Summary
Tumor metabolism is reorganized to support proliferation in the face of growth-related stress. Unlike the widespread profiling of changes to metabolic enzyme levels in cancer, comparatively less attention has been paid to the substrates/products of enzyme-catalyzed reactions, small-molecule metabolites. We developed an informatic pipeline to concurrently analyze metabolomics data from over 900 tissue samples spanning 7 cancer types, revealing extensive heterogeneity in metabolic changes relative to normal tissue across cancers of different tissues of origin. Despite this heterogeneity, a number of metabolites were recurrently differentially abundant across many cancers, such as lactate and acyl-carnitine species. Through joint analysis of metabolomic data alongside clinical features of patient samples, we also identified a small number of metabolites, including several polyamines and kynurenine, which were associated with aggressive tumors across several tumor types. Our findings offer a glimpse onto common patterns of metabolic reprogramming across cancers, and the work serves as a large-scale resource accessible via a web application (http://www.sanderlab.org/pancanmet).
Keywords: cancer metabolism, metabolomics, genomics, clinical data, meta-analysis
eTOC blurb

Reznik et al. develop a computational approach for integrative analysis of metabolomics data and apply it to data from 900 tissue samples spanning 7 different cancer types. They identify recurrent metabolic changes associated with tumor initiation and progression to aggressive disease.
Introduction
Profiling the genomes, epigenomes, and proteomes of large cohorts of tumors has generated a detailed census of hallmark molecular alterations evident across many tumor types and likely to drive malignancy (Ciriello et al., 2013). These findings have contributed to a holistic understanding of cancer and have elucidated novel and potentially targetable vulnerabilities. In contrast, while altered metabolism is a well-accepted hallmark of cancer (Hanahan and Weinberg, 2011; Vander Heiden et al., 2009), the technical challenges of measuring the abundance of metabolites (e.g. using mass spectrometry) have limited the use of metabolomic profiling in studies of cancer tissues. Instead, much of the work in cancer metabolism has focused on examination of changes in the expression of metabolic enzymes, and the identification of patterns of metabolic gene expression common to many cancer types (Gatto et al., 2014; Nilsson et al., 2014; Hu et al., 2013). In contrast, metabolomic studies of cancer have been largely limited to in vitro flux studies of central carbon metabolism, and larger metabolomic studies focused on individual cancer types (Sullivan et al., 2016; Possemato et al., 2011; Mullen et al., 2012; Hensley et al., 2016; Lu et al., 2012, 2013), with notable and ever-more frequent exceptions (Mayers et al., 2016; Goveia et al., 2016).
Tumors grow and divide in the presence of stress imposed by proliferation and augmented by cytotoxic and targeted therapy. To survive, the activity of metabolic pathways is modulated to meet energetic demands, produce suitable levels of biosynthetic precursors, sustain redox potential and maintain epigenetic integrity. As a whole, these metabolic alterations are implemented via changes in the levels of intracellular metabolites, enzymes, and transporters (DeBerardinis and Chandel, 2016; Pavlova and Thompson, 2016). As with genetic alterations, these metabolic alterations may differ substantially according to, among other factors, the tissue-of-origin and stage of disease. Furthermore, many functional and common genetic alterations in cancer involve metabolic enzymes (e.g. IDH, SDH, FH) or regulators of metabolism (e.g. MTOR, VHL) (Jalbert et al., 2017; Haider et al., 2016; Grabiner et al., 2014). In contrast, in this study, we seek to derive some organizing principles for how tumors alter their metabolomes, the repertoire of intracellular, small molecule metabolites constituting the cell.
Our work focuses on analysis of published mass-spectrometry (MS)-based metabolomic profiling data of cancer tissues. Integration of MS measurements from different labs is inherently difficult because of incomplete/inconsistent measurement conditions and normalization procedures between different labs. To overcome this limitation, we use meta-analysis, i.e. metabolomic data is analyzed separately within each dataset, and the results of the individual analyses are subsequently combined. This approach enables us to determine if different cancers exhibit common patterns of metabolic changes between tumor and normal tissues. By additionally integrating clinical features into our analysis (e.g. pathological grade of the tumor), we are able to examine the association between tissue metabolite levels and the clinically aggressive features of a tumor. Unlike other recent analyses, this approach produces a single merged dataset with standardized nomenclature. This dataset, the informatic pipeline used to produce it, and all analysis code is made publicly available at https://github.com/dfci/pancanmet_analysis for future work. An accompanying website (http://www.sanderlab.org/pancanmet/) is provided to allow readers to explore the dataset, including 1) univariate and bivariate change in metabolite levels and 2) clinical associations by metabolite and by study. The Supplementary Data also contains tables facilitating mapping metabolites to various database identifiers.
Results
Assembly of a Cross-Cancer Compendium of Metabolomics Data
The first step of our analysis was to develop a computational framework to jointly analyze metabolomic data produced from different laboratories. We obtained published cancer tissue metabolomics data from 11 studies covering 7 distinct cancer types (see Figure 1, Figure S1, and Supplementary Table 1: Merged Metabolomics). Data for all studies was collected by the original investigators using mass spectrometry. Three cancer types (breast, prostate, and pancreatic ductal adenocarcinoma) were represented by at least 2 different datasets, enabling us to evaluate the consistency of findings across different studies. In total, our dataset encompasses 928 tissue samples.
Figure 1. Metabolomics data (928 samples, 7 cancer types, 11 studies) analyzed in this study.
Data from eleven distinct metabolomics studies were aggregated. Due to incomplete coverage of the metabolome, many metabolites were profiled in a small proportion of studies. The number of tumor/normal samples varied from study to study. All but one study conducted on gliomas contained normal samples. In some, but not all cases, tumor/adjacent-normal tissue samples were collected from the same patient (i.e. “matched” or “paired” samples, terms used interchangeably). This matched data is indicated in Supplementary Table 2.
Usage of metabolomics data for the purpose of meta-analyses involves unique technical and informatic challenges. Unlike genomic sequencing, which frequently covers the full breadth of the exome or genome, metabolomics relies on the identification of a small (on the order of hundreds) library of compounds. Each individual metabolomic study is likely to profile distinct assortments of these molecules (Cho et al., 2014). Therefore, molecules profiled in one study may partially overlap the set of molecules profiled in other studies, and inconsistent use of standard nomenclature for metabolites makes their “alignment” across studies challenging. For example, lactate is synonymously referred to as lactic acid and (S)-2-Hydroxypropanoate, and is reported along-side many other database identifiers (e.g. CHEBI, KEGG, HMDB, and PubChem IDs) (Kim et al.,2016; Wishart et al., 2007; Kanehisa et al., 2016).
Technical limitations and inconsistent use of common standards render infeasible the direct comparison of raw MS-based data of metabolite levels from different laboratories. Nevertheless, there is a clear missed opportunity for analyzing such data jointly, e.g. for discovering common metabolic changes across cancer types. To overcome the technical issues associated with normalizing raw mass spectra from different labs, we elected to use pre-normalized, published data, and to apply a series of tools to make comparison between them possible. Each of the datasets we used provided metabolite abundances normalized by the original investigators. In 3/11 studies, the acquired data was not imputed or was un-standardized (here, by un-standardized we specifically mean the data was on a scale unsuitable for visualization, not that the data was not appropriately un-normalized). For metabolomic data that was already imputed and standardized by the original investigators, we left the data unaltered. For the studies where this was not the case, we imputed any missing measurements for a compound with the minimum measured value of that compound. To simplify data visualization, we then divided all measurements of the compound by the median abundance of that compound within the study (for further details, see Figure 2, Methods and code at https://github.com/dfci/pancanmet_analysis). All downstream analysis of the processed metabolomics data (e.g. differential abundance tests) then relied on (1) use of non-parametric statistical methods, which do not make assumptions on the underlying distribution of the data, and (2) the use of normal tissues as a “reference” by which to identify cancer-specific changes in metabolite abundance. Importantly, while our pipeline is robust to variation in the underlying distribution of the data, erroneous normalization by the original producers of the data will (obviously) lead to erroneous results from our pipeline.
