Abstract
The ATR kinase is crucial for DNA damage and replication stress responses. Here we describe a surprising role of ATR in mitosis. Acute inhibition or degradation of ATR in mitosis induces whole-chromosome missegregation. The effect of ATR ablation is not due to altered CDK1 activity, DNA damage responses, or unscheduled DNA synthesis, but to loss of an ATR function at centromeres. In mitosis ATR localizes to centromeres through Aurora A-regulated association with CENP-F, allowing ATR to engage RPA-coated centromeric R loops. As ATR is activated at centromeres, it stimulates Aurora B through Chk1, preventing formation of lagging chromosomes. Thus, a mitosis-specific and R loop-driven ATR pathway acts at centromeres to promote faithful chromosome segregation, revealing functions of R loops and ATR in suppressing chromosome instability.
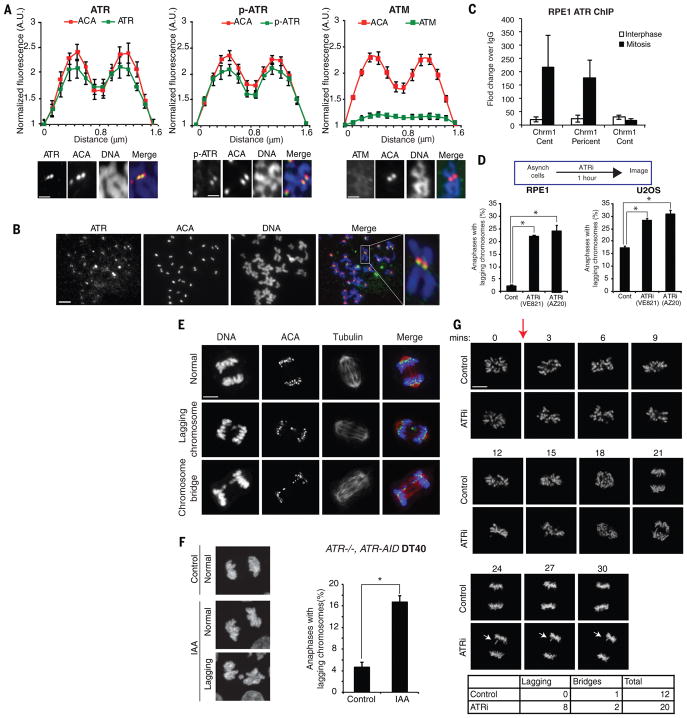
ATR is a master checkpoint kinase safeguarding the genome (1). Loss of ATR during DNA replication increases genomic instability in S phase and subsequent mitosis (2–4). While analyzing the localization of ATR in diploid RPE1 cells across the cell cycle, we surprisingly found that ATR localized to all centromeres detected by ACA (anti-centromere antibody) on mitotic chromosomes (Fig. 1A–B, S1A). Autophosphorylated ATR (p-ATR T1989) also colocalized with ACA (Fig. 1A, S1B), suggesting that ATR is active at centromeres. The staining of ATR and p-ATR was eliminated by ATR siRNA (Fig. S1C–D), confirming the specificity of the antibodies. Colocalization of ATR and CENP-B was detected by the proximity ligation assay in unperturbed mitotic cells, but not in interphase cells (Fig. S2A–B). Chromatin immunoprecipitation (ChIP) of ATR confirmed that ATR was enriched at the centromere of chromosome 1 compared to a non-centromere locus in mitotic RPE1 and U2OS cells (Fig. 1C, S2C). In contrast to ATR, another master checkpoint kinase ATM was not detected at centromeres (Fig. 1A).
Figure 1. ATR localizes to centromeres in mitosis and promotes accurate whole-chromosome segregation.
(A) Line scan analysis (top) and representative images (bottom) of fluorescence of ATR, phospho-ATR S1989 (p-ATR), ATM, and ACA at centromeres in chromosome spreads of RPE1 cells. Cells were arrested with nocodazole for 5 h. Centromeric fluorescence intensities of the indicated proteins are normalized to the background fluorescence near centromeres (see Methods). Scale bar, 2 μm. (B) Chromosome spreads of unsynchronized mitotic RPE1 cells were stained for ATR and ACA. Asynchronously growing cells were treated with nocodazole for 15 min to facilitate spreading of chromosomes. Scale bar, 10 μm. (C) Quantitative PCR of ATR ChIP in mitotic and interphase RPE1 cells. (D) Percentage of anaphase RPE1 and U2OS cells (>300 anaphases analyzed per condition) with lagging chromosomes after mock or ATRi (10 μM VE-821, 1 h) treatments. (E) Representative images of anaphase RPE1 cells in (D). Scale bar, 5 μm. (F) ATR−/−, AID-ATR DT40 cells were untreated or treated with 0.5 mM indole-3-acetic acid (IAA) for 30 min to induce AID-ATR degradation. Representative maximum projection images of anaphase cells (left) and quantification of cells with lagging chromosomes (right) are shown (>300 anaphases analyzed per condition). Scale bar, 5 μm. (G) Time-lapse imaging of H2B-GFP expressing RPE1 cells untreated or treated with ATRi. Red arrow signifies time of treatment. A lagging chromosome is demarcated by white arrows. Scale bar, 5 μm. Error bars in all panels represent SEM. *P ≤ 0.01, two-tailed t-test.
The presence of ATR at centromeres during mitosis prompted us to investigate if ATR functions in chromosome segregation. Short (1h) treatments of asynchronously growing RPE1 and U2OS cells with two structurally distinct ATR inhibitors (ATRi), VE-821 and AZ20, increased the rate of lagging chromosomes in anaphase (Fig. 1D–E), indicating a defect in whole-chromosome segregation (5, 6). Rapid degradation of AID degron-tagged ATR in mitotic ATR−/−, AID-ATR DT40 cells also increased lagging chromosomes (Fig. 1F, S3A) (7). In contrast to lagging chromosomes, anaphase bridges typically arise from S-phase problems persisting into mitosis but not whole-chromosome missegregation (8). Short ATRi treatments did not induce anaphase bridges (Fig. S3B), suggesting that the S-phase cells affected by ATRi had not entered mitosis. As ATRi-treated mitotic cells progressed into the next cell cycle, an increased rate of aneuploidy was observed (Fig. S3C). These results suggest that ATR has a previously uncharacterized role in chromosome segregation distinct from its functions in S phase.
Replication interference in S phase may affect mitosis indirectly by inducing persistent DNA synthesis in mitotic cells (Fig. S4A–B) (9). To investigate if ATR has a replication-independent role in mitosis, we arrested RPE1 and U2OS cells in G2 with CDK1 inhibitor and then released them into mitosis in the absence or presence of ATRi (Fig. S4A–B). Post-replication ATRi treatment did not induce DNA synthesis or anaphase bridges, but increased lagging chromosomes (Fig. S4B–D). To unequivocally separate the functions of ATR in S phase and mitosis, we treated live prometaphase cells with ATRi and followed chromosome segregation. ATRi treatment of cells already in mitosis increased the rate of lagging chromosomes in anaphase (Fig. 1G), demonstrating that ATR acts in mitosis to ensure faithful chromosome segregation.
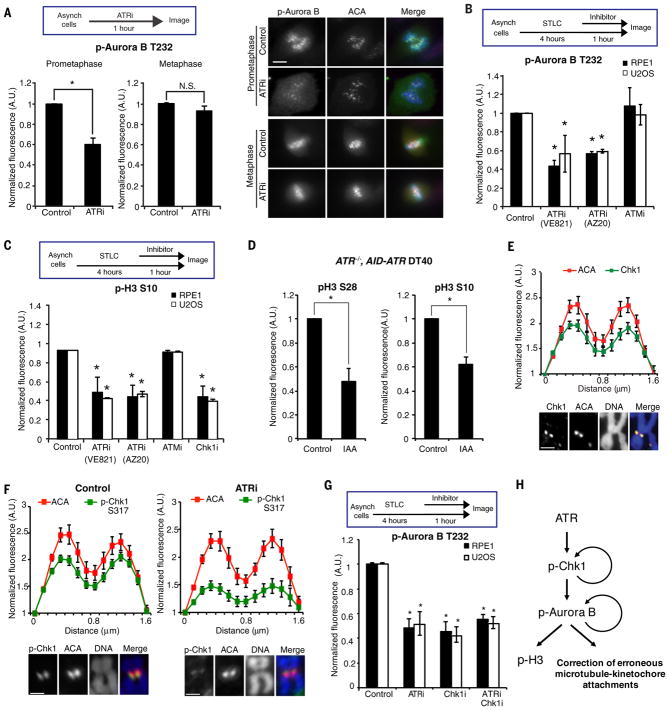
