Abstract
The genetic evaluation of dilated cardiomyopathy (DCM) has been challenging, owing in large part to marked genetic heterogeneity. However, lower costs from next generation sequencing have enabled gene discovery and the expansion of genetic testing panels. These advances have improved molecular diagnostics and predictive testing in DCM. Here we provide a rationale and recommendation for clinical genetic testing in all of DCM.
Introduction
Molecular genetic sequencing, made cost-effective by next generation sequencing, has provided an enormous opportunity to transform the practice of cardiovascular medicine. The most tractable genetic conditions for clinical practice are those known to be familial, the cardiomyopathies, channelopathies and aortopathies. Here we focus on idiopathic dilated cardiomyopathy (DCM), a pan-ethnic condition, characterized by left ventricular enlargement (LVE) and systolic dysfunction.
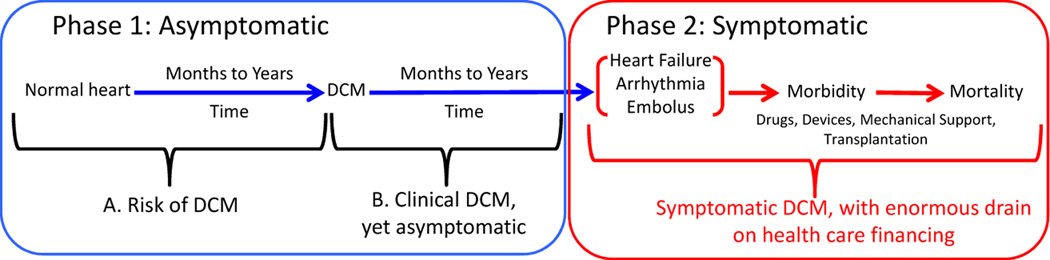
DCM is the most common heritable cardiomyopathy, with an estimated prevalence of 1/200 to 1/500.1 DCM is also the most common indication for heart transplantation in pediatric and adult populations.2 Understanding DCM genetics, and skillfully using genetic testing for all cases of DCM, both familial and non-familial (a recommendation that currently exceeds published guidelines,3 although our preliminary data suggests that the frequency of relevant variants in familial and non-familial DCM is similar4) presents enormous opportunity to prevent the morbidity and mortality that eventually accompanies most cases. This is because DCM usually presents with heart failure, arrhythmia, stroke, or sudden cardiac death, which are very late phase aspects of disease (Figure 1).5 However, an asymptomatic phase precedes symptomatic DCM, which can be divided into periods of risk. In the first period an individual carries a predisposing rare variant but the clinical evidence of DCM is not present. During the second period, diagnostic structural or functional changes of DCM are present, but there are no symptoms. DCM may continue in this asymptomatic phase for years.
Figure 1. The Asymptomatic and Symptomatic Phases of DCM.
Phase 1 includes two periods, both asymptomatic. In the first period (1A), individuals who harbor one or more rare DCM variants have risk of developing DCM over time. During this phase, genetic information identifies the individuals who would benefit from periodic clinical screening to detect early clinical disease. In Phase 1B, DCM is present, but asymptomatic, at times for years, thus evading detection without periodic efforts to detect it. With detection of asymptomatic clinical disease (Phase 1B), medical therapy can be initiated in an effort to prevent progression to Phase 2. In Phase 2, late stage disease becomes symptomatic with heart failure, arrhythmia or embolus, the presenting features of DCM. As noted in the text, this construct is useful principally in the adult population.
During the first phase, genetic risk can only be identified by molecular genetic testing (Figure 1). If a pathogenic variant is identified, the individual should then undergo cardiovascular screening at intervals to identify early disease onset. With the earliest evidence of DCM, medical therapy can be started to prevent symptomatic, advanced disease. All efforts to prevent or ameliorate heart failure, by definition a symptomatic condition, is a laudable goal.
In most cases, individuals with DCM only come to clinical attention when symptoms appear, usually ages 30–60. Symptomatic presentation may also occur at younger or older ages, from neonatal onset to the elderly. A subset of DCM, peripartum or pregnancy associated cardiomyopathy (PPCM/PACM), presents during pregnancy or the post-partum period.
DCM may also occur in the fetal, neonatal, and pediatric population,6 and while fetal and neonatal and early pediatric DCM phenotypically is similar to the adult population reviewed above, including evidence of rare variant cause,7, 8 the DCM epidemiology above is primarily applicable to adults.
