Abstract
Objective
Body mass index (BMI) is commonly used to assess obesity, which is associated with numerous diseases and negative health outcomes. BMI has been shown to be a heritable, polygenic trait, with close to 100 loci previously identified and replicated in multiple populations. We aim to replicate known BMI loci and identify novel associations in a trans-ethnic study population.
Subjects
Using eligible participants from the Population Architecture using Genomics and Epidemiology (PAGE) consortium, we conducted a trans-ethnic meta-analysis of 102,514 African Americans, Hispanics, Asian/Native Hawaiian, Native Americans and European Americans. Participants were genotyped on over 200,000 SNPs on the Illumina Metabochip custom array, or imputed into the 1000 Genomes Project (Phase I). Linear regression of the natural log of BMI, adjusting for age, sex, study site (if applicable), and ancestry principal components, was conducted for each race/ethnicity within each study cohort. Race/ethnicity-specific, and combined meta-analyses using fixed-effects models.
Results
We replicated 15 of 21 BMI loci included in the Metabochip, and identified two novel BMI loci at 1q41 (rs2820436) and 2q31.1 (rs10930502) at the Metabochip-wide significance threshold (p<2.5x10−7). Bioinformatic functional investigation of SNPs at these loci suggests a possible impact on pathways that regulate metabolism and adipose tissue.
Conclusion
Conducting studies in genetically diverse populations continues to be a valuable strategy for replicating known loci and uncovering novel BMI associations.
Introduction
Obesity is a heritable risk factor for a large number of serious health conditions(1–4). It already imposes an enormous burden on the public health system and will continue to impact the cost of medical care through the predicted rise in diseases linked to chronic obesity(5–7). In US ethnicities, obesity rates vary in African Americans (36.2%), Hispanics/Latinos (31.5%), Native Americans (41.2%), European Americans (27.9%), and Asians (9.9%)(8). Body mass index (BMI) heritability studies estimate that up to 70% of BMI variability may be attributed to genetic factors(9–11). While this might suggest that genetic traits contribute to racial/ethnic differences in rates of obesity, the relative importance of genetics compared with diet, behavior, and socioeconomic factors is under continued investigation(12). However, it is indisputable that many minority groups have been disproportionately affected by the obesity epidemic and obesity research in minorities must remain a public health priority.
Genome-wide association studies (GWAS) in European ancestry populations have successfully identified numerous genetic variants associated with BMI, firmly establishing the importance of genetic factors on obesity(13–15). However, examining genetic associations in minority groups may reveal previously unidentified BMI loci and help to pinpoint causal variants. Conducting analyses in underrepresented minority populations has been shown to improve the statistical power to detect novel loci by increasing allele frequency and the variance of allele counts for some genetic variants(16–18). A recent fine-mapping study in African Americans benefitted from the lower linkage disequilibrium (LD) patterns when identifying independent signals in known BMI loci, and also found two novel loci, presumably aided by the gain in power due to the higher minor allele frequencies of these variants in those with African genetic ancestry(16). GWAS restricted to minority populations have had similar successes, uncovering additional BMI loci previously unidentified in studies of exclusively European ancestry(19–23). To date, the largest and most comprehensive BMI GWAS included individuals of both European and non-European descent, confirmed 41 known and found 56 novel BMI-associated loci(24). The results from these studies highlights the feasibility and benefits of using diverse human populations as a strategy to broaden our knowledge of BMI genetics.
To identify additional BMI loci, we leveraged the multi-ethnic design of the Population Architecture using Genomics and Epidemiology (PAGE) consortium to conduct a discovery meta-analysis in up to 102,514 individuals. Using this approach, we identified two novel BMI-associated loci, rs2820436 (1q41) and rs10930502 (2q31.1).