Figure 2. Workflow for the aggregation and comparison of metabolite profiles across studies.
Metabolomics data collected in different laboratories is difficult to directly compare. Our approach is to analyze data from each metabolomics dataset separately (e.g. comparing tumor to normal samples) and then aggregate these results across cancer types. Typically, for each metabolite, abundance is reported relative to the median of all measurements of a metabolite within a study. Comparisons between different metabolites profiled in the same study is not directly possible (leftmost red oval). Furthermore, comparisons between the same metabolite profiled across different studies is not possible (topmost red oval). Throughout our analysis, we frequently examine the change in abundance of a single metabolite across different subsets of tissue samples (e.g. tumor/normal samples) from the same study (green oval). Correlative analysis with variables of interest (e.g. clinical data) is also feasible.
The product of our computational pipeline is a merged metabolomics dataset containing data and samples from all studies and additional metadata (Figure S1, Supplementary Table 1 Merged Metabolomics) including mappings to identifiers of several key databases (i.e. CHEBI, KEGG, HMDB, and PubChem IDs) found in Supplementary Table 1: Merged IDs. Because ambiguous nomenclature from each individual dataset is resolved, metabolites which are measured across many studies are aligned and can be compared with meta-analysis. Access to the study data is available for interactive data exploration at the project website http://www.sanderlab.org/pancanmet. On this website, users may explore the data by visualizing univariate and bivariate relationships, changes in abundance between tumor and normal samples across user-selected sets of metabolites, associations between the available clinical data and metabolite levels, and links between drug compounds that target enzymatic processes used to synthesize metabolites of interest.
Analysis of Metabolic Variation in Tumors and Normal Tissue
We first sought to understand the extent to which each cohort of tumors differed from normal tissue. In analogy to prior studies of genetic changes across cancers, we expected that some tumor types may have relatively few metabolic changes compared to normal tissues, while others may have many.
Therefore, on a study-by-study basis, we examined which metabolites were differentially abundant between tumor and normal samples (Mann-Whitney U test, Benjamini-Hochberg (BH)-corrected p-value < 0.05). For the three cancer types for which we had replicate studies, changes in abundance across the studies were in good agreement (Figure S2). To further corroborate whether our differential abundance findings (and the data in general) reflect true biological differences or are artifacts of technical noise, we took advantage of detailed knowledge on metabolic network structure. We asked whether the levels of metabolites that are adjacent in the metabolic network (i.e. are substrates/products for a common enzyme) are more likely to be correlated to each other than non-adjacent metabolites. We expected that, if technical noise in the data was not excessive, we would see an enrichment for high positive correlations in adjacent metabolite pairs. Applying this analysis to each data set in our study, we found that in all but one data set (Bladder), adjacent metabolites were significantly more correlated than non-adjacent metabolites (Figure S2, SI Table 1: Covariation_Results).
The proportion of differentially abundant metabolites varied substantially from one cancer type to the next (Figure 3A). When using the multiple-hypothesis-adjusted Mann-Whitney p-value as the only criterion for significance (i.e. ignoring the magnitude of change), over 60% of metabolites in both breast cancer studies and the kidney cancer study were differentially abundant. In contrast, pancreatic cancers had characteristically fewer differentially abundant metabolites (< 20% in 2/3 studies, Supplementary Table 1: Differential Abundance Summary). We tested whether the large variation in propensity for differential abundance arose due to sample-size effects, and we did not find evidence to support this hypothesis (Figure S3).
Figure 3. Differential metabolite abundances in tumor versus normal across cancer types.
(A and B) In total, 10/11 studies contained adjacent-normal tissue for comparison and were included in this analysis (in total, 482 tumor samples and 378 normal samples). Analysis was done without regard to whether tumor/normal samples were paired. (A) Proportion of metabolites in each study which were differentially abundant in tumor vs. normal samples (BH-corrected p-value < 0.05). (B) The frequency of differential abundance across all metabolites. Most metabolites are rarely differentially abundant, in part because they may only be measured in a small number of studies. A small number of metabolites are often differentially abundant. (C) Variability of metabolite levels for each study. MAD ratio distributions show the inherent variability of metabolite levels; distributions shifted to the right of zero show a greater degree of variability in tumors compared to normal tissue, while those distributions shifted to the left show less. A total of 343 pairs of tumor/normal samples from 8 studies (Bladder, Breast Terunuma, Breast Tang, Kidney, Pancreatic Kamphorst, Pancreatic Zhang1, Pancreatic Zhang2, Prostate Priolo, and Prostate Sreekumar) were used.
This analysis revealed a cancer-type-dependent trend towards unequal proportions of metabolites that increased or decreased between tumor and normal tissues. In both breast cancer studies, we found that nearly all metabolites deemed differentially abundant were at higher levels in tumor tissue, compared to normal tissue. Similar results were reported in the original publications for these two datasets (Terunuma et al., 2014; Tang et al., 2014). While biases were evident in other studies (e.g. differentially abundant metabolites in pancreatic tumors were more frequently depleted than accumulated in tumors), the effect was particularly striking for breast cancer. Although it is not possible for us to determine the source of this effect, we speculate it could arise from the disproportionate extent of data imputation for metabolites in normal breast tissue, compared to the extent of imputation in tumor tissue (Figure S1). Because the effect is replicated in both breast cancer studies, it is possible that metabolites are broadly at higher concentrations in breast tumor compared to benign breast tissue.
It is also possible that tumor and normal tissues differ in the inherent variability of metabolite levels, without differential abundance. To test this possibility, we compared the median absolute deviation (MAD, a non-parametric measure of the variability of a collection of measurements) of a metabolite for tumor and normal samples in each study. To control for potential biases in the relative numbers of tumor and normal samples, only data with matched tumor and adjacent-normal samples from the same patient was used. The distribution of MAD ratios over all metabolites for a given cancer type indicates whether metabolite levels were inherently more variable in tumors (distribution shifted to the right of zero) or less variable in tumors (distribution shifted to the left) compared to normal tissue (Figure 3C). We observed significant differences in variability of metabolite levels across tumor types: metabolite levels in breast and kidney tumors were significantly more variable than in their normal tissue counterparts (Mann-Whitney p < 0.05). In contrast, in 2/3 pancreatic tumor datasets, tumors were significantly less variable than their normal tissue counterparts. Together with the results above, these results reflect major differences in both absolute metabolite levels and in metabolic variability associated with each individual cancer type.
Common Patterns of Metabolic Alterations Across Cancers
Cancers share common genetic alterations despite inherent physiological and molecular differences in their cell of origin. Describing these commonalities across cancer types has expanded our understanding of the operation of molecular processes and the coordination of disparate cellular pathways (Lawrence et al., 2014; Zhang et al., 2017). Furthermore, it has changed our perspective on how patients are treated and recruited to clinical trials (Conley and Doroshow, 2014). Previously, because of a lack of data, it has not been possible to assess whether metabolic alterations, manifested through shifts in the abundances of metabolites from tumors to normal tissue, also had common patterns across different cancer types. With the dataset organized for this work, we can begin to address this question.
