and informed its content.
Abbreviations
- GI
gastrointestinal
- RNAi
RNA interference
Funding Statement
NIH (grant number R01GM095372). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Provenance: Commissioned; externally peer reviewed.
References
- 1.Douglas AE. Fundamentals of Microbiome Science. Princeton, NJ: Princeton University Press; 2018. [Google Scholar]
- 2.Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann Intern Med. 2016;165(9):609–16. Epub 2016/11/01. doi: 10.7326/M16-0271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. New Eng J Med. 2013;368(5):407–15. Epub 2013/01/18. doi: 10.1056/NEJMoa1205037 . [DOI] [PubMed] [Google Scholar]
- 4.Jiggins FM. The spread of Wolbachia through mosquito populations. PLoS Biol. 2017;15(6):e2002780 doi: 10.1371/journal.pbio.2002780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt TL, Barton NH, Rasic G, Turley AP, Montgomery BL, Iturbe-Ormaetxe I, et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 2017;15(5):e2001894 doi: 10.1371/journal.pbio.2001894 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–51. Epub 2016/05/18. doi: 10.1038/nrg.2016.49 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallick H, Ma S, Franzosa EA, Vatanen T, Morgan XC, Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18(1):228 Epub 2017/12/01. doi: 10.1186/s13059-017-1359-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balint M, Bahram M, Eren AM, Faust K, Fuhrman JA, Lindahl B, et al. Millions of reads, thousands of taxa: microbial community structure and associations analyzed via marker genes. FEMS Microbiol Rev. 2016;40(5):686–700. Epub 2016/07/01. doi: 10.1093/femsre/fuw017 . [DOI] [PubMed] [Google Scholar]
- 9.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. Epub 2011/04/22. doi: 10.1038/nature09944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. Epub 2010/03/06. doi: 10.1038/nature08821 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. Epub 2012/06/16. doi: 10.1038/nature11234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486(7402):215–21. Epub 2012/06/16. doi: 10.1038/nature11209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1(1):14 Epub 2014/01/24. doi: 10.1186/2049-2618-1-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapp J. Evolution by Association: A History of Symbiosis. Oxford, UK: Oxford University Press; 1994. [Google Scholar]
- 15.Douglas AE. The Symbiotic Habit. Princeton, NJ: Princeton University Press; 2010. [Google Scholar]
- 16.Joy JB. Symbiosis catalyses niche expansion and diversification. Proc Biol Sci. 2013;280(1756):20122820 Epub 2013/02/08. doi: 10.1098/rspb.2012.2820 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner GD, Cornwell WK, Sprent JI, Kattge J, Kiers ET. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun. 2014;5:4087 Epub 2014/06/11. doi: 10.1038/ncomms5087 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redman RS, Sheehan KB, Stout RG, Rodriguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298(5598):1581 Epub 2002/11/26. . [DOI] [PubMed] [Google Scholar]
- 19.Lowe CD, Minter EJ, Cameron DD, Brockhurst MA. Shining a light on exploitative host control in a photosynthetic endosymbiosis. Curr Biol. 2016;26(2):207–11. Epub 2016/01/11. doi: 10.1016/j.cub.2015.11.052 . [DOI] [PubMed] [Google Scholar]
- 20.Sudakaran S, Retz F, Kikuchi Y, Kost C, Kaltenpoth M. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 2015;9(12):2587–604. Epub 2015/05/30. doi: 10.1038/ismej.2015.75 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas AE. The molecular basis of bacterial-insect symbiosis. J Mol Biol. 2014;426(23):3830–7. Epub 2014/04/17. doi: 10.1016/j.jmb.2014.04.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFall-Ngai M. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 2014;12(2):e1001783 doi: 10.1371/journal.pbio.1001783 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci U S A. 2015;112(33):10169–76. Epub 2015/02/26. doi: 10.1073/pnas.1421388112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27(7):701–18. Epub 2013/04/18. doi: 10.1101/gad.212522.