Materials and Methods
Study Population
The Population Architecture using Genomics and Epidemiology (PAGE) consortium is funded by the National Human Genome Research Institute to investigate the epidemiologic architecture of well-replicated genetic variants associated with human diseases or traits(25). The PAGE-I study, initiated in 2008, consists of a coordinating center and four consortia, each with access to large, diverse population-based studies. The four consortia are: Epidemiologic Architecture for Genes Linked to Environment (EAGLE), which is based on data from Vanderbilt University Medical Center’s biorepository linked to de-identified electronic health records (EAGLE-BioVU); the Multiethnic Cohort Study (MEC); the Women’s Health Initiative (WHI); and Causal Variants Across the Life Course (CALiCO), a consortium of five cohort studies: the Atherosclerosis Risk in Communities (ARIC) study, Coronary Artery Risk Development in Young Adults (CARDIA), the Cardiovascular Health Study (CHS), the Hispanic Community Health Study/Study of Latinos (SOL), and the Strong Heart Study(25). The PAGE-II study, initiated in 2013, added the Charles Bronfman Institute for Personalized Medicine at Mount Sinai Medical Center, BioMe™ BioBank (MSSM). For specific analyses in this paper, PAGE reached out to additional studies, including GenNet and the Hypertension Genetic Epidemiology Network (HyperGen) to increase the African American sample size. The Supporting Information includes detailed descriptions of each study.
African American, Hispanic, Asian/Native Hawaiian, Native American and European participants from the ARIC, EAGLE-BioVU, CHS, CARDIA, MEC, SOL, WHI, GenNet, and HyperGen were eligible for inclusion in this study (S1 Table). Race/ethnicity was self-reported in most studies except for EAGLE-BioVU, where race/ethnicity was administratively-reported and recorded in the electronic health record(26, 27). All studies were approved by Institutional Review Boards at their respective sites, and all study participants save EAGLE-BioVU provided informed consent. The Vanderbilt University Internal Review Board has determined that data contained within EAGLE-BioVU are considered limited datasets as defined by the Health Insurance Portability and Accountability Act (HIPAA) and are in accordance with provisions of Title 45, Code of Federal Regulations, part 46 (45 CFR 46) that define criteria for “non-human subjects” research.
The final sample of minorities from PAGE included 35,606 African American, 26,048 Hispanic/Latino, 22,466 Asian/Native Hawaiian, 17,859 European American, and 535 Native American participants (S1 Table).
Anthropometric measurements
BMI was calculated by taking the ratio of the weight (kg) and height squared (m2). For ARIC, CHS, CARDIA, HyperGEN, GenNet and WHI, BMI was calculated from height and weight measured at the time of study enrollment. In EAGLE-BioVU and MSSM, the median height and weight was calculated across all complete medical histories. MEC used self-reported height and weight. A validation study within MEC was conducted to assess the validity of these measures and showed that self-reported BMI was sometimes underestimated, but the difference was small (<1 BMI unit) compared to the findings from national surveys(28). To reduce the influence of outliers on the analysis, individuals who were underweight (BMI<18.5 kg/m2) and extremely overweight (BMI>70 kg/m2) were excluded, and BMI values were natural log transformed to correct for the right-skewed distribution of BMI.
Genotyping and Imputation
Genotyping was performed using the Metabochip, whose design has been described elsewhere(29). In brief, the Metabochip is a custom Illumina iSelect genotyping array of nearly 200,000 SNP markers and was designed to cost-effectively analyze putative association signals identified through GWAS meta-analyses of many obesity-related metabolic and cardiovascular traits. Imputation of Metabochip SNPs was conducted in MEC African Americans and Hispanics, MSSM African Americans and Hispanics, and WHI African Americans (SHARe) and Europeans. Study-specific reference samples(30), or reference samples from 1000 Genomes Phase I(31) were used. The programs MaCH and minimac were used for phasing and imputation, respectively(32–34). A summary of genotyping and imputation performance for each participating study has been published previously (35) and reproduced in S2 Table.
Within each race/ethnicity, related participants were identified within and between studies using PLINK(36). Identity by descent was estimated and when apparent first-degree relative pairs were identified, the member with the lower call rate was excluded from further analyses, with the exception of GenNet, SOL, and HyperGen. These studies accounted for family structure using linear mixed models (GenNet, HyperGen) or with generalized estimated equations which incorporate clusters of first degree relative pairs/household members (SOL)(37). In the remaining studies, participants with an inbreeding coefficient F>0.15 were excluded. Ancestry principal components were generated using the Eigensoft software(38, 39) using either an unrelated subset, or in the 1000 Genomes reference populations, which were then projected into the study sample. Ancestral outliers were excluded from further analyses, as described previously(40). Additional information is included in the Supporting Information.