Unlike the prior analysis, we now focused on analyzing common metabolomic changes across cancers of different tissues of origin. We identified 27 metabolites differentially abundant (tumor vs. normal tissue) in at least 6 studies (BH-corrected p-value < 0.05, Figure 3B and Figure 4, Supplementary Table 1: Differential Abundance Summary). Among these, a single metabolite, taurine, was differentially abundant in nine of the ten studies in which it was measured (increased in tumors in 6 studies, decreased in tumors in 3 studies, Figure 3B). Only a handful of metabolites were uniformly elevated in 5 or more studies (with no findings of significant depletion in any tumor type): four acyl-carnitine species (acetyl-, butyryl-, hexanoyl-, and octanoyl-), carnitine itself, beta-alanine, 5,6-dihydrouracil, citrulline, kynurenine, and lactate (Supplementary Table 1: DifferentialAbundanceSummary). Comparatively fewer metabolites had a tendency towards recurrent depletion in cancers. Four metabolites, including three medium chain fatty acids (pelargonate, caprate, and laurate), were depleted in 4 different studies. These results were in good agreement with a recent meta-analysis (see Figure S4) (Goveia et al., 2016) (e.g. recurrent elevation of lactate and taurine, depletion of myo-inositol, and a general elevation (rather than depletion) of metabolites in tumors compared to normal tissue). Interestingly, Goveia and colleagues (Goveia et al., 2016) observed substantially more frequent elevation of various amino acids (e.g. serine, arginine) than we do here, which may in part be due to their broader coverage of cancer types.
Figure 4. Differential abundance across all cancer types.
Blue circles indicate metabolites depleted in tumors relative to normal tissue, red circles indicate metabolites increased in tumors relative to normal tissue). Only metabolites differentially abundant in 6 or more studies are displayed.
Among the metabolites exhibiting recurrent differential abundance, several have been previously implicated in the development or progression of cancer. Perhaps, the best studied example is lactate, the terminal product of the high-flux, energy- and precursor-producing pathway of glycolysis, and the metabolite originally described by Otto Warburg as elevated in cancerous tissues (Vander Heiden et al., 2009). In contrast to the changes in lactate, which are likely associated with a fundamental rearrangement of energy metabolism in cancer cells, the elevation of kynurenine in many cancer cells is likely not a reflection of changes to energetic demands. Instead, kynurenine is a derivative of tryptophan which is well-known to have immunosuppressive properties through pro-apoptotic effects when secreted into the extracellular milieu, and its elevation suggests recurrent changes to the tumor microenvironment across many cancers (Platten et al., 2012; Belladonna et al., 2007; Chen and Guillemin, 2009).
Pathway-Scale Metabolic Alterations
An alternative approach to understanding the broad patterns of metabolic alterations is through analysis of pathways into which metabolites may be organized. For each study, we mapped metabolites onto KEGG pathways (Supplementary Table 1: Merged IDs), and calculated an aggregate differential abundance score for each pathway (see Methods, Figure S5, Supplementary Table 1: AllPathway). In total, 373 metabolites were identified as belonging to any given KEGG pathway and from a collection of 246 KEGG pathways 179 (73%) were represented by at least one metabolite. Additionally, by jointly comparing KEGG and DrugBank databases as stored in the Pathway Commons database using the paxtoolsr R package (Luna et al., 2016), we were able to identify 70 enzymes (involved in 35 KEGG pathways) producing or consuming substrates in the current study that are targetable by 57 approved or investigational compounds present in DrugBank (Supplementary Table 1: kegg_drugbank_results) (Luna et al., 2016; Cerami et al., 2011; Law et al., 2014). While a large number of pathways are represented minimally, the overall coverage of KEGG remains low with only 373 out of 3330 (11%) KEGG metabolites being represented. Because of low coverage across pathways and with many metabolites missing from each pathway (i.e. a high degree of missing data and low statistical power), we did not assign statistical significance to pathway-level differential abundance scores (in line with prior work (Hakimi et al., 2016)).
We restricted our analysis to well-represented metabolic pathways for which at least 5 metabolites in the pathway were profiled across 6 or more studies for a total of 34 pathways. Pathways associated with sugar metabolism (e.g. fructose/mannose metabolism, glycolysis/gluconeogenesis, and amino sugar and nucleotide sugar metabolism) were generally elevated. Many tumor types also had increases in glutathione metabolism, which produces the critical cellular antioxidant reduced glutathione (GSH) (Singh et al., 2012). A single pathway, fatty acid biosynthesis, had weak, but recurrent depletion of its constituent metabolites across cancers and mirrors the increase of various acyl-carnitines across many cancer types (Figure 3). Acyl-carnitines are generated by beta-oxidation of fatty acids and are used to refuel the TCA cycle with acetyl-CoA. Our data suggests that the general elevation of acyl-carnitines and the concurrent depletion of medium chain fatty acids in tumors may reflect an increased reliance on fatty acid catabolism to replenish the TCA cycle in tumor cells.
A central problem in metabolism research is understanding the connection between metabolite levels and the fluxes of reactions involving those metabolites as substrates/products. In general, flux through a metabolic reaction is dependent (in a complex, nonlinear fashion) on (1) the levels of substrates/products of the reaction, (2) the levels of the enzyme catalyzing the reaction, and (3) the network context, (i.e. the kinetics of the reactions surrounding the flux of interest in the metabolic network). For example, it is not necessarily true that an increase in the levels of a substrate will increase flux through the corresponding substrate-utilizing enzyme (e.g. if transcription of the enzyme is downregulated). Due to this complexity, the inference of fluxes from metabolite levels remains unresolved and has been the focus of numerous studies from our group and others (Reznik et al., 2013; Gerosa et al., 2015; Reaves et al., 2013). Despite these challenges, drawing inferences on changes in flux from the pattern of metabolite changes is instructive for understanding high-level changes in metabolism, and critical for generating experimentally testable hypotheses regarding metabolic flux.
With the above caveats in mind, we began to dissect the patterns of metabolite changes in glycolysis and the TCA cycle, which in addition to producing NADH/FADH2/ATP generate high volumes of metabolic building blocks for biosynthesis (Figure 5). We first noted that while lactate levels are commonly elevated across most cancers, the remaining metabolites in glycolysis were in general not elevated. To evaluate if the changes in metabolite abundances were synchronized across samples, we calculated the correlation between metabolites in glycolysis and lactate. We observed that levels of metabolites in upper glycolysis (e.g. glucose 6-phosphate (G6P) and fructose 6-phosphate (F6P)) and 3-phosphoglycerate were frequently correlated to levels of lactate across most cancer types. In contrast, 2-phosphoglycerate and phosphoenolpyruvate were commonly not positively correlated or negatively correlated to levels of lactate. Taken together, the (1) prevalence of coordinated changes in metabolites from upper glycolysis with lactate, (2) increase in lactate levels in 6/9 studies, and (3) increases in levels of phosphorylated glucose (G6P) in 4/8 studies, suggests that increases in lactate levels may be attributed to increases in glucose uptake and phosphorylation, and increases in glycolytic flux and excretion of lactate. As others have pointed out (Vander Heiden et al., 2009), such an increase in glycolytic flux may provide additional carbon (via overflow into peripheral biosynthetic pathways) needed for growth. Interestingly, one cancer type (clear cell kidney) had a strong (Spearman rho -.6, p-value < 10−14) anti-correlation between levels of citrate and lactate. Clear cell kidney tumors are known to be highly HIF-activated, which can induce phosphorylation and inactivation of pyruvate dehydrogenase (PDH) via the action of pyruvate dehydrogenase kinase (PDK) and lead to preferential shunting of pyruvate into lactate rather than into the TCA cycle.
Figure 5. Central carbon metabolism across cancer types.
