Abstract
Oral microbiome is essential for maintenance of oral cavity health. Imbalanced oral microbiome causes periodontal and other diseases. It is unknown whether oral microbiome affect oral stem cells function. In this study, we used a common clinical anti-biotic treatment approach to alter oral microbiome ecology and examine whether oral mesenchymal stem cells (MSCs) are affected. We found that altered oral microbiome resulted gingival MSCs deficiency, leading to a delayed wound healing in male mice. Mechanistically, oral microbiome release LPS that stimulates the expression of microRNA-21 (miR-21) and then impair the normal function of gingival MSCs and wound healing process through miR-21/Sp1/TERT pathway. This is the first study indicate that interplay between oral microbiome and MSCs homeostasis in male mice.
Keywords: oral microbiota, mesenchymal stem cells, proliferation, wound healing
INTRODUCTION
The oral cavity is colonized by a complex microbial community that grows as diverse biofilms on mucosal and dental surfaces. The oral microbiome are found naturally in health, and forms an ecosystem that carries a broad range of functions indispensable for the health of the host [1]. Oral diseases such as gingivitis, periodontitis and oral mucositis are suggested to be associated with the shifts in the microbial ecosystem from a healthy state to a diseased state [2,3]. Wounds are common in oral cavity, caused by either diseases or surgery. At wound sites, both the oral epithelium and connective tissue response to microorgansims. Mesenchymal stem cells belong to a primitive cell type originating from the mesodermal germ layer, and are characterized by their capacities for self-renewal and differentiation into multiple cell lineages. The multilineage differential potential and paracrine property of MSCs facilitates them to be an ideal tool for regenerative medicine [4–6]. It is widely accepted that MSCs play a critical role in wound healing and tissue repair [7, 8]. However, the effects of oral mucosa-microbe interactions on oral MSCs function and wound healing remains poorly defined.
Antibiotic treatment is commonly used in various inflammatory diseases. Studies have revealed that dysregulation of microbiota through antibiotic use is linked with diseases in multiple organs both in animals and humans [9, 10]. Because of the important link between oral microbiota and oral health, we hypothesize that ecological balance of oral microbiota plays important role in maintaining gingival MSCs function. Gingival MSCs are a population of precursor cells from gingiva and exhibit stem cell-like properties as MSCs derived from bone marrow. The characteristics include expression of mesenchymal stem cell surface markers, colony-forming ability, self-renew, and multi-lineage differentiation capacity [11–13]. Compared with MSCs derived from other dental tissues, gingival MSCs exhibit a higher proliferative ability and can be easily expanded ex vivo [11,12], therefore these cells might serve as a unique source of stem cells for regenerative medicine. To test the effect of oral microbiota on oral MSCs, we used a common clinical anti-biotic treatment approach to alter oral microbiome ecology in mice and examined whether gingival MSCs function are affected. We also established a critical size of wound on mouse palate to examine the alteration in oral microbiota and investigated how the alteration impact wound healing process. We found altered oral microbiome release LPS that stimulates the expression of micro RNA-21 (miR-21) and then impairs gingival MSCs normal function and wound healing capability through miR-21/Sp1/TERT pathway. This is the first study which indicates the interplay between oral microbiome and MSCs homeostasis in male mice, and the results suggests antibiotics-induced derangement of the oral microbiota as a potential environmental risk factor for MSC-mediated tissue repair.
MATERIALS AND METHODS
Mice
Eight-week-old wild-type C57BL/6 male mice were used. Mice were kept at standard conditions according to the institutional guidelines of the Animal Care and Welfare Committee of the School of Stomatology, Capital Medical University for the use and care of live animals. All animal experiments were performed under the institutionally approved protocols for the use of animal research.
Antibiotic treatment
For ablation of oral bacteria in mice, an antibiotic cocktail of 1300 mg/L of metronidazole and 660 mg/L of levofloxacin was administered orally in the drinking water for 2 weeks, followed by 1 days without antibiotics and then the subsequent experiments were conducted.
16S rRNA sequencing and analysis
The oral mucosal samples were obtained from a total of 14 mice, 7 for each group. Oral bacteria were washed from the surfaces of teeth and oral mucosa using PBS. Microbial genomic DNA was extracted using the E.Z.N.A.® DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s instruction. The V3–V4 hypervariable regions of the 16S rRNA gene were subjected to high-throughput sequencing by Beijing Allwegene Tech, Ltd (Beijing, China) using the Illumina Miseq PE300 sequencing platform (Illumina, Inc., CA, USA). The extraction of high-quality sequences was firstly performed with the QIIME package (Quantitative Insights Into Microbial Ecology) (v1.2.1). The unique sequence set was classified into operational taxonomic units (OTUs) under the threshold of 97% identity using UCLUST. The Student’s t-test was used to calculate alpha- and beta-diversity. Significance of categorical variables was determined using the Kruskal-Wallis Test.
Wounding and Morphometric Analysis of Palatal Wound Tissue
Mice were anesthetized with an injection of chloral hydrate (40 mg/g i.p.) in physiological saline, after which a region of mucosa approximately 2.5 mm by 3.0 mm was excised from the hard palate between the upper molars by means of a surgical blade. To determine the healing process, maxillary tissues were harvested 5, 8, and 14 days after wounding. The images of the maxillae were captured using a stereoscopic microscope (Olympus, SZX12, Tokyo, Japan). The unhealing area of the wound was measured at various time points after the initial wounding. Maxillary tissues were then fixed in 4% paraformaldehyde and embedded in paraffin, cut into 5-μm sections in the sagittal plane through the midline of the palate, and stained with haematoxylin and eosin. Each group included 6 mice and the experiments were repeated three times.