In mitotic cells, ATRi did not affect baseline levels of DNA synthesis and γH2AX, and the canonical ATR pathway was not activated by replication inhibitor hydorxyurea (Fig. S5A–C). ATRi did not significantly alter mitotic progression or affect the inhibitory phosphorylation of CDK1 at Y15 (Fig. S6A–C). Furthermore, ATRi did not increase the rate of multipolar spindles in prometaphase or lead to premature mitotic exit (Fig. S6D–E). However, ATRi reduced the autophosphorylation of Aurora B at T232 (p-Aurora B) in prometaphase (Fig. 2A–B, S7A) (10). ATRi did not affect Aurora B activity in vitro (Fig. S7B), ruling out off-target effects on Aurora B. In contrast to ATRi, ATM inhibitor (ATMi) did not affect p-Aurora B (Fig. 2B). While ATRi reduced p-Aurora B, it did not affect the centromeric localization of Aurora B and other components of the chromosome passenger complex (Fig. S7C). Moreover, the activation of Aurora A and PLK1 was not affected by ATRi (Fig. S7D). The Aurora B-dependent phosphorylation of histone H3 at S10 and S28 was reduced by ATRi (Fig. 2C, S8A–B). When AID-ATR was rapidly degraded in mitotic ATR−/−, AID-ATR DT40 cells, p-H3 S10 and p-H3 S28 was also reduced (Fig. 2D, S8C). Nonetheless, ATRi did not significantly affect the localization of BubR1 to kinetochores (Fig. S8D), suggesting that Aurora B retains some of its functions (11). Partial inhibition of Aurora B by AurBi increased the rate of lagging chromosomes (Fig. S9A–B) (12), recapitulating the effects of ATRi. Combination of ATRi with low concentrations of AurBi further decreased p-Aurora B and increased lagging chromosomes (Fig. S9A–B). Thus, ATR is required for the full activation of Aurora B at centromeres, which is critical for correction of erroneous microtubule attachments at kinetochores (Fig. 2H) (12, 13).
Figure 2. ATR promotes Aurora B activation at centromeres.
(A) Fluorescence intensity of phospho-Aurora B T232 (p-Aurora B) at centromeres in prometaphase or metaphase RPE1 cells untreated or treated with ATRi for 1 h (left). Representative images of untreated and ATRi-treated cells (right). Scale bar, 5 μm. (B–C) Fluorescence intensities of centromeric p-Aurora B (B) and overall p-H3 S10 (C) in S-Trityl L-cysteine (STLC)-arrested prometaphase cells after mock treatments or treatments with the indicated inhibitors for 1 h. (D) Fluorescence intensities of p-H3 S28 and p-H3 S10 in ATR−/−, AID-ATR DT40 cells untreated or treated with IAA for 30 min. (E) Line scan analysis of Chk1 and ACA at centromeres in chromosome spreads of RPE1 cells. Cells were arrested with nocodazole for 5 h. Scale bar, 2μm. (F) Line scan analysis of p-Chk1 and ACA at centromeres in chromosome spreads of RPE1 cells. Cell were arrested with nocodazole for 4 h and then mock or ATRi treated for 1 h. Scale bar, 2μm. (G) Fluorescence intensity of centromeric p-Aurora B in STLC-arrested prometaphase cells after mock treatments or treatments with the indicated inhibitors. Error bars in all panels represent SEM. *P ≤ 0.01, two-tailed t-test. (H) A schema of the ATR-Chk1-Aurora B pathway in mitosis. This pathway is required for full Aurora B activation, which is necessary for H3 phosphorylation and proper kinetochore-microtubule attachment.
To determine how ATR regulates Aurora B in mitosis, we analyzed the ATR effector kinase Chk1, which is implicated in Aurora B activation (14). Like ATR, Chk1 and its ATR- and auto-phosphorylated forms (p-Chk1 S317, S345, S296) were detected at all centromeres in mitotic cells (Fig. 2E–F, S10A–B). ATRi reduced p-Chk1 at centromeres (Fig. 2F), suggesting that Chk1 is phosphorylated by ATR. Similar to ATRi, Chk1 inhibitor (Chk1i) did not affect Aurora B activity in vitro, but reduced p-Aurora B, p-H3 S10, and p-H3 S28 in mitotic cells (Fig. 2C, 2G, S8B, S10C). Furthermore, the combination of ATRi and Chk1i did not further reduce p-Aurora B compared to ATRi or Chk1i alone (Fig. 2G), suggesting that ATR promotes Aurora B activation through Chk1 (Fig. 2H).