A total of 88,505 individuals were genotyped with the Metabochip, and an additional 14,009 with GWAS data were imputed into the 1000 Genomes Project(31) or study-specific reference samples(30). For individuals with imputed data, only the Metabochip genetic variants were included. Genotype data was cleaned by standard quality control procedures as described in the Supporting Information.
Analysis
As has been done in previous publications (16, 41), BMI values were natural log transformed to account for the right-skewed distribution. Extreme BMI values less than 18.5 kg/m2 or greater than 70 kg/m2 were excluded from the analysis, with the assumption that these outliers could be attributable to data coding errors or an underlying rare condition outside the scope of this investigation. Given that CARDIA participants were generally younger, and young adults may have naturally low BMI measurements, the 18.5 kg/m2 exclusion criteria was waived for this cohort. The analyses were restricted to adults 20 years or older.
The population was stratified by study and self-identified race/ethnicity, with each subgroup analyzed separately. Multivariable linear regressions for each study-specific minority group were adjusted for age, sex, study site (if applicable), and ancestry principal components (S2 Table). A sex*age interaction term was included in all models (except WHI, which only includes women) to account for possible effect modification by sex. The sex*age interaction term was intended to account for potential sex-specific effects on BMI that vary by age, given that obesity risk and body composition are known to vary by age, and our study population includes both elderly participants and young adults older than 20 years of age. The results from each ethnicity, and for all ethnicities combined, were meta-analyzed using an inverse-variance weighted fixed-effects model in METAL(42). No inflation was observed in this meta-analysis (inflation factor λ=0.97).
The SNP with the smallest p-value within a locus was considered the lead SNP. BMI associations were considered statistically significant if the p-value surpassed the Bonferroni corrected threshold of significance (p<2.5×10−7), correcting for approximately 200,000 SNPs included on the Metabochip array. The locus was considered novel if the lead SNP was not in LD (r2<0.1 in any 1000 Genomes population) of a previously published known BMI loci. The list of known BMI loci was obtained by extracting records from the GWAS Catalog of the National Human Genome Research Institute (http://www.ebi.ac.uk/gwas/, accessed 4/26/2016), and through a literature search (April 2017) identifying publications based on high-throughput genotyping arrays that are not genome-wide (and thus, excluded from the GWAS Catalog)(16), BMI studies examining GxE associations(43, 44), and internal publications from collaborators that we expect to be published within the next year (Turcot V, in progress). Bioinformatic functional follow-up was performed for the most significant index SNP and all SNPs in high LD with the index (r2>=0.8 in African 1000 Genomes Population) at the four loci. HaploReg v4(45) and the UCSC Genome Browser from the Roadmap epigenomics project were used to assess whether variants in each of these loci were positioned in a putative enhancer or promoter specific to adipose tissue. GTEx expression data was also used to assess whether any of the loci overlapped eQTL results.
Results
The Metabochip array contains high density genetic variants at 21 previously published GWAS-identified BMI loci. We first assessed these known loci to evaluate the reproducibility of these loci in a multi-ethnic study population. Our study confirmed 15 of the 21 previously known BMI loci, significant at p<5.8×10−5, an approximate Bonferroni multiple testing correction for the average 866 SNPs at each BMI locus (S3 Table). Among the Metabochip previously known BMI loci that failed to replicate, the meta-analysis p-values approached significance, with most in the 10−4 range.
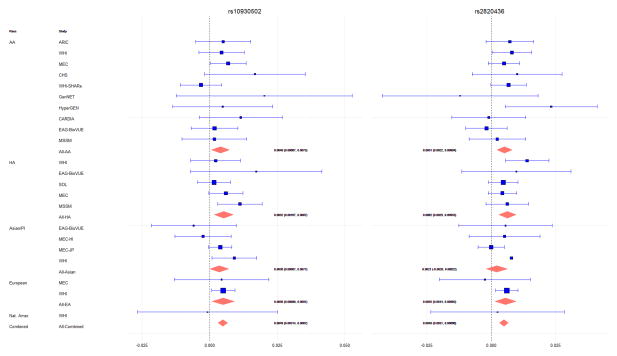
When we examined the remaining Metabochip content, we found an additional 14 loci associated with BMI, which achieved a Metabochip-wide significance level of p<2.5×10−7, correcting for approximately 200,000 SNPs on the Metabochip array (S4 Table). Eleven of these loci (or SNPs in high LD, r2>0.8, with these loci) were in LD (r2>0.1 in any 1000 Genomes population) with BMI loci previously identified since the development of the Metabochip (14, 21–24, 46–49). A twelfth SNP (rs11927381) no longer achieved Metabochip-wide significance after conditioning on a nearby SNP (rs1516725) that had previously been associated with BMI (24). Thus, we discovered two novel BMI-associated loci: 1q41 (rs2820436) and 2q31.1 (rs10930502) (Table 1). No evidence of heterogeneity was observed across studies at these two loci, with Cochran’s Q heterogeneity p-values of 0.65 and 0.94, for rs2820436 and rs10930502, respectively (Table 1, Fig 1).