(A) Differential abundance (tumor vs. normal) in glycolysis and the TCA cycle across many cancer types. Bars indicate whether the metabolite was higher/lower/unchanged in tumor samples compared to normal tissue; the absence of a bar means the data was missing. BRCA-T: breast data from (Tang et al., 2014), PAAD-H1/2: pancreatic data from (Zhang et al., 2013), PRAD-P: prostate data from (Priolo et al., 2014). Data from 10/11 studies (LGG excluded for no normal samples, in total, 482 tumor samples and 378 normal samples). (B) Sorted log2 ratio of lactate levels in matched tumor/normal samples from 5 cancer types. Approximately 26% of samples have less lactate in tumors compared to normal tissue. In both (A) and (B), analysis was done without regard to whether tumor/normal samples were paired. (C) Frequency of elevation for each metabolite across all matched tumor-normal pairs (343 pairs in total). Kynurenine is elevated in 85% of matched samples.
Unexpected patterns related to tissue-of-origin emerged when contrasting the pattern of metabolomic changes in central carbon metabolism across cancer types. For example, both prostate cancer studies support a drop in citrate levels in prostate tumors compared to normal tissue, a feature that is not evident in any other cancer type. This is in agreement with prior research showing that while normal prostate glandular epithelium secretes large amounts of citrate, depletion of zinc (an inhibitor of mitochondrial aconitase) relieves a “metabolic bottleneck” and leads to an increase in the oxidation of citrate/aconitate to isocitrate (Singh et al., 2006). Similarly, clear cell kidney tumors are the only cancer type to show an increase in metabolites upstream of SDH in the TCA cycle (succinate, citrate, aconitate) and depletion of metabolites downstream of SDH (fumarate and malate). These tumors are known to be highly depleted of mitochondrial DNA and RNA (Reznik et al., 2016, 2017), which suppresses the activity of the mitochondrial respiratory chain and consequently Complex II/SDH. We propose that the sudden shift in metabolite patterns at SDH in clear cell kidney tumors may therefore arise from a bottleneck associated with suppression of the mitochondrial respiratory chain.
In some ways, the depiction of metabolomic changes in Figure 5A may be misleading; while an increase in the abundance of a metabolite may be evident in aggregate across all samples, there may remain individual tumors which are depleted of that metabolite. To better understand the nature of elevated lactate levels in tumors, we leveraged a unique part of our data: 343 matched pairs of tumor/adjacent-normal tissue samples (i.e. each such pair was derived from the same patient). While both a paired and unpaired analyses are informative, a paired analysis normalizes for potential patient-to-patient variation (independent of tumor-to-normal variation) in the levels of a metabolite. Despite elevation of lactate in 6/9 studies, 82/319 (26%) paired tumor/normal samples contained lower levels of lactate than their matched normal tissue counterpart. While most cancer types were enriched for tumors with increased levels of lactate, prostate tumors were the outliers: the majority of prostate tumors had reduced levels of lactate relative to paired adjacent normal tissue. Nevertheless, all studies had > 10% of samples with depleted lactate levels relative to normal tissue (e.g. 32/138 ≈23% of kidney cancer pairs). This heterogeneity in lactate levels compared to normal tissue may partially explain the cancer-type-specific utility of FDG-PET imaging, which relies on elevated glucose uptake (and potentially an increased rate of lactate production) to visualize tumors (Menzel et al., 2013). Interestingly, when repeating the paired analysis above on all metabolites, we found that lactate was the third most frequently elevated metabolite, behind glutamate and kynurenine (Figure 5C).
Metabolic Indicators of Tumor Progression
Tumors become clinically aggressive by acquiring genetic, physiological, and morphological features that enable them to invade foreign tissues and metastasize. Although clinical data was sparsely reported in the studies we examined, five studies reported data on clinical stage or grade, with a greater number of data points available for tumor grade (Supplementary Table 1: ClinFeatures). Importantly, while tumor grading systems vary from one cancer type to the next, the principle underlying grading (examining the abnormal appearance of a cell) is common across all cancer types: higher grade tumors are more aggressive. Therefore, we examined our data for metabolic signals associated with progression of tumors to higher grade, a histological measure of the extent of abnormal appearance of tumor cells. Because of the decreased statistical power associated with looking for associations between metabolite levels and subsets of our tumor data, we adopted a meta-analysis approach. We identified metabolites which had recurrent increases/decreases in metabolite levels with increasing tumor grade across several cancer types. As before, our meta-analysis approach relies strictly on non-parametric methods (that do not make assumptions about the underlying distribution of the data) and is suitable for the metabolomics data used here. Furthermore, we designed our analysis to account for the frequency of imputed data for each metabolite under consideration (see detailed description in Methods).
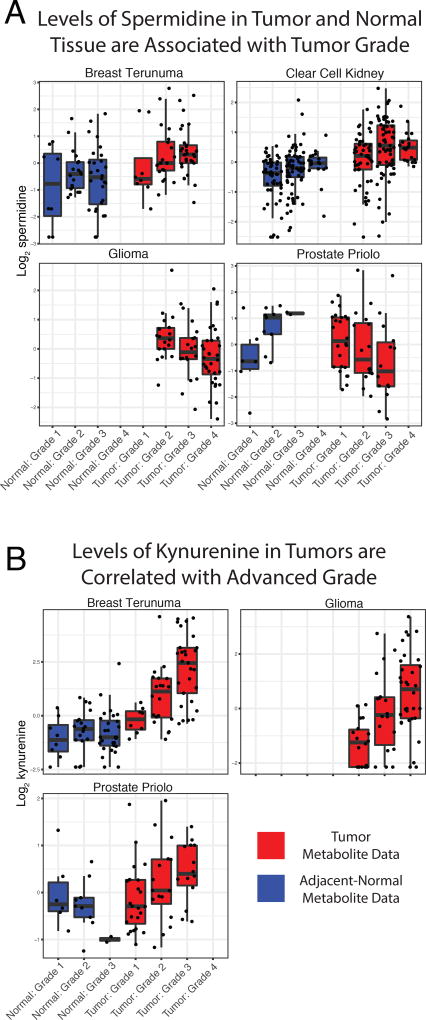
In total, we found 174 tumor tissue metabolites whose abundance were significantly correlated to tumor grade (BH-adjusted meta-p-value < 0.05, Supplementary Table 2). Filtering these post-hoc results further to extract metabolites significantly associated with clinical features across many tumor types, we found 4 metabolites (spermidine, uracil, 3-aminoisobutyrate, and 2-hydroxyglutarate) associated with tumor grade in at least 4 studies, and 57 metabolites associated with tumor grade in 3 studies. However, the number of metabolites significantly associated with grade in each study (post-hoc) was correlated to the number of samples in the study with clinical data, suggesting that this analysis may be biased toward studies with larger samples sizes (Figure S3). Notably, spermidine had a significant association with tumor grade across four different cancer types (breast, kidney, brain, and prostate cancer) (Figure 6A). Spermidine is a polyamine; a class of molecules essential for eukaryotic cell growth as well as tumorigenesis (Mandal et al., 2013). Furthermore, spermidine has been previously reported as a potential biomarker for prostate cancer aggressiveness (Giskeødegård et al., 2013). In many cases the direction of association for a single metabolite often varied between cancer types. For example, 2-hydroxyglutarate was associated with increased grade in breast and kidney tumors, but decreased grade in glioma and prostate tumors. We speculate that differences both in genetic background, as well as in enantiomeric identity, may explain the discordant associations between 2-HG and tumor grade. Specifically, in gliomas, the D-enantiomer of 2-HG is produced by neomorphic IDH mutations, which are themselves associated with a better prognosis. In contrast, the more potent (in terms of inhibitory capacity) L-enantiomer of 2-HG, whose production is induced by hypoxia (Intlekofer et al., 2017), is likely the enantiomer produced by HIF-driven clear-cell renal cell carcinomas (Shim et al., 2014). To evaluate whether differences in the stereoisomeric identity of 2-HG actually account for the differences in association with grade, further enantiomer-specific metabolite profiling will be required.
Figure 6. Metabolic correlation with tumor progression.
A total of 638 of the tissue samples had some type of associated clinical data (392 tumors and 246 normal tissues from 7 studies, including Breast Terunuma, Breast Tang, Kidney, Glioma, Ovarian, Prostate Priolo, and Prostate Sreekumar). Analysis was done without regard to whether tumor/normal samples were paired. Meta-analysis identified 174 metabolites whose abundance in tumors was significantly associated to tumor grade in 1 or more metabolomics studies. (A) Spermidine, a polyamine involved in cell proliferation, had a significant association with tumor grade across four different cancer types (breast, kidney, brain, and prostate cancer). (B) Association of kynurenine levels with tumor grade across several cancers; kynurenine is part of the tryptophan degradation pathway with pro-apoptotic effects on immune cells.
We observed that kynurenine levels in tumor samples were associated with increased tumor grade in three studies (Figure 6B, Breast Terunuma, Glioma, and Prostate Sreekumar, uncorrected p-value < 0.05, meta p-value < 10−4), and were trending toward significance in the other prostate study (uncorrected p-value 0.097). Kynurenine is a metabolic by-product of the degradation of tryptophan by two groups of enzymes: tryptophan dioxygenases and indoleamine 2,3-dixoygenases (Platten et al., 2012). Binding of kynurenine to aryl hydrocarbon receptors (AHRs) suppresses the activity of T-effector cells, as well as indirectly activating regulatory pro-tumorigenic T cells (Nguyen et al., 2014). Kynurenine was particularly unique in our meta-analysis because it was found to be elevated (compared to normal tissue) in 8/10 studies. In these studies, it was more than double the abundance of that found in normal tissue; thus, kynurenine seems to be not only a recurrent differentiating feature of tumor tissue, but also a potential marker of disease progression in cancer (Chuang et al., 2014; Lim et al., 2017; Heng et al., 2016).
The normal tissue and microenvironment surrounding the tumor plays a key role in the development of cancers (Chang et al., 2015; Place et al., 2011; Quail and Joyce, 2013). It is therefore possible that the metabolic demands of a tumor can drive a physiological change in the metabolism of nearby normal tissue. Therefore, we repeated our meta-analysis above, focusing on the association between metabolite levels in the adjacent normal tissue and the pathological grade of the matched tumor. We identified 4 metabolites whose abundance in normal tissue was associated with tumor grade: putrescine, galactose, and 2 fatty acids (margarate and stearate) (uncorrected individual study p-value < 0.05 in at least 2 studies, BH-corrected meta-p-value < 0.1). For putrescine, another polyamine, increased levels in normal tissue were associated with higher grade in both kidney and prostate cancers. Notably, 2-difluoromethylornithine (DFMO) is an enzymatic inhibitor of ornithine decarboxylase, an enzyme necessary for the production of both spermidine and putrescine (Casero and Marton, 2007), and there is some evidence that it may be useful in treatment of a variety of cancers (colorectal and melanoma) as well as a part of combinatorial therapy for brain cancers (Gerner and Meyskens, 2004). Interestingly, clinical associations in normal tissue were sometimes, but not always, mirrored in the analogous tumor tissue (Figure S6A). While many metabolites (e.g. choline) had common patterns of association in kidney tumor and normal tissue (and less frequently in breast and prostate tissues), other metabolites had opposing associations (e.g. spermidine in prostate tumors/normal tissues, Figure 6).
What is a plausible interpretation of the connection between normal tissue metabolism and aggressive tumor features? One explanation is that the nearby tissue changes physiologically in the course of tumor progression, for example in response to cytotoxic therapies. Another, more intriguing, possibility is that there may some interaction between tumor cells and the surrounding normal tissue. To evaluate if this may be the case, we again used metabolite data on 343 matched tumor/normal pairs from the same patient, and evaluated (study-by-study) whether metabolite levels in tumor and normal tissue were correlated. Of the 205 metabolites measured in at least 5/8 studies with paired samples, we identified 8 metabolites that were recurrently correlated between tumor and normal tissues in at least 5 studies (Supplementary Table 1: Pairscores). Of these, a number were metabolites which circulate physiologically through the body (e.g. 1,5-anhydroglucitol, 3-hydroxybutyrate, urea). Thus, our data is suggestive of many correlations between tumor and normal metabolite levels arising because of broad changes to the physiology of the patient. Interestingly, however, alanine was significantly correlated between tumor and normal tissues in the Pancreatic Kamphorst study and this correlation approached statistical significance in a second pancreatic study (Pancreatic Zhang 1). This possibly supports the recent report of shuttling of alanine from nearby pancreatic stellate cells to tumor cells (Sousa et al., 2016).
Discussion
In spite of the diversity of genetic alterations across cancers, it is possible that the development of cancer may place common demands on cellular metabolism, irrespective of the tissue of origin. In this study, we offer a glimpse onto the common patterns and distinguishing features of metabolic reprogramming across cancers.
Our approach to analyzing a compendium of small molecule data derived from mass spectrometry has inherent assumptions, which we addressed to the extent possible. For example, we relied on the normalization and standardization procedures undertaken by the original researchers of a study. While all the data in our study was pre-normalized, three studies contained data that was not imputed or standardized; in those cases, we applied a simple imputation/standardization pipeline. We also implemented a bioinformatic pipeline to align metabolites identified using disparate nomenclature, the code repository for which is publicly available on GitHub (https://github.com/dfci/pancanmet_analysis). Finally, to compare data across studies following unknown and potentially various statistical/distributional properties, we relied heavily on non-parametric statistics that make no assumptions on underlying distributions, and either (1) used changes in abundance relative to normal tissue as the unit of comparison or (2) non-parametrically associated changes in metabolite levels with clinical data. We expect that this pipeline, through collaboration with others in the metabolomics community, can be expanded and improved to more fully address systematic biases and differences in data acquisition.
Our findings illustrate that cancers of widely different types share some common patterns of changes to metabolite levels. A subset of metabolites, including acyl-carnitines, lactate, kynurenine, and taurine, were recurrently higher in abundance across many tumor types (when compared to normal tissue). Comparatively fewer metabolites, including some fatty acids, were recurrently depleted across these same cancer types. These findings echo those from a recent meta-analysis of published cancer metabolomics data (Goveia et al., 2016) (which did not directly analyze the metabolomics data, but aggregated results from individual studies into a meta-analysis). Importantly, the metabolites displaying clear, consistent shifts in abundance across cancers are in the minority. The majority of metabolites, both in central carbon metabolism and in secondary metabolic pathways, had heterogeneous changes relative to normal tissues across tumor types.
We also found that the metabolism of tumors is linked to their clinical presentation and, in particular, to their grade. While it should come as no surprise that tumor metabolism necessarily accommodates and supports the development of aggressive disease and metastasis, it was intriguing to find that some metabolites were indicative of aggressive disease across several cancer types. Our analysis also indicated that the metabolism of adjacent-normal tissue, which is physiologically linked to the tumor by physical location and shared vasculature, is remodeled in patients with high-grade tumors. One potential explanation for the association between metabolite levels in tumor/normal tissue and tumor grade is due to the variation in the degree of infiltrating immune or stromal cells between tumors of different grades. Such infiltration could also, for example, enrich or dilute the abundance of metabolites specific to either tumor or immune/stromal cells. Because one study (Tang et al., 2014) used TCGA-profiled tissue samples, we were able to recover estimates of stromal and immune infiltration for 23 samples from this study and investigate if infiltration was correlated with metabolite levels in this study. Interestingly, while no metabolites reached statistical significance upon multiple-hypothesis correction, several metabolites of high interest (e.g. NADH) were close to significance (Figure S6). Given the relatively small sample size of this dataset (23 samples), we hypothesize that improved statistical power may indeed yield metabolites indicative of or enriched in various components of the tumor microenvironment.
In some tumor types and, in particular, in clear-cell renal cell carcinomas, many metabolites had changes in abundance in normal tissue which were associated with the grade of the patient’s tumor. Much remains to be understood of this phenomenon, including in particular the possibility that tumor and normal tissue may be shuttling nutrients between each other in support of tumor cell proliferation (Sousa et al., 2016). Importantly, observing (or failing to observe) a correlation between steady-state metabolite levels in tumor/normal tissues is not sufficient evidence to conclude nutrient shuttling between the two tissues. Instead, the precise mechanism of shuttling could lead to different kinds of correlations between tumor/normal metabolites; for example, a positive correlation may arise if excess production of a metabolite in the normal tissue is shuttled to the tumor passively by blood vessels. Similarly, a negative correlation could arise if a fixed amount of metabolite produced in the normal tissue is actively transported to the tumor, e.g. by overexpression of a nutrient exporter in the normal tissue. Thus, tumor/normal metabolite correlations should be interpreted with care and would make an interesting subject for future investigation.
One outcome of our work is a merged, standardized metabolomics dataset which can be used by others in the future with an accompanying website (http://www.sanderlab.org/pancanmet/) allowing readers to explore 1) metabolite data stratified by metadata (e.g. HMDB classifications) and by study, 2) univariate and bivariate changes by metabolite and by study, and 3) clinical associations by metabolite and by study. The analysis in this paper has focused on non-parametric, univariate analysis of changes in metabolite levels between tumor and normal samples. Yet, other analyses that respect the limitations of the data (Figure 2) are possible. One example is a correlation analysis, akin to those proposed in several publications using gene expression data (Reznik and Sander, 2015). It is possible that, by examination of changes in covariation patterns between metabolites between different subsets of samples (e.g. tumor/normal, high grade/low grade tumors), the identification of putatively “rewired” metabolic pathways may be possible. Expanding covariation analyses to include other data types (e.g. transcriptomics, proteomics) may yield further insights.
Finally, given the proliferation of clinical genomic sequencing of cancer samples, it seems natural to propose that connections be drawn between genetic alterations and the metabolic landscape of tumors. Several prominent examples now exist of molecular alterations which manifest with, among other things, distinct metabolic phenotypes: KRAS mutations in pancreatic adenocarcinomas, which induce macropinocytotic scavenging of extracellular nutrients (Kamphorst et al., 2015) and hotspot IDH1 and IDH2 mutations which produce 2-hydroxyglutarate and induce DNA hypermethylation via inhibition of α -ketoglutarate-dependent DNA demethylases (Lu et al., 2012; Xu et al., 2011). Furthermore, recent work has demonstrated that metabolic changes arising from a single common genetic lesion are tissue-specific, and can vary substantially from one cancer type to the next (Mayers et al., 2016). Thus, it is likely that other mutation-associated metabolic effects, perhaps subtle but nevertheless impacting tumor viability, remain to be uncovered.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chris Sander (sander.research@gmail.com).
Quantification and Statistical Analysis
Data Imputation and Standardization
All data used in this study was pre-normalized (i.e. no raw mass spectra were analyzed). For data which was already imputed and standardized, we used the data as reported by the original authors. This includes both breast studies (Terunuma et al., 2014; Tang et al., 2014), glioma (Chinnaiyan et al., 2012), kidney (Hakimi et al., 2016), ovary, (Fong et al., 2011), 2 of the pancreatic studies (Zhang et al., 2013), and one of the prostate studies (Priolo et al., 2014). For the bladder study (Putluri et al., 2011), data was obtained normalized after log transformation, with no further imputation required. For this study, normalized data was simply re-exponentiated. For two studies, PAAD (Kamphorst et al., 2015) and PRAD (Sreekumar et al., 2009), for which no imputation was completed, we applied the following imputation and standardization procedure. For each metabolite, imputed values were set equal to the minimum measured abundance of that metabolite; additional statistics on imputed metabolites are shown in Figure S1. Then, all measurements of the metabolite were divided by the median abundance of that metabolite. The entire data import pipeline can be replicated using the code provided in the GitHub repository.
Because all subsequent statistical analyses were non-parametric, no assumptions were made on the underlying distribution of the data.
Metabolite ID Mapping
In addition to data standardization, a bioinformatic challenge in the current work was the identification of metabolites profiled across multiple studies. Unlike other high-throughput technologies which can measure the abundance of all relevant species in a sample (e.g. RNA sequencing), metabolomic profiling samples only a fraction of all compounds in the metabolite. More importantly, metabolites are referred to by different synonymous names and database identifiers. To address this issue, we used the Chemical Translation Service (Wohlgemuth et al., 2010) to retrieve synonymous identifiers of each metabolite. These identifiers were then used to assemble a meta-dataset of all metabolomics data, thus “aligning” metabolites sharing common identifiers. Manual inspection of the aligned dataset confirmed that our method correctly matched metabolites across different studies. All code for data standardization and alignment is provided on GitHub (https://github.com/dfci/pancanmet_analysis).
These mapped identifiers were used in conjunction with the paxtoolsr R Bioconductor package (https://www.bioconductor.org/packages/release/bioc/html/paxtoolsr.html) to map identifiers onto the Pathway Commons Version 8 database, which includes both KEGG and DrugBank interactions to identify drug targetable pathways.
Differential Abundance Tests
Differential abundance was calculated using the ratio of the average abundance of a metabolite in tumor tissue to the average abundance of a metabolite in normal tissue. As with other analysis in the manuscript, statistical significance was assessed using non-parametric Mann-Whitney U-tests. P-values were adjusted for multiple-hypothesis testing using the Benjamini-Hochberg procedure.
Pathway Differential Abundance Score
The differential abundance (DA) score for a pathway was defined as DA = (I-D)/S, where I is the number of measured metabolites in a pathway that increased in abundance relative to normal tissue, D is the number which decreased, and S is the total number of measured metabolites. A DA score of 1 indicates that all metabolites increased in abundance, whereas a score of −1 indicates that all metabolites decreased in abundance, relative to normal tissue.
Calculation of Metabolic Variation (MAD Score)
Metabolic variation analysis was completed for each study with paired tumor/adjacent-normal samples from the same patient. For each study, we restricted analysis to such paired data and partitioned the metabolomics data into measurements from tumor and normal tissue samples. Then, for each metabolite m with measurements from n samples, X = X1, X2, …, Xn, a normalized median absolute deviation (MAD) was calculated according to the formula
The MAD was calculated for each metabolite m separately for tumor (MADT) and normal (MADN) samples. The ratio of these two MAD scores for each metabolite m, MADr , was then calculated. The distribution of MADr scores indicates whether tumor samples show significantly different levels of variation than normal tissues (i.e. if the distribution of MADr scores is skewed to be significantly less than 1, this indicates that tumor samples exhibit less variation than normal samples). The results of the MAD analysis were invariant to the exclusion of highly-imputed metabolites.
Clinical Features
In general, clinical data was imported from the datasets reported by original investigators. For prostate tumors specifically (Prostate Priolo and Prostate Sreekumar studies), we converted Gleason score to tumor grade as follows (3+3 = Grade 1; 3+4,4+3 = Grade 2; 4+4,5+4 = Grade 3).