BrdU injection and immunostaining
After 3 days of wounds were made, animals received intraperitoneal injections of BrdU at a concentration of 100 mg/kg body weight once a day for 3 consecutive days. Animals then were sacrificed. Wound tissues were collected, fixed and embedded. 5-μm sections were cut and routinely prepared for immunohistochemical staining. Anti-BrdU antibody (1:10, Novus, USA) and anti-CD146 antibody (1:200, abcam, USA) were used as primary antibody and Polymer-HRP&AP Double Staining Kit (GBI Labs, Bothell, USA) was used to detect BrdU and CD146.
Gingival MSCs isolation and culture
Gingival tissues were isolated from mice treated with or without antibiotics. The tissues were minced into fragments and digested at 37°C for 1 h in sterile PBS containing 3 mg/ml collagenase I (Sigma-Aldrich) and 4 mg/ml dispase II(Sigma-Aldrich). The dissociated cell suspension was filtered through a 70-μm cell strainer (Falcon), plated on 10-cm petri dishes (Corning) with complete α-MEM (Invitrogen) containing 10% FBS (Invitrogen), 100 U/ml penicillin/100 μg/ml streptomycin (Invitrogen), and 2 mM L-glutamine, and cultured at 37°C in a humidified tissue culture incubator with 5% CO2. Cells from 2nd passage were used in the experiments.
Western Blot Analysis
Gingival tissues and cells were lysed in RIPA buffer. The method for western blot was described previously [14]. Proteins of interest were detected using anti-TERT (1:1000, Santa Cruz), anti-SP1 (1:2000, Novus). β-actin was used as control and detected with anti-β-actin antibody (1:2000, Abcam).
CFU-F assay
MSCs isolated from gingival tissues were seeded on 25-cm culture flasks. After 14 days, the cells were washed with PBS and stained with a mixture of 0.1% toluidine blue (Sigma, USA) and 2% paraformaldehyde solution. Total colony numbers were counted per flask and only colonies containing >50 cells were considered as single colony clusters.
EdU assay for cell proliferation
Cells were cultured in 6-well plates for 2–3 days. Then the cells were harvested and incubated with EdU solution (1:1000, Invitrogen, Thermo Fisher Scientific) for 24 hours and stained with a EdU Assay Kit (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instruction. The cells were analyzed with a flow cytometer (BD Inmmunocytometry Systems, San Jose, CA). The number of EdU-positive cells was indicated as a percentage to the total cell number. The assay was done in duplicate in each group from at least three dependent experiments.
CFSE analysis for cell proliferation
CFSE proliferation assay was performed according to the manufacturer’s instruction. (CFSE, Invitrogen, USA). The method was described previously [14].
Determination of percentage of apoptotic cells
To detect apoptotic cells, we utilized the Annexin V Apoptosis Detection Kit FITC (eBioscience, San Diego, CA) according to the manufacturer’s instructions.
Cell Migration Assay
Cells were cultured as confluent monolayers and then wounded by using 200-μl sterile pipette tip to scratch the monolayers. After wounding, cells were washed with PBS and incubated in culture medium for 36 h. Average rates of wound closure were calculated from 3 independent experiments. The assay was done in duplicate in each group from at least three dependent experiments.
LPS determination
LPS was quantified using a commercially available ELISA Kit (CUSABIO, Wuhan, China), according to the manufacturer’s protocol. Protein was extracted from gingival tissues and serum was obtained from blood sample by eyeball extirpating. Samples were collected from 6 animals for each group. The experiments were repeated three times.
MicroRNA mimics and inhibitor transfection
MiR-21 mimics (sense 5′-UAGCUUAUCAGACUGAUGUUGA-3′ and anti-sense 5′-AUCGAAUAGUCAGACUACAACU-3′), miR-21 inhibitor (5′-AUCGAAUAGUCUGACUACAACU-3′), control mimics (sense 5′-UUUGUACUACACAAAAGUACUG-3′ and anti-sense 5′-AAACAUGAUGUGUUUUCAUGAC-3′), and contol inhibitor (5′-AAACAUGAUGUGUUUUCAUGAC-3′), were purchased from RiboBio (Guangzhou, China). The transfection was performed using Lipofectamine™ 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Twenty-four or 48 h after transfection, the cells were harvested for further experiments.
Reverse Transcriptase-polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from Gingival MSCs using Trizol reagents (Invitrogen, USA) and reverse transcribed into cDNA using PrimeScript™RT reagent Kit with gDNA Eraser (Takara, Dalian, China). The real-time PCR reactions were performed using the SYBR Premix Ex Taq™ II (Takara, Dalian, China) and IcycleriQ Multi-color Real-time PCR Detection System. The primers were synthesized by Sangon Biotech: Sp1 (5′-TGAGACAGCAGGTGGAGAAG-3′, 5′-GGCTCTTCCCTCACTGTCTT-3′); tert (5′-TGCTGGACACTCAGACTTTGGA-3′, 5′-TTCAACCGCAAGACCGACA-3′); GAPDH (5′-TGAAGCAGGCATCTGAGGG-3′, 5′-CGAAGGTGGAAGAGTGGGAG-3′). Primers for miR-21 and U6 are purchased from RiboBio (Guangzhou, China).
Chromatin Immunoprecipitation Assays
Cells were seeded on 10 cm dishes and fixed by addition of 1% paraformaldehyde to the culture medