Abstract
Epigenetic inheritance of resistance to exogenous nucleic acids via small interfering (si) RNA is well established in animal models. Rechavi et al. demonstrate epigenetic inheritance of a starvation-induced pattern of gene silencing caused by endogenous siRNAs and resulting in an increased longevity in the third generation progeny. Combined with recent findings in prokaryotes, these results suggest that Lamarckian-type inheritance of acquired traits is a major evolutionary phenomenon.
Various forms of epigenetic inheritance are now firmly established as an important complement to the vertical transmission of genetic information in diverse organisms including plant and animal models (Heard and Martienssen, 2014). In particular, double-stranded (ds)RNA-mediated gene silencing in the nematode Caenorhabditis elegans can be maintained over dozens of generations via transgenerational inheritance of distinct pools of small interfering (si) RNA (Grishok, 2013; Rechavi, 2014). A similar mechanism provides heritable immunity to viruses and transposons (Grishok, 2013; Rechavi, 2014). So far, however, all well characterized epigenetic mechanisms based on vertical transmission of siRNAs involved the response either to artificially introduced RNA or to mobile genetic elements. So is epigenetic inheritance limited to defense against exogenous nucleic acids or is it a general adaptive phenomenon? In this issue of Cell, the latter possibility is strongly suggested by the work of Rechavi et al. (2014) who showed that the changes in the levels of endogenous siRNAs that are involved in starvation-induced developmental arrest in C. elegans are inherited at least across several generations. The affected siRNAs primarily target genes involved in nutrition, affirming the biological relevance of the discovered phenomenon. Furthermore, the heritability of the siRNA-based starvation response depends on the dsRNA-binding protein RDE4 and the germline-expressed argonaute protein HRDE1, indicating that a specific RNAi pathway is involved.
The transgenerational transmission of the siRNA-based starvation response clearly comes across as inheritance of an acquired trait, or Lamarckian evolution. The essential features of the Lamarckian mechanism of evolution can be formulated as follows: (1) an environmental factor directly causes heritable changes, (2) the induced changes are targeted to a limited set of cellular components that are functionally relevant for the challenge, (3) the changes provide specific adaptation to the original challenge (Koonin and Wolf, 2009). The transgenerational inheritance of the starvation-induced endogenous siRNA, as well as that of antivirus siRNAs, perfectly meets all these criteria, with one key qualification: the inheritance is epigenetic and hence (almost) by definition short-lived.
The work of Rechavi et al. (2014) suggests the dramatic possibility that, at least in multicellular eukaryotes, epigenetic adaptation is a general strategy to cope with all types of environmental challenges. Moreover, similar to their eukaryotic homologs, prokaryotic argonaute proteins contribute to the defense against viruses and plasmids by surveilling the bacterial transcriptome with endogenous siRNAs, allowing for the recognition and inactivation of foreign DNA via exogenous siRNAs (Olovnikov et al., 2013) or small guide DNAs (Swarts et al., 2014). These discoveries raise the intriguing possibility that adaptive epigenetic inheritance is a fundamental property of all cellular life.
The heritability of the epigenetic adaptations defies the common belief that “evolution has no forecast.” Indeed, the transgenerational persistence of epigenetic changes would be selected precisely because lineages capable of predicting that stress conditions typically last longer than a single generation span and maintaining the adaptation across generations would have advantage over clueless rivals. Still, the limitation remains that, at least at face value, this highly efficient Lamarckian adaptation strategy does not lead to long-term evolutionary change.