Association with Tumor Grade
Different statistical tests were applied to identify correlation between metabolites and clinical features depending on the level of censoring (i.e. imputation) in the data. If there was less than 20% censoring within the tested set, the Jonckheere-Terpstra test (a non-parametric test which uses permutations to calculate the p-value) was used to examine ordered differences among classes. If there was more than 20% but less than 80% censored data, an exact log-rank trend test was used with left censored data. Both tests assume a metabolite should increase/decrease monotonically with stage/grade. If there is more than 80% censoring, no test was used, and the metabolite was ignored. To summarize the tests across cohorts Fisher’s method was used to combine p-values. Note that resulting combined p-value is parametric. Since, we were interested in finding associations of a common sign (e.g. consistently positive correlation across multiple studies), one-sided p-values were calculated first, aggregated into a combined p-value using Fisher’s method (Loughin, 2004), and then transformed to a two-sided combined p-value.
Data and Software Availability
All code associated with the analysis is available at https://github.com/dfci/pancanmet_analysis/. This code replicates both the merging of metabolomics data (in the import/folder) and the analysis (in the analysis/) folder.
Additional Resources
Interactive visualization of the data and associated analysis is available at https://www.sanderlab.org/pancanmet.
Supplementary Material
Supplementary Table 1 - Merged Metabolomics Data, related to Figure 1 (merged_metabolomics.csv)
Supplementary Table 1 - Tumor/Normal Pairs, related to Figure 1 (tumor_normal_pairs.csv) Supplementary Table 1 - Clinical Data, related to Figure 1 (clinfeatures.csv)
Supplementary Table 1 - Differential Abundance Results, related to Figure 3 (DifferentialAbundanceSummary.csv)
Supplementary Table 1 - Metabolite Classifications, related to STAR Methods (hmdbClassification.csv)
Supplementary Table 1 - Figure S5 Data: Differential Abundance Scores, related to Figure 4 (AllPathway_diffAbundanceBig.csv)
Supplementary Table 1 - KEGG/DrugBank Pharmacology Coverage Data, related to STAR Methods (kegg_drugbank_results.txt)
Supplementary Table 1 - Metabolite ID Dictionary, related to STAR Methods (Sept152015/merged_IDs.csv)
Supplementary Table 1 - Tumor/Normal Correlation Scores, related to Figure 5 (pairscores.csv)
Supplementary Table 1 - Adjacent Metabolite Analysis, related to Figure 3 (Covariation_Results.csv)
Supplementary Table 2 - Clinical Data Analysis, related to Figure 6 (updated results stage and grade Jan 2017 max1.xlsx)
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Breast: Pre-normalized metabolomics data collected in tumor samples | Tang et al., 2014 | N/A |
| Breast: Pre-normalized metabolomics data collected in tumor samples | Terunuma et al., 2014 | N/A |
| Glioma: Pre-normalized metabolomics data collected in tumor samples | Chinnaiyan et al., 2012 | N/A |
| Kidney: Pre-normalized metabolomics data collected in tumor samples | Hakimi et al., 2016 | N/A |
| Ovary: Pre-normalized metabolomics data collected in tumor samples | Fong et al., 2011 | N/A |
| Pancreatic: Pre-normalized metabolomics data collected in tumor samples (2-datasets) | Zhang et al., 2013 | N/A |
| Prostate: Pre-normalized metabolomics data collected in tumor samples | Priolo et al., 2014 | N/A |
| Bladder: Pre-normalized metabolomics data collected in tumor samples | Putluri et al., 2011 | N/A |
| Pancreatic: Pre-normalized metabolomics data collected in tumor samples | Kamphorst et al., 2011 | N/A |
| Prostate: Pre-normalized metabolomics data collected in tumor samples | Sreekumar et al., 2009 | N/A |
| Human Metabolome Database: Metabolite classifications | http://www.hmdb.ca | Version 3.0 |
| DrugBank: Drug compound classifications | http://drugbank.ca | Version 5.0.3 |
| Software and Algorithms | ||
| Data standardization, nomenclature alignment, and analysis pipeline | This paper, https://github.com/dfci/pancanmet_analysis | N/A |
| Chemical Translation Service | http://cts.fiehnlab.ucdavis.edu/ | N/A |
| Pathway Commons Database accessed through paxtoolsr R package | https://www.bioconductor.org/packages/release/bioc/html/paxtoolsr.html | Pathway Commons: Version 8 |
| Data exploration site | This paper,http://www.sanderlab.org/pancanmet | N/A |
| KEGG Pathways accessed through KEGGREST | https://www.bioconductor.org/packages/release/bioc/html/KEGGREST.html | N/A |
Highlights.
-
-
Computational pipeline to integrate cancer metabolomics data across studies
-
-
Free, open-source pipeline, data, and online visualization portal available
-
-
Recurrent patterns of differentially abundant metabolites across cancer types
-
-
Discovery of metabolites associated with aggressive disease across cancer types
Acknowledgments
We would like to thank each of the original investigators and their teams who contributed their data to this work and kindly offered us their feedback, including but not limited to James Hsieh (kidney), Stefan Ambs (breast), Jeffrey R. Marks (breast), Max Loda (prostate), Arun Sreekumar (bladder), Josh Rabinowitz (pancreatic), Prakash Chinnaiyan (glioma), Arul Chinnaiyan (prostate), S. Perwez Hussain (pancreatic), and Sham Kakar (ovarian). The work was supported by Ruth L. Kirschstein National Research Service Award (grant no. F32 CA192901 to AL), and the National Resource for Network Biology (NRNB) from the National Institute of General Medical Sciences (NIGMS) (grant no. P41 GM103504 to CS). CJC was supported by NIH grant CA125123. ER, AAH, and CS were supported by National Cancer Institute (P30-CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ER, AL, and CS conceived the project. ER, AL, BAA, EML, KL, IO, CJC, AAH, and CS contributed to analysis. ER, AL, and CS wrote the manuscript. All authors approved of the manuscript.