Table 1.
Novel BMI-Associated Loci
| Metabochip Loci | Chr:BP | Gene | A1/A2 | Population | N | CAF | Beta | SE | P-value | HetP |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2820436 (1q41) | 1:219640680 | LYPLAL1/ZC3H11B | A/C | Combined | 94255 | 0.3876 | 0.0049 | 0.0009 | 3.79E-08 | 0.65 |
| AA | 35606 | 0.4782 | 0.0051 | 0.0015 | 8.34E-04 | NA | ||||
| HA | 26046 | 0.4395 | 0.0062 | 0.0017 | 1.61E-04 | NA | ||||
| AS | 14210 | 0.1952 | 0.0021 | 0.0021 | 3.20E-01 | NA | ||||
| EA | 17859 | 0.3446 | 0.0024 | 0.0132 | 8.56E-01 | NA | ||||
| NA | 534 | 0.3446 | 0.0024 | 0.0132 | 8.56E-01 | NA | ||||
| rs10930502 (2q31.1) | 2:172890588 | METAP1D | A/G | Combined | 94256 | 0.6794 | 0.0048 | 0.0009 | 1.35E-07 | 0.94 |
| AA | 35599 | 0.7000 | 0.004 | 0.0017 | 1.70E-02 | NA | ||||
| HA | 26043 | 0.6555 | 0.0043 | 0.0018 | 1.40E-02 | NA | ||||
| AS | 14220 | 0.3327 | 0.0056 | 0.0018 | 1.45E-03 | NA | ||||
| EA | 17859 | 0.6971 | 0.0056 | 0.0021 | 8.89E-03 | NA | ||||
| NA | 535 | 0.6794 | 0.0102 | 0.0137 | 4.56E-01 | NA |
Chr: chromosome, BP: base pair position hg19/GRCh37, A1: coded allele, A2: non-coded allele, CAF: coded allele frequency, StdErr=Standard Error, AS=Asian, AA=African American, HA=Hispanic American, EA=European American, NA=Native American, HetP: heterogeneity p-value,
fine-mapping regions for other traits (HDL, T2D, WHR).
Figure 1. Combined and Study-Specific Associations in Novel BMI-Associated Loci.
AA=African American, HA=Hispanic American, PI=Pacific Islander, ARIC=Atherosclerosis Risk in Communities Study, WHI=Women’s Health Initiative, MEC=Multiethnic Cohort, CHS=Cardiovascular Health Study, SHARe=WHI SNP Health Association Resource, GenNET=GenNet study, HyperGEN=Hypertension Genetic Epidemiology Network, CARDIA=Coronary Artery Risk Development in Young Adults study, EAG-BioVUE=Epidemiologic Architecture for Genes Linked to Environment accessing Vanderbilt University Medical Center BioVU, MSSM= The Charles Bronfman Institute for Personalized Medicine at Mount Sinai Medical Center, BioMe™ BioBank, SOL= The Hispanic Community Health Study / Study of Latinos.
The minor allele frequencies of these SNPs differed across the different ethnic groups (Table 1). rs2820436 was most frequent among PAGE African Americans (CAF=0.48), and least frequent in Asians (CAF=0.20), with the strongest association seen in the African Americans (p=8.34E-04) and Hispanic/Latinos (p=1.61E-04). While rs10930502 was also most frequent among African Americans (CAF=0.70) and European Americans (CAF=0.70), and least frequent among Asians (CAF=0.33), the association was strongest among the Asians (p=1.45E-03) and European Americans (p=8.89E-03). Generally, the observed allele frequencies in our own study population were similar to those from the same ethnic groups in the 1000 Genomes populations. Both of these SNPs were analyzed in the most recent and largest BMI GWAS study to date (p(rs2820436)=1.02E-02; p(rs10930502)=2.91E-04) (24), and were directionally consistent with our own results, providing additional support for these variants.