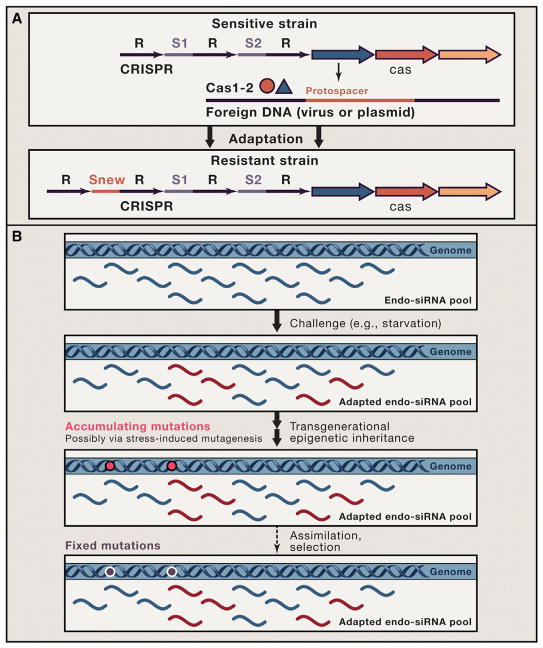
So can Lamarckian inheritance have a long-lasting effect in evolution? The possibilities lie in two areas (Figure 1). The first one is the direct genetic Lamarckian inheritance that is most spectacularly manifest in the prokaryotic system of antivirus defense known as CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated proteins) (Koonin and Wolf, 2009; Makarova et al., 2011). This prokaryotic adaptive immunity mechanism functions by inserting pieces of foreign DNA into the CRISPR cassettes and using the transcripts of these unique spacers to attack and inactivate the cognate virus or plasmid (Figure 1A). The genome modification mediated by CRISPR-Cas is short lived on the evolutionary scale but still can persist for many thousands of generations in contrast to the epigenetic inheritance that lasts for tens of generations at best. Moreover, if a given agent is a permanent challenge to a microbe, the spacers can be replenished such that the acquired resistance persists indefinitely. Several other forms of genomic changes such as some routes of horizontal gene transfer in prokaryotes that leads, among many other adaptations, to antibiotic resistance or the utilization, by the animal germline cells, of transposon sequences to produce defensive piRNAs, can be considered at least “quasi-Lamarckian” (Koonin and Wolf, 2009).
Figure 1. The Epigenetic versus Genetic Routes of Lamarckian Evolution.
(A) The genetic mechanism. This route of Lamarckian evolution is illustrated using the case of the CRISPR-cas system. R, repeat; S, spacer (denoted protospacer in foreign DNA). The insertion of a new spacer is accompanied by repeat duplication. Cas1-Cas2, complex of two Cas protein that is involved in the excision of the protospacer from theforeign DNA and itsinsertion between repeats at the end of the CRISPR cassette. (B) The epigenetic mechanism. Assimilation is denoted by a broken arrow to emphasize that this is not a well-characterized process.
The second possible route of long-term Lamarckian evolution is linked to the epigenetics and involves the phenomenon known as assimilation whereby an epigenetically inherited trait becomes genetically inherited through the fixation of mutations with the same phenotypic effect as the respective epigenetic adaptation (Figure 1B) (Koonin, 2012). A direct experimental demonstration of assimilation has been reported for the prion-mediated epigenetic effects in yeast (Halfmann et al., 2012). How common is assimilation is presently not clear at all, but the mechanism appears realistic and certainly worthy of investigation, especially considering that epigenetic adaptation on many occasions could be accompanied by stress-induced mutagenesis. Under the assimilation scenario, evolutionary adaptation is a two-phase process, with the first phase being the Lamarckian epigenetics and the second phase Darwinian selection of mutations with the appropriate phenotypic effect (Figure 1). Assimilation can be viewed as an elaborate evolutionary strategy that involves “probing the waters” with the short-lived epigenetic adaptation followed by the long-term genetic inheritance of the same adaptation should the challenge prove to be long-lasting. Clearly, unlike the deterministic epigenetic adaptation, assimilation involves a major stochastic component, but the outcome is the same, namely inheritance of an acquired trait.
One of the notable results in the work of Rechavi et al. (2014) is the demonstration of the increased lifespan in the third generation descendants of the starvation-stressed worms. Thus, traits that are of obvious, immediate interest to humanity can be transmitted across generations via epigenetic adaptation. Lamarck was particularly keen about exercise of organs as a cause of evolution, an idea that has been ridiculed ad nauseam. However, exercise certainly causes epigenetic effects, so could it be that Lamarck was not entirely wrong?
The realization that Lamarckian evolution is reality rather than myth is only dawning on biologists, and the study of its impact and generality is just beginning. Nevertheless, it seems clear that Lamarck is back, and perhaps with a vengeance.
Acknowledgments
E.V.K. is funded through the intramural funds of the US Department of Health and Human Services (to National Library of Medicine).
References
- Grishok A. Adv Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Biol Direct. 2012;7:27. doi: 10.1186/1745-6150-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Mol Cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O. Trends Cell Biol. 2014;24:212–220. doi: 10.1016/j.tcb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Cell. 2014;158(this issue):277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, van der Oost J. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]