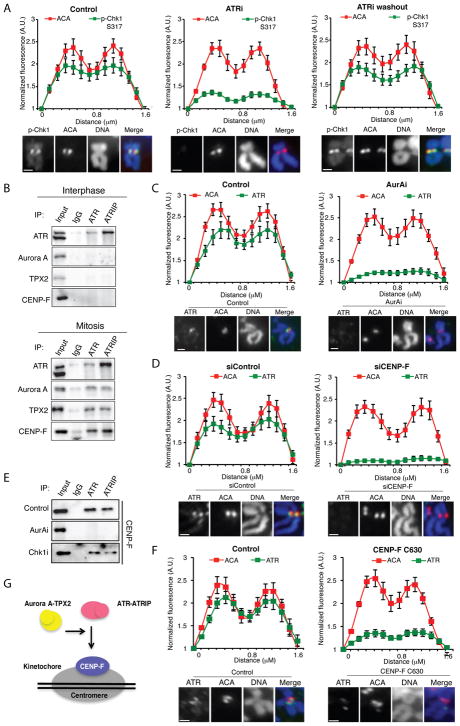
The unexpected role of ATR at centromeres begs the question as to how ATR is activated. To distinguish whether ATR is activated in or prior to mitosis, we tested if ATR can be reactivated in mitotic cells after transient inhibition. At 2 μM, the ATRi VE-821 inhibited ATR in a reversible manner in interphase cells (Fig. S11A). On mitotic chromosomes, the p-Chk1 at centromeres was reduced by 2 μM ATRi but quickly recovered after ATRi washout (Fig. 3A, S11B), showing that ATR is reactivated. Similarly, in mitotic cells p-Aurora B and p-H3 S10 also recovered after ATRi washout (Fig. S11C–D), confirming de novo activation of ATR in mitosis.
Figure 3. ATR localizes to centromeres in an Aurora A and CENP-F-dependent manner.
(A) Line scan analysis (top) and representative images (bottom) of centromeric p-Chk1 and ACA in chromosome spreads of RPE1 cells. Mitotic cells were isolated by shake off after 4h of nocodazole treatment. Cells were then mock treated (control) or treated with 2 μM VE-821 (ATRi) for 1 h in the presence of nocodazole. The ATRi washout sample was released from VE-821 for 1 h in the presence of nocodazole. (B) Immunoprecipitates of endogenous ATR and ATRIP from interphase (top) or mitotic (bottom) RPE1 cell extracts. Input = 5%. (C) Line scan analysis (top) and representative images (bottom) of centromeric ATR in chromosome spreads of RPE1 cells. Cells were arrested with nocodazole for 4h and then mock or AurAi treated for 1 h. (D) Line scan analysis of centromeric ATR and ACA in chromosome spreads of RPE1 cells. Cells were treated with control or CENP-F siRNA and arrested with nocodazole for 5 h. (E) CENP-F in the immunoprecipitates of endogenous ATR and ATRIP from mitotic RPE1 cell extracts. Cells were treated with AurAi and Chk1i as indicated. Input = 5%. (F) Line scan analysis (top) and representative images (bottom) of centromeric ATR and ACA in U2OS cells uninfected or infected with CENP-F C630 expressing retrovirus. Scale bar in all panels, 2 μm. Error bars in all panels represent SEM. *P ≤ 0.01, two-tailed t-test. (G) A model in which ATR is recruited to the vicinity of centromeres through an interaction with CENP-F. This interaction is dependent on Aurora A activity and occurs specifically in mitosis.