The variant rs10930502 was included on the Metabochip to follow-up on significant and suggestive signals from the largest available GWAS meta-analysis on BMI, while rs2820436 was included on the array for fine-mapping regions associated with waist-to-hip ratio (WHR). Given that rs2820436 was included on the Metabochip due to its previously published association with a non-BMI trait, we evaluated whether the associations with BMI were independent using individuals where WHR data was available (n=53,481). When the association between rs2820436 and BMI was adjusted by WHR, the overall association did not noticeably change. Conversely, when the association between rs2820436 and WHR was examined, adjusting for BMI, this p-value also achieved Metabochip-wide Bonferroni significance (p=3.09E-10). These findings suggest that this loci may influence multiple phenotypes related to body composition.
Functional investigation of the SNPs supports their likely involvement in lipid metabolism. We found that rs2820436 strongly tagged (r2=0.94 in 1000 Genomes Phase I Africans) a putative enhancer variant, rs2605096, positioned in an eQTL for the gene lysophospholipase-like 1 (LYPLAL1) previously associated with adiponectin(50), adiposity(51), cholesterol, T2D, and WHR(52).
Although rs10930502 was positioned in an eQTL for a lincRNA in adipose tissue, it did not strongly tag a putative regulatory variant. However, it was in moderate LD (r2=0.48, D′=0.85 in 1000 Genomes Phase I Africans) with variant rs34636594 at 2q31.1, which was positioned in a transcription factor binding enhancer in adipose tissue. LincRNAs are highly tissue specific and typically co-expressed with neighboring genes and thus we hypothesize that the 2q31.1 association may exert its effects on the candidate gene SLC25A12, through regulation of lincRNA.
Discussion
This trans-ethnic meta-analysis replicated 15 of 21 previously known BMI loci included on the Metabochip. Of the six loci that did not reach statistical significance in our own study, two of these had lead SNPs that were very rare, with CAF<0.01 in 1000 Genomes populations and PAGE racial/ethnic subgroups. Since most of these loci were originally discovered in GWAS studies with much larger sample sizes (13–15, 24, 53–55), our smaller study was likely insufficiently powered to replicate the rarer variants (Supporting Information). Other loci that we failed to replicate had lead SNPs that were more frequent in Europeans than in non-Europeans. Given that only 17% of our study sample consisted of those with European ancestry, insufficient power may also have contributed to our inability to replicate some of these loci, especially if these were European-specific associations.
Interestingly, both of the novel loci we identified, rs2820436 (1q41) and rs10930502 (2q31.1), are common in those with European ancestry, with a frequency of 0.68 and 0.31 in 1000 Genomes Europeans, respectively. Previous large, European-based BMI GWAS studies may have failed to detect these associations due to population-specific GxG interactions, or GxE interactions linked to cultural, socioeconomic, or behavior risk factors, resulting in a more pronounced effect on BMI in minority groups compared to Europeans. For both novel SNPs reported here, the largest betas in our study occurred in a non-European subgroup, suggesting that the genotypes might have a greater effect on BMI among non-Europeans. Should non-European population-specific effects exist, our large sample of minority subjects may have yielded more power to detect those associations compared to previous GWAS studies that may have been underpowered to detect population-specific effects related to a certain minority group.
Another possible explanation for why these associations were not detected in previous GWAS efforts is that these SNPs may be a poor proxy for the underlying causal SNP in European populations, but are a better proxy for the causal SNP in non-European populations. LD patterns are known to differ by genetic ancestry. It is possible that these SNP are in poor LD with the causal SNP in those with European ancestry, but in high LD with the causal SNPs in those with non-European ancestry. This would cause the association to be weaker or non-significant in Europeans due to exposure misclassification, where the tag SNP is an inaccurate indicator for the presence of the causal SNP. Given that the Metabochip was designed to facilitate fine-mapping in non-Europeans, it is not surprising that some of the Metabochip tag SNPs may perform better at estimating causal genotype-phenotype associations in a predominantly non-European study population.