Genetics of DCM
Clinical Genetics
A study comparing family history and cardiovascular screening of relatives of DCM probands found that family history information can reveal familial dilated cardiomyopathy (FDC) in only 5% of cases while cardiovascular screening can detect FDC in 20%.9 Subsequent studies found an FDC prevalence of 25–35%.10–12 Considering that isolated left ventricular enlargement may precede DCM, and thus may be taken as early evidence of disease, FDC was estimated to be present in 48% of family members.11 Most families show an autosomal dominant pattern of inheritance, where first degree relatives have a 50% probability of having the condition. However, not all affected family members have the same degree of severity (variable expressivity).1 Further complicating the assessment of FDC is the observation of some family members who carry the DCM causing mutation but escape the condition (reduced penetrance).1
Molecular Genetics
Research studies have shown that mutations in approximately 40 genes (locus heterogeneity) of diverse ontology segregate in approximately 40% of these families.1 When a genetic cause is known, the two most frequently implicated genes in DCM are TTN (truncating variants, 20%) and LMNA (6%). Preliminary studies have shown a similar yield of genetic variants among individuals with FDC and those with a negative family history,4 suggesting that most idiopathic DCM may have a genetic basis. A complicating factor is that almost all mutations are unique to individual families and thus require unique sets of molecular data and functional validation to be proven pathogenic.
Clinical Impact
Genetic testing panels can identify a pathogenic variant in about 20% of cases. A pathogenic variant is one that, based on previous cases, family studies, and/or functional data, among others, is unequivocally expected to cause DCM (Table 1). If a pathogenic variant is identified in an affected individual, a genetic diagnosis of DCM is confirmed. Moreover, finding a pathogenic variant in LMNA or DES in an affected individual may have prognostic significance when identified in an affected individual because of the increased risk of sudden cardiac death.3, 13 In addition, for patients with DCM and conduction system disease (often caused by LMNA mutations), early ICD implantation may be recommended to avoid syncope or sudden cardiac death.3, 13
Table 1.
Variant Terminology
| Pathogenic | The variant causes the patient’s phenotype. |
| Likely pathogenic | There is a > 90% probability that the variant is pathogenic. |
| Variant of uncertain significance | The variant cannot be classified. |
| Likely benign | There is a > 90% probability that the variant is benign. |
| Benign | The variant is not the cause of the patient’s phenotype. |
This follows terminology recommended by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology.14
Testing for the variant in unaffected, at-risk family members helps to realize the enormous potential of cardiovascular genetic medicine. If an at-risk family member is found to carry the pathogenic variant, this individual is expected to have an increased lifetime risk of developing DCM. Such individuals have been recommended to pursue cardiovascular clinical screening every 1–3 years consisting of echocardiogram, ECG, and cardiovascular-directed exam.3 At-risk family members in whom the variant is not identified are no longer considered to be at increased risk, and thus may be discharged from cardiac surveillance.
If, on the other hand, an affected individual undergoes panel testing and a variant is not identified, a genetic etiology cannot be ruled out, as the detection rate of genetic testing for DCM is not 100%. These individuals may benefit from follow up genetic testing in the future as more genes are discovered and added to panels. Importantly, first-degree relatives of these individuals remain at risk for DCM. Therefore, a screening ECG and some measure of ventricular size and function, usually by echocardiography, is recommended every 3–5 years.3 The same guidelines apply for family members of individuals with DCM who are unable to pursue or decline genetic testing. Regardless of genetic testing, screening of family members who are at risk can facilitate early diagnosis and treatment.
Genetic testing performance is not only limited by an imperfect detection rate; in our experience, the probability of finding a variant of uncertain significance (VUS) is about 20%. A variant may be classified as such if it has not been previously observed or if the associated data is conflicting (Table 1).14 Consequently, a VUS cannot be used for predictive risk testing in at-risk relatives. Segregation analyses, when a variant is tracked among affected family members to correlate its presence with the phenotype, can be highly instrumental when adjudicating variants. This approach is not always possible, as the proband may not have affected family members. Importantly, though, cardiovascular screening of the family members can identify silent (asymptomatic) DCM. Such affected individuals contribute to segregation studies, should a variant of uncertain significance be identified. While an affecteds-only approach is the only way to assess segregation, evaluating unaffected family members for a variant of uncertain significance may provide insights regarding penetrance. Because penetrance is age-dependent, informative unaffected relatives must be old enough to have a high probability that they have escaped disease (typically > 70 years old). For this reason, pre-test counseling in familial cases should include a discussion about the need for family communication and involvement, should co-segregation studies be indicated. All of the above considerations are based upon a genetic DCM paradigm where a single highly penetrant variant causes disease. We have recently reviewed an alternative multi-variant paradigm for DCM.1
At the population level, aggregating all DCM genotype data can also help sorting out variants. Currently, all sequencing data is stored in individual clinical genetic laboratories, data repositories, and public and private databases that use different methods to categorize variants. Through ClinVar, a publically available database (http://www.ncbi.nlm.nih.gov/clinvar/), ClinGen (an international effort aimed at data sharing and developing standards for variant interpretation) aims to ameliorate this problem by categorizing every known variant.15 ClinGen has adopted recently published standard terminology to categorize variants (Table 1).14
Practical Considerations
Genetic testing for DCM should be considered at any age in idiopathic cases, that is, after other usually detectable clinical causes have been excluded, many of which are age-dependent (e.g., metabolic disease in the neonatal period, coronary artery disease in mid- and later life). If a pathogenic variant is identified, surveillance screening recommendations vary depending on age. If the proband is an adult and predictive testing is considered for a child or adolescent, during counseling it should be communicated that the probability of detecting cardiac abnormalities at this age is low, thus supporting a recommendation for deferring testing adult-onset disease to adults. However, with onset of DCM in childhood, adolescence or early adulthood, the decision to test at risk relatives who are children has much greater merit. Regardless, genetic testing in children requires provider judgment and ample discussion with parents. Providers must balance evidence from family history (to assess for early onset cases), parental concern, and the availability of pediatric cardiology that can accurately evaluate cardiac pediatric data.