To understand how ATR localizes to centromeres in mitosis, we tested if ATR interacts with mitotic regulators and centromere proteins. Immunoprecipitations of ATR and its regulatory partner ATRIP from mitotic cell extracts, but not interphase cell extracts, captured Aurora A, TPX2 and CENP-F (Fig. 3B, S12A). ATR-ATRIP did not capture several other mitotic regulators (Fig. S12B), suggesting that ATR transiently but specifically interacts with the Aurora A-TPX2 complex and CENP-F in mitosis. The levels of ATR at centromeres were reduced by AurAi and CENP-F siRNA (Fig. 3C–D, S12C). The p-ATR at centromeres was diminished by two independent AurAi, but not by inhibitors of ATM, Chk1, Aurora B, MPS1 and the proteasome (MG132) (Fig. S12D). Importantly, AurAi disrupted the interaction between ATR-ATRIP and CENP-F without altering the levels and localization of CENP-F (Fig. 3E, S12E), suggesting that Aurora A promotes the localization of ATR to centromeres by enabling ATR-ATRIP to bind CENP-F (Fig. 3G). Consistent with the role for CENP-F in ATR localization, a CENP-F fragment that binds to kinetochores exerted dominant negative effects on ATR and p-H3 S10 at centromeres (Fig. 3F, S13) (15). Thus, Aurora A and CENP-F likely bring ATR-ATRIP to the vicinity of centromeres in mitosis, facilitating its activation at centromeres (Fig. 3G).
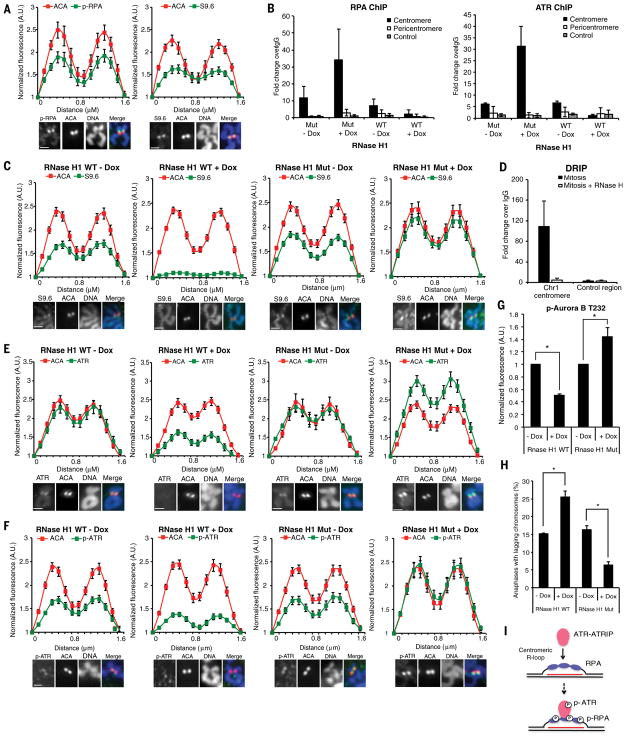
We next investigated how ATR is activated at centromeres in mitosis. The single-stranded DNA (ssDNA) binding protein RPA directly interacts with ATRIP and plays a key role in ATR activation at sites of DNA damage and stalled replication forks (16). Surprisingly, although DNA damage and DNA synthesis were undetectable at centromeres in mitosis (9), p-RPA staining was detected at all centromeres in synchronized and unsynchronized mitotic cells (Fig. 4A, S14A–B). The RPA at centromeres was phosphorylated at S33, a substrate site of ATR. Like ATR, RPA was detected at the centromere of chromosome 1 but not a non-centromere locus by ChIP (Fig. 4B, S14C). ssDNA is not only generated by DNA repair and DNA replication, but also by R loops, which contain DNA-RNA hybrids and displaced ssDNA (17, 18). In yeast R loops are found at centromeres and linked to H3 S10 phosphorylation and chromatin compaction (19). We recently showed that RPA is a sensor of R loops (20), raising the possibility that RPA is recruited to centromeres by the ssDNA in R loops.
Figure 4. ATR activation at centromeres is driven by R loops.