Our findings demonstrate the value of conducting GWAS in non-European populations, both when replicating findings previously discovered in large, often European-centric GWAS, and for discovering novel associations which may be population-specific, or have stronger effects in those with non-European ancestry. Finally, the functional findings provide additional evidence for the biological relevance of these new loci in the BMI phenotype, which warrant further investigation. While these results are intriguing, additional replication is needed, especially using study populations that include underrepresented individuals. Both of these SNPs are most frequent in those with African ancestry, and our association in rs10930502 appears to be the strong in those with Asian ancestry.
Many genetic studies of BMI with larger sample sizes have been published and comparatively, we were underpowered to detect and replicate weaker associations, especially in less frequent variants. It is possible that additional novel, or population-specific loci may be found in a larger, trans-ethnic study population. However, we assembled one of the largest and most diverse non-European study populations and were still able to confirm 15 of the 21 known BMI loci included on the Metabochip. While the Metabochip was designed to replicate and fine-map loci known to be associated with 23 disease-related traits, its content is not genome-wide and non-Metabochip loci were not evaluated in this study. Yet, the inclusion of strong and well-established metabolically-related loci allowed us to identify a potential pleiotropic association with WHR. Studies that replicate our findings are advised to isolate the association that contributes specifically to BMI, given that our associations with BMI remain significant after adjusting for WHR. Through accompanying research efforts, we will benefit from the Metabochip’s increased marker density to fine-map these associations and further describe the relationship between these loci, BMI, and related phenotypes(35).
Certainly there are challenges associated with multi-ethnic genetic studies, but there are also legitimate benefits which may help explain more of the BMI heritability. The dearth of studies that include underrepresented populations only sustains disparities in genetic research, inhibits our ability to identify population-specific genetic risk factors, and hinders the development and application of genetic findings in real-world clinical settings(56–58). Our findings are promising and perhaps more importantly, demonstrate the need to conduct additional genetic studies of complex traits in non-European individuals.
Supplementary Material
Acknowledgments
KKN was supported by a National Cancer Institute training grant: Cancer Prevention Training in Nutrition, Exercise and Genetics (R25CA094880). LFR was supported by the Cardiovascular Disease Epidemiology Training Grant from the National Heart, Lung, and Blood Institute (T32HL007055) and the American Heart Association (AHA) predoctoral grant (13PRE16100015).
The Population Architecture Using Genomics and Epidemiology (PAGE I) program is funded by the National Human Genome Research Institute (NHGRI), supported by U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (MEC), U01HG004790 (WHI), and U01HG004801 (Coordinating Center), and their respective NHGRI ARRA supplements.
The Population Architecture Using Genomics and Epidemiology (PAGE II) program is funded by the National Human Genome Research Institute (NHGRI), with co-funding from the National Institute on Minority Health and Health Disparities (NIMHD), supported by U01HG007416 (CALiCo), U01HG007417 (ISMMS), U01HG007397 (MEC), U01HG007376 (WHI), and U01HG007419 (Coordinating Center).
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The complete list of PAGE members can be found at PAGE website (http://www.pagestudy.org). The data and materials included in this report result from a collaboration between the following studies:
The “Epidemiologic Architecture for Genes Linked to Environment (EAGLE)” is funded through the NHGRI PAGE program (U01HG004798-01 and its NHGRI ARRA supplement). The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
The Multiethnic Cohort study (MEC) characterization of epidemiological architecture is funded through NHGRI (HG004802, and HG007397) and the NHGRI PAGE program (U01 HG007397, U01HG004802 and its NHGRI ARRA supplement). The MEC study is funded through the National Cancer Institute (CA164973, R37CA54281, R01 CA 063464, P01CA33619, U01CA136792, and U01CA98758). The datasets used for the analyses described in this manuscript were obtained from dbGaP under accession phs000220.