References
- Belladonna ML, Puccetti P, Orabona C, et al. Immunosuppression via tryptophan catabolism: the role of kynurenine pathway enzymes. Transplantation. 2007;84(1 Suppl):S17–20. doi: 10.1097/01.tp.0000269199.16209.22. [DOI] [PubMed] [Google Scholar]
- Casero RA, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature Reviews Drug Discovery. 2007;6(5):373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39(Database issue):D685–90. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Qiu J, O’Sullivan D, et al. Metabolic Competition in the Tumor Microen-vironment Is a Driver of Cancer Progression. Cell. 2015;162(6):1229–41. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guillemin GJ. Kynurenine pathway metabolites in humans: disease and healthy States. International journal of tryptophan research : IJTR. 2009;2:1–19. doi: 10.4137/ijtr.s2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan P, Kensicki E, Bloom G, et al. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer research. 2012;72(22):5878–88. doi: 10.1158/0008-5472.CAN-12-1572-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, Mahieu NG, Johnson SL, Patti GJ. After the feature presentation: technologies bridging untargeted metabolomics and biology. Current opinion in biotechnology. 2014;28:143–8. doi: 10.1016/j.copbio.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang S-C, Fanidi A, Ueland PM, et al. Circulating Biomarkers of Tryptophan and the Kynurenine Pathway and Lung Cancer Risk. Cancer Epidemiology Biomarkers & Prevention. 2014;23(3):461–468. doi: 10.1158/1055-9965.EPI-13-0770. [DOI] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nature genetics. 2013;45(10):1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley BA, Doroshow JH. Molecular Analysis for Therapy Choice: NCI MATCH. Seminars in Oncology. 2014;41(3):297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Science advances. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong MY, McDunn J, Kakar SS. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PloS one. 2011;6(5):e19963. doi: 10.1371/journal.pone.0019963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1319196111. 1319196111–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nature Reviews Cancer. 2004;4(10):781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Gerosa L, Haverkorn van Rijsewijk BR, Christodoulou D, et al. Pseudo-transition Analysis Identifies the Key Regulators of Dynamic Metabolic Adaptations from Steady-State Data. Cell Systems. 2015;1(4):270–282. doi: 10.1016/j.cels.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Giskeødegård GF, Bertilsson H, Selnæs KM, et al. Spermine and Citrate as Metabolic Biomarkers for Assessing Prostate Cancer Aggressiveness. PLoS ONE. 2013;8(4):e62375. doi: 10.1371/journal.pone.0062375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goveia J, Pircher A, Conradi L-C, et al. Meta-analysis of clinical metabolic profiling studies in cancer: challenges and opportunities. EMBO molecular medicine. 2016;8(10):1134–1142. doi: 10.15252/emmm.201606798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer discovery. 2014;4(5):554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, McIntyre A, van Stiphout RGPM, et al. Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia. Genome Biology. 2016;17(1):140. doi: 10.1186/s13059-016-0999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi A, Reznik E, Lee C-H, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell. 2016;29(1):104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Heng B, Lim CK, Lovejoy DB, et al. Understanding the role of the kynurenine pathway in human breast cancer immunobiology. Oncotarget. 2016;7(6):6506–20. doi: 10.18632/oncotarget.6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31(6):522–9. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Wang B, Liu H, et al. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nature Chemical Biology. 2017;13(5):494–500. doi: 10.1038/nchembio.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalbert LE, Elkhaled A, Phillips JJ, et al. Metabolic Profiling of IDH Mutation and Malignant Progression in Infiltrating Glioma. Scientific reports. 2017;7:44792. doi: 10.1038/srep44792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer research. 2015;75(3):544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic acids research. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Thiessen PA, Bolton EE, et al. PubChem Substance and Compound databases. Nucleic Acids Research. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research. 2014;42(Database issue):D1091–7. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qiu B, Lee DSM, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CK, Bilgin A, Lovejoy DB, et al. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Scientific Reports. 2017;7:41473. doi: 10.1038/srep41473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughin TM. A systematic comparison of methods for combining p-values from independent tests. Computational Statistics & Data Analysis. 2004;47(3):467–485. [Google Scholar]
- Lu C, Venneti S, Akalin A, et al. Induction of sarcomas by mutant IDH2. Genes & Development. 2013;27(18):1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Babur Ö, Aksoy BA, Demir E, Sander C. PaxtoolsR: pathway analysis in R using Pathway Commons. Bioinformatics. 2016;32(8):1262–1264. doi: 10.1093/bioinformatics/btv733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proceedings of the National Academy of Sciences. 2013;110(6):2169–2174. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304) doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel MI, Farrell EV, Janich MA, et al. Multimodal assessment of in vivo metabolism with hyperpolarized [1-13C]MR spectroscopy and 18F-FDG PET imaging in hep-atocellular carcinoma tumor-bearing rats. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2013;54(7):1113–9. doi: 10.2967/jnumed.112.110825. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, Nakahama T, Le DH, et al. Aryl hydrocarbon receptor and kynurenine: recent advances in autoimmune disease research. Frontiers in immunology. 2014;5:551. doi: 10.3389/fimmu.2014.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature communications. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metabolism. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast cancer research : BCR. 2011;13(6):227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer research. 2012;72(21):5435–40. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo C, Pyne S, Rose J, et al. AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer research. 2014;74(24):7198–204. doi: 10.1158/0008-5472.CAN-14-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putluri N, Shojaie A, Vasu VT, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer research. 2011;71(24):7376–86. doi: 10.1158/0008-5472.CAN-11-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine. 2013;19(11):1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves ML, Young BD, Hosios AM, Xu Y-F, Rabinowitz JD. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature. 2013;500(7461):237–41. doi: 10.1038/nature12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, Mehta P, Segrè D. Flux Imbalance Analysis and the Sensitivity of Cellular Growth to Changes in Metabolite Pools. PLoS Computational Biology. 2013;9(8):e1003195. doi: 10.1371/journal.pcbi.1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, Miller M, Ådđenbabaoħlu Y, et al. Mitochondrial DNA copy number variation across human cancers. eLife. 2016;5 doi: 10.7554/eLife.10769. FEBRUARY2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, Sander C. Extensive Decoupling of Metabolic Genes in Cancer. PLoS Computational Biology. 2015;11(5) doi: 10.1371/journal.pcbi.1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, Wang Q, La K, Schultz N, Sander C. Mitochondrial respiratory gene expression is suppressed in many cancers. eLife. 2017;6 doi: 10.7554/eLife.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E-H, Livi CB, Rakheja D, et al. L-2-Hydroxyglutarate: An Epigenetic Modifier and Putative Oncometabolite in Renal Cancer. Cancer Discovery. 2014;4(11):1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Desouki MM, Franklin RB, Costello LC. Mitochondrial aconitase and citrate metabolism in malignant and nonmalignant human prostate tissues. Molecular cancer. 2006;5:14. doi: 10.1186/1476-4598-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Khan AR, Gupta AK. Role of glutathione in cancer pathophysiology and therapeutic interventions. Journal of experimental therapeutics & oncology. 2012;9(4):303–16. [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–83. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Sullivan LB, Gui DY, Vander Heiden MG. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nature reviews. Cancer. 2016 doi: 10.1038/nrc.2016.85. [DOI] [PubMed] [Google Scholar]
- Tang X, Lin C-C, Spasojevic I, et al. A joint analysis of metabolomics and genetics of breast cancer. Breast cancer research : BCR. 2014;16(4):415. doi: 10.1186/s13058-014-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. The Journal of clinical investigation. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, N.Y.) 2009;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, et al. HMDB: the Human Metabolome Database. Nucleic acids research. 2007;35(Database issue):D521–6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth G, Haldiya PK, Willighagen E, Kind T, Fiehn O. The Chemical Translation Service-a web-based tool to improve standardization of metabolomic reports. Bioin-formatics (Oxford, England) 2010;26(20):2647–8. doi: 10.1093/bioinformatics/btq476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of a -ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, He P, Tan H, et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(18):4983–93. doi: 10.1158/1078-0432.CCR-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwok-Shing Ng P, Kucherlapati M, et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell. 2017;31(6):820–832. doi: 10.1016/j.ccell.2017.04.013. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 - Merged Metabolomics Data, related to Figure 1 (merged_metabolomics.csv)
Supplementary Table 1 - Tumor/Normal Pairs, related to Figure 1 (tumor_normal_pairs.csv) Supplementary Table 1 - Clinical Data, related to Figure 1 (clinfeatures.csv)
Supplementary Table 1 - Differential Abundance Results, related to Figure 3 (DifferentialAbundanceSummary.csv)
Supplementary Table 1 - Metabolite Classifications, related to STAR Methods (hmdbClassification.csv)
Supplementary Table 1 - Figure S5 Data: Differential Abundance Scores, related to Figure 4 (AllPathway_diffAbundanceBig.csv)
Supplementary Table 1 - KEGG/DrugBank Pharmacology Coverage Data, related to STAR Methods (kegg_drugbank_results.txt)
Supplementary Table 1 - Metabolite ID Dictionary, related to STAR Methods (Sept152015/merged_IDs.csv)
Supplementary Table 1 - Tumor/Normal Correlation Scores, related to Figure 5 (pairscores.csv)
Supplementary Table 1 - Adjacent Metabolite Analysis, related to Figure 3 (Covariation_Results.csv)
Supplementary Table 2 - Clinical Data Analysis, related to Figure 6 (updated results stage and grade Jan 2017 max1.xlsx)