(A) Line scan analysis (top) and representative images (bottom) of centromeric phospho-RPA32 S33 (p-RPA), S9.6, and ACA in chromosome spreads of RPE1 cells. Scale bar, 2 μm. (B) Quantitative PCR of RPA (right) or ATR (left) ChIP in HeLa-derived RNaseH1 WT/MUT inducible cell lines. Cells were synchronized in G2 with CDK1i, uninduced or induced with Dox for 4 h, and released into mitosis in the presence of nocodazole for 1 h (see Methods). (C,E,F) Line scan analysis (top) and representative images (bottom) of centromeric S9.6 (C), ATR (E), p-ATR (F), and ACA in RNaseH1 WT/MUT inducible cell lines. Cells were treated as in (B). Scale bar, 2 μm. (D) Quantitative PCR of DRIP in mitotic RPE1 cells. (G) Relative fluorescence intensity of p-Aurora B in RNaseH1 WT/MUT inducible cell lines. Cells were treated as in (B). (F) Percentage of anaphase cells with lagging chromosomes. Cells were treated as in (B). Scale bar for all panels, 2μm. Error bars in all panels represent SEM. *P ≤ 0.01, two-tailed t-test. (I) A model in which ATR-ATRIP is recruited to and activated by RPA-coated centromeric R loops in mitosis. The red line depicts centromeric RNA hybridized with DNA.
To test if R loops are present at centromeres in human cells, we analyzed mitotic chromosomes in RPE1 cells with the monoclonal antibody S9.6, which specifically recognizes DNA-RNA hybrids (18). Indeed, S9.6 staining was detected at all centromeres in synchronized and unsynchronized mitotic cells (Fig. 4A, S15A–B). S9.6 staining was enriched at centromeres compared to telomeres and chromosome arms (Fig. S15C). To confirm that centromeric S9.6 signals arise from DNA-RNA hybrids, we conditionally expressed RNaseH1, an enzyme that cleaves the RNA in DNA-RNA hybrids, in HeLa cells. The centromeric S9.6 staining was reduced by induction of wild-type RNaseH1 (RNaseH1WT) (Fig. 4C). In contrast, the catalytically inactive RNaseH1 D210N mutant (RNaseH1MUT) (21), which is dominant negative over endogenous RNaseH1, increased S9.6 staining at centromeres (Fig. 4C). RNaseA, which cleaves single- and double-stranded RNA, did not affect S9.6 staining at centromeres (Fig. S15D). Consistent with S9.6 staining, R loops were detected at the centromere of chromosome 1 but not a non-centromere locus by DNA-RNA immunoprecipitation (DRIP) (Fig. 4D). The DRIP signal at the centromere was abolished by RNaseH1. Thus, like ATR, R loops are specifically enriched at centromeres in mitosis.
Next, we tested if R loops regulate RPA and ATR at centromeres in mitosis. The ChIP signals of RPA and ATR at the centromere of chromosome 1 were reduced by RNaseH1WT but enhanced by RNaseH1MUT (Fig. 4B). Furthermore, the ATR staining at centromeres was decreased by RNaseH1WT but increased by RNaseH1MUT (Fig. 4E). The centromeric p-ATR and p-RPA staining was also diminished by RNaseH1WT but enhanced by the RNaseH1MUT (Fig. 4F, S16A), suggesting that R loops are important for ATR activation at centromeres (Fig. 4I). The levels of CENP-F at kinetochores were only slightly affected by RNaseH1 (Fig. S16B), suggesting that R loops largely act downstream of CENP-F to promote ATR activation. RNA polymerase II (RNAPII)-mediated transcription occurs at centromeres in mitosis and is required for accurate chromosome segregation (22, 23). Inhibition of RNAPII but not RNA polymerase III in mitotic cells reduced the ATR and p-RPA at centromeres (Fig. S16C), suggesting that centromeric R loops are generated through RNAPII-mediated transcription during mitosis.
The role of R loops in ATR activation at centromeres predicts that R loops are important for Aurora B activation and suppression of lagging chromosomes. Indeed, p-Aurora B was reduced by RNaseH1WT but increased by RNaseH1MUT (Fig. 4G). Furthermore, the rate of lagging chromosomes was increased by RNaseH1WT but reduced by RNaseH1MUT (Fig. 4H). Notably, RNaseH1MUT increased p-H3 S10 and reduced lagging chromosomes, and both of these effects were reversed by ATRi (Fig. S17A–B), suggesting that stabilization of R loops facilitates Aurora B activation and chromosome segregation through ATR. Together these results strongly suggest that the R loop-ATR circuitry is important for accurate chromosome segregation.