Funding support for the “Epidemiology of putative genetic variants: The Women’s Health Initiative” study is provided through the NHGRI PAGE program (U01HG004790 and its NHGRI ARRA supplement). Funding support for the “Exonic variants and their relation to complex traits in minorities of the WHI” study is provided through the NHGRI PAGE program (U01HG007376, U01HG004790). The WHI program is funded by the National Heart, Lung, and Blood Institute; NIH; and U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. WHI PAGE II is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. The datasets used for the analyses described in this manuscript were obtained from dbGaP under accession phs000227. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
Funding support for the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) program was provided through the NHGRI PAGE program (U01HG007416, U01HG004803 and its NHGRI ARRA supplement). The following studies contributed to this manuscript and are funded by the following agencies:
The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
The Coronary Artery Risk Development in Young Adults (CARDIA) study is supported by the following National Institutes of Health, National Heart, Lung and Blood Institute contracts: N01-HC-95095; N01-HC-48047; N01-HC-48048; N01-HC-48049; N01-HC-48050; N01-HC-45134; N01-HC-05187; and N01-HC-45205. CARDIA is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of Alabama at Birmingham (HHSN268201300025C & HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging.
The Hispanic Community Health Study/Study of Latinos (SOL) was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). Additional support was provided by 1R01DK101855-01 and 13GRNT16490017. The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Center on Minority Health and Health Disparities, the National Institute of Deafness and Other Communications Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements.
The Cardiovascular Health Study (CHS) is supported by contracts HHSN268201200036C, HHSN268200800007C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants HL080295 and HL087652 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/PI.htm. CHS GWAS DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by the National Center for Research Resources, grant UL1RR033176, and is now at the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124; in addition, the National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
The Strong Heart Study (SHS) is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, U01 HL65521 and R01 HL109301. The datasets used for the analyses described in this manuscript were obtained from dbGaP under accession phs000223 (ARIC), phs000236, (CARDIA), phs000301 (CHS), phs000555 (HCHS/SOL). The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Indian Health Service.
GenNet is one of four networks in the Family Blood Pressure Program, established in 1995 and supported by a series of agreements with the NIH National Heart, Lung and Blood Institute.
Samples and data of The Charles Bronfman Institute for Personalized Medicine (IPM) BioMe Biobank used in this study were provided by The Charles Bronfman Institute for Personalized Medicine at the Icahn School of Medicine at Mount Sinai (New York). Phenotype data collection was supported by The Andrea and Charles Bronfman Philanthropies. Funding support for the Population Architecture Using Genomics and Epidemiology (PAGE) IPM BioMe Biobank study was provided through the National Human Genome Research Institute (U01 HG007417). The datasets used for the analyses described in this manuscript were obtained from dbGaP under accession phs000925.
The Hypertension Genetic Epidemiology Network (HyperGEN) study was supported by National Heart, Lung, and Blood Institute contracts HL086694 and HL055673.
Assistance with phenotype harmonization, SNP selection and annotation, data cleaning, data management, integration and dissemination, and general study coordination was provided by the PAGE Coordinating Center ((U01HG007419, U01HG004801-01 and its NHGRI ARRA supplement). The National Institutes of Mental Health also contributes to the support for the Coordinating Center. The authors gratefully acknowledge Dr. Ben Voight for sharing the Metabochip SNP linkage disequilibrium and minor allele frequency statistics estimated in the Malmö Diet and Cancer Study. The PAGE consortium thanks the staff and participants of all PAGE studies for their important contributions.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supplementary information is available at the International Journal of Obesity’s website.