Although there are no physical risks or ethical dilemmas associated with genetic testing for DCM in adults, the potential ramifications should be carefully examined before testing proceeds. A close collaboration between an experienced cardiologist and a genetics professional can be fruitful to establish a well-defined phenotype as well as to choose an appropriate genetic testing strategy. Genetic counselors can also take responsibility for discussing risk and technical information, as well as disclosing results, dealing with complex genotypes, and providing family support.
In our experience, almost all patients with DCM are interested in pursuing genetic testing, the greatest issue (in the US) being the availability of insurance and/or out-of-pocket costs. In the US a patient’s out-of-pocket cost ranges from $0 to several thousand dollars, depending on the patient’s insurance situation. Genetic testing laboratories commonly investigate out-of-pocket charges before testing begins, a valuable component to the patient’s decision-making.
Our experience has also shown that most cardiologists do not conduct or refer patients with DCM for a genetic evaluation. This is an important issue that mandates an intensive research effort. Understanding the key factors influencing cardiologists’ knowledge, attitudes, and barriers to incorporating genetic evaluations into practice patterns is imperative to fulfill the promise of precision medicine for genetic DCM.
Conclusion
Genetic testing for DCM should be offered to every individual with non-ischemic DCM of any age, including those with PPCM/PACM, who provide consent. If used appropriately, genetic testing for DCM can be a powerful tool: it can establish an etiological diagnosis that may be helpful to establish a prognosis and to guide treatment. Also, cascade variant-specific testing when positive in at-risk family members can guide surveillance clinical screening and allow early therapy to prevent or ameliorate advanced disease. Having a well-rounded team and an appropriate work flow to support this service is key.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piran S, Liu P, Morales A, Hershberger RE. Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure. J Am Coll Cardiol. 2012;60:283–289. doi: 10.1016/j.jacc.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Towbin JA, Lowe AM, Colan SD, et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA. 2006;296:1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 7.Rampersaud E, Siegfried JD, Norton N, Li D, Martin E, Hershberger RE. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog Pediatr Cardiol. 2011;31:39–47. doi: 10.1016/j.ppedcard.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindel SJ, Miller EM, Gupta R, et al. Pediatric cardiomyopathy: importance of genetic and metabolic evaluation. J Card Fail. 2012;18:396–403. doi: 10.1016/j.cardfail.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 10.Grunig E, Tasman JA, Kucherer H, Franz W, Kubler W, Katus HA. Frequency and phenotypes of familial dilated cardiomyopathy [see comments] J Am Coll Cardiol. 1998;31:186–194. doi: 10.1016/s0735-1097(97)00434-8. [DOI] [PubMed] [Google Scholar]
- 11.Baig MK, Goldman JH, Caforio AP, Coonar AS, Keeling PJ, McKenna WJ. Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J Am Coll Cardiol. 1998;31:195–201. doi: 10.1016/s0735-1097(97)00433-6. [DOI] [PubMed] [Google Scholar]
- 12.McKenna C, Codd M, McCann H, Sugrue D. Idiopathic dilated cardiomyopathy: familial prevalence and HLA distribution. Heart. 1997;77:549–552. doi: 10.1136/hrt.77.6.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Priori SG, Wilde AA, Horie M, et al. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Heart Rhythm. 2013;10:e85–e108. doi: 10.1016/j.hrthm.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehm HL, Berg JS, Brooks LD, et al. ClinGen - The Clinical Genome Resource. N Engl J Med. 2015 doi: 10.1056/NEJMsr1406261. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]