Our results revealed a mitosis-specific ATR pathway that is distinct from the canonical ATR pathway operating in S phase and the DNA damage response. The activation of the mitotic ATR pathway is independent of DNA damage and DNA replication, but dependent on the key mitotic regulator Aurora A and R loops at centromeres (Fig. S18). In an Aurora A-dependent manner, ATR interacts with CENP-F in mitosis and is brought to the vicinity of centromeres. The R loops at centromeres may present RPA-coated ssDNA, a key platform for ATR activation. Unlike the canonical ATR pathway, the mitotic ATR pathway avoids the suppression by centromeric DNA loops behind replication forks (24). The requirement of both the Aurora A-CENP-F axis and the R loop-RPA axis for ATR activation may ensure that ATR is specifically activated in prometaphase at centromeres. The local activation of ATR at centromeres does not appear to affect CDK1 in mitosis, possibly due to the restriction of p-Chk1 to centromeres and the stabilization of CDC25A by CDK1 (25, 26). Instead, ATR activates Chk1 locally at centromeres to promote Aurora B activation, ensuring accurate chromosome segregation in mitosis.
This study uncovers an R loop-driven signaling pathway that promotes accurate cell division. While R loops cause genomic instability in S phase, they are necessary for faithful mitosis. The opposite effects of R loops on genomic stability during S phase and mitosis create a demand for cell cycle-regulated ATR responses. Through two distinct ATR pathways, ATR suppresses R loop-associated genomic instability in S phase, but mediates the centromeric function of R loops in mitosis (Fig. S18). The activation and function of ATR at DNA breaks and replication forks are regulated by DNA repair and replication proteins. The unique ability of the ATR circuitry to rewire in a context-dependent manner may allow it to play an integral role at centromeres in mitosis distinct from its other functions. Both the genomic instability in S phase (replication stress) and the chromosome instability in mitosis (CIN) are hallmarks of cancer and vulnerabilities of cancer cells (27–29). The dual functions of ATR in countering replication stress and CIN may make ATR a particularly attractive therapeutic target in cancers with both vulnerabilities.
Supplementary Material
Acknowledgments
We thank Dr. D. Compton for H2B-GFP-expressing RPE1 cells, Dr. N. Lowndes for the ATR−/−, AID-ATR DT40 cell line expressing degron-ATR, Dr. S. Taylor for the CENP-F C630 expression plasmid, Dr. N. Dyson and members of Zou and Dyson laboratories for discussions. L.K., H.D.N., and R.B. are supported by the NIH fellowships 5T32CA009216-35, T32DK007540, and 1K99CA212154, respectively. L.Z. is the James and Patricia Poitras Endowed Chair in Cancer Research, and supported by the Jim and Ann Orr Massachusetts General Hospital Research Scholar Award. This work is supported by the NIH grants (GM076388 and CA197779) to L.Z.
Footnotes
References and Notes
- 1.Yazinski SA, Zou L. Functions, Regulation, and Therapeutic Implications of the ATR Checkpoint Pathway. Annu Rev Genet. 2016;50:155–173. doi: 10.1146/annurev-genet-121415-121658. [DOI] [PubMed] [Google Scholar]
- 2.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 3.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 4.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eykelenboom JK, et al. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Rep. 2013;5:1095–1107. doi: 10.1016/j.celrep.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev. 2014;26:1–5. doi: 10.1016/j.gde.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Minocherhomji S, et al. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 10.Yasui Y, et al. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J Biol Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 11.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petsalaki E, Akoumianaki T, Black EJ, Gillespie DA, Zachos G. Phosphorylation at serine 331 is required for Aurora B activation. J Cell Biol. 2011;195:449–466. doi: 10.1083/jcb.201104023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussein D, Taylor SS. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J Cell Sci. 2002;115:3403–3414. doi: 10.1242/jcs.115.17.3403. [DOI] [PubMed] [Google Scholar]
- 16.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.Hamperl S, Cimprich KA. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 19.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen HD, et al. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell. 2017;65:832–847. e834. doi: 10.1016/j.molcel.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Lima WF, Crooke ST. Investigating the structure of human RNase H1 by site-directed mutagenesis. J Biol Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 22.Chan FL, et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gent JI, Dawe RK. RNA as a structural and regulatory component of the centromere. Annu Rev Genet. 2012;46:443–453. doi: 10.1146/annurev-genet-110711-155419. [DOI] [PubMed] [Google Scholar]
- 24.Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol. 2016;18:684–691. doi: 10.1038/ncb3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mailand N, et al. Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 2002;21:5911–5920. doi: 10.1093/emboj/cdf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zachos G, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–1143. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 30.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.