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Berg M, Dierking K, Felix MA, Shapira M, Samuel BS, et al. Caenorhabditis elegans as a model for microbiome research. Front Microbiol. 2017;8:485 Epub 2017/04/08. doi: 10.3389/fmicb.2017.00485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trinder M, Daisley BA, Dube JS, Reid G. Drosophila melanogaster as a high-throughput model for host-microbiota interactions. Front Microbiol. 2017;8:751 Epub 2017/05/16. doi: 10.3389/fmicb.2017.00751 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melancon E, De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 2017;138:61–100. Epub 2017/01/29. doi: 10.1016/bs.mcb.2016.11.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015;8(3):476–86. Epub 2014/12/11. doi: 10.1038/mi.2014.113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. SeminarsImmunol. 2007;19(2):59–69. Epub 2006/11/23. doi: 10.1016/j.smim.2006.10.002 . [DOI] [PubMed] [Google Scholar]
- 30.Hill JH, Franzosa EA, Huttenhower C, Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife. 2016;5 Epub 2016/12/14. doi: 10.7554/eLife.20145 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inamine H, Ellner SP, Newell PD, Luo Y, Buchon N, Douglas AE. Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. mBio. 2018;9(1). Epub 2018/01/11. doi: 10.1128/mBio.01453-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, et al. The mouse intestinal bacterial collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1(10):16131 Epub 2016/09/28. doi: 10.1038/nmicrobiol.2016.131 . [DOI] [PubMed] [Google Scholar]
- 33.Lim B, Zimmermann M, Barry NA, Goodman AL. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell. 2017;169(3):547–58 e15 Epub 2017/04/22. doi: 10.1016/j.cell.2017.03.045 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arrieta MC, Walter J, Finlay BB. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19(5):575–8. Epub 2016/05/14. doi: 10.1016/j.chom.2016.04.014 . [DOI] [PubMed] [Google Scholar]
- 35.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. Epub 2015/01/07. doi: 10.1242/dmm.017400 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–93. Epub 2012/06/26. doi: 10.1016/j.cell.2012.04.037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill DR, Spence JR. Gastrointestinal organoids: understanding the molecular basis of the host-microbe interface. Cell Mol Gastroenterol Hepatol. 2017;3(2):138–49. Epub 2017/03/10. doi: 10.1016/j.jcmgh.2016.11.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535 Epub 2016/05/12. doi: 10.1038/ncomms11535 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110(9):3229–36. Epub 2013/02/09. doi: 10.1073/pnas.1218525110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171(5):1015–28 e13 Epub 2017/10/24. doi: 10.1016/j.cell.2017.09.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winans NJ, Walter A, Chouaia B, Chaston JM, Douglas AE, Newell PD. A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol Ecol. 2017;26(17):4536–50. Epub 2017/07/02. doi: 10.1111/mec.14232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532(7600):512–6. Epub 2016/04/21. doi: 10.1038/nature17655 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham DM. A walk on the wild side. Lab Anim (NY). 2017;46(11):423–7. Epub 2017/10/28. doi: 10.1038/laban.1372 . [DOI] [PubMed] [Google Scholar]
- 44.Murillo-Rincon AP, Klimovich A, Pemoller E, Taubenheim J, Mortzfeld B, Augustin R, et al. Spontaneous body contractions are modulated by the microbiome of Hydra. Sci Rep. 2017;7(1):15937 Epub 2017/11/23. doi: 10.1038/s41598-017-16191-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio. 2013;4(2). Epub 2013/04/04. doi: 10.1128/mBio.00167-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79(19):6117–23. Epub 2013/07/31. doi: 10.1128/AEM.01543-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife. 2017;6 Epub 2017/08/23. doi: 10.7554/eLife.27014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32(23):3017–28. Epub 2013/10/22. doi: 10.1038/emboj.2013.224 . [DOI] [PMC free article] [PubMed] [Google Scholar]