References
- 1.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–91. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 2.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 3.Miller WM, Nori-Janosz KE, Lillystone M, Yanez J, McCullough PA. Obesity and lipids. Curr Cardiol Rep. 2005;7(6):465–70. doi: 10.1007/s11886-005-0065-8. [DOI] [PubMed] [Google Scholar]
- 4.Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann N Y Acad Sci. 2012;1271:37–43. doi: 10.1111/j.1749-6632.2012.06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cawley J, Meyerhoefer C. The medical care costs of obesity: an instrumental variables approach. J Health Econ. 2012;31(1):219–30. doi: 10.1016/j.jhealeco.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Danaei G, Ding EL, Mozaffarian D, Taylor B, Rehm J, Murray CJ, et al. The preventable causes of death in the United States: comparative risk assessment of dietary, lifestyle, and metabolic risk factors. PLoS Med. 2009;6(4):e1000058. doi: 10.1371/journal.pmed.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olshansky SJ, Passaro DJ, Hershow RC, Layden J, Carnes BA, Brody J, et al. A potential decline in life expectancy in the United States in the 21st century. N Engl J Med. 2005;352(11):1138–45. doi: 10.1056/NEJMsr043743. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: national health interview survey, 2012. Vital Health Stat. 2014;10(260):1–161. [PubMed] [Google Scholar]
- 9.Hjelmborg J, Fagnani C, Silventoinen K, McGue M, Korkeila M, Christensen K, et al. Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity (Silver Spring) 2008;16(4):847–52. doi: 10.1038/oby.2007.135. [DOI] [PubMed] [Google Scholar]
- 10.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–51. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 11.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–4. [PubMed] [Google Scholar]
- 12.McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, et al. Ten putative contributors to the obesity epidemic. Crit Rev Food Sci Nutr. 2009;49(10):868–913. doi: 10.1080/10408390903372599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong J, Schumacher F, Lim U, Hindorff LA, Haessler J, Buyske S, et al. Fine Mapping and Identification of BMI Loci in African Americans. Am J Hum Genet. 2013;93(4):661–71. doi: 10.1016/j.ajhg.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulit SL, Voight BF, de Bakker PI. Multiethnic genetic association studies improve power for locus discovery. PLoS One. 2010;5(9):e12600. doi: 10.1371/journal.pone.0012600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Stram DO. The role of local ancestry adjustment in association studies using admixed populations. Genet Epidemiol. 2014;38(6):502–15. doi: 10.1002/gepi.21835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, Keating BJ, et al. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum Mol Genet. 2013;22(1):184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45(6):690–6. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44(3):302–6. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–11. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23(20):5492–504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, et al. The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol. 2011;174(7):849–59. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010;12(10):648–50. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall JB, Dumitrescu L, Dilks HH, Crawford DC, Bush WS. Accuracy of administratively-assigned ancestry for diverse populations in an electronic medical record-linked biobank. PLoS One. 2014;9(6):e99161. doi: 10.1371/journal.pone.0099161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connor Gorber S, Tremblay MS. The bias in self-reported obesity from 1976 to 2005: a Canada-US comparison. Obesity (Silver Spring) 2010;18(2):354–61. doi: 10.1038/oby.2009.206. [DOI] [PubMed] [Google Scholar]
- 29.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu EY, Buyske S, Aragaki AK, Peters U, Boerwinkle E, Carlson C, et al. Genotype imputation of Metabochip SNPs using a study-specific reference panel of ~4,000 haplotypes in African Americans from the Women’s Health Initiative. Genet Epidemiol. 2012;36(2):107–17. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Rhodes L, Gong J, Haessler J, Franceschini N, Graff M, Nishimura KK, et al. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci. Hum Genet. 2017;136(6):771–800. doi: 10.1007/s00439-017-1787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin DY, Tao R, Kalsbeek WD, Zeng D, Gonzalez F, 2nd, Fernandez-Rhodes L, et al. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2014;95(6):675–88. doi: 10.1016/j.ajhg.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, et al. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 2012;7(4):e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fesinmeyer MD, North KE, Ritchie MD, Lim U, Franceschini N, Wilkens LR, et al. Genetic risk factors for BMI and obesity in an ethnically diverse population: results from the population architecture using genomics and epidemiology (PAGE) study. Obesity (Silver Spring) 2013;21(4):835–46. doi: 10.1002/oby.20268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Justice AE, Winkler TW, Feitosa MF, Graff M, Fisher VA, Young K, et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat Commun. 2017;8:14977. doi: 10.1038/ncomms14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, et al. Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet. 2017;13(4):e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melen E, Granell R, Kogevinas M, Strachan D, Gonzalez JR, Wjst M, et al. Genome-wide association study of body mass index in 23 000 individuals with and without asthma. Clin Exp Allergy. 2013;43(4):463–74. doi: 10.1111/cea.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stergiakouli E, Gaillard R, Tavare JM, Balthasar N, Loos RJ, Taal HR, et al. Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring) 2014;22(10):2252–9. doi: 10.1002/oby.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warrington NM, Howe LD, Paternoster L, Kaakinen M, Herrala S, Huikari V, et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol. 2015;44(2):700–12. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015;11(10):e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 54.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22(17):3597–607. doi: 10.1093/hmg/ddt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475(7355):163–5. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–66. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haga SB. Impact of limited population diversity of genome-wide association studies. Genet Med. 2010;12(2):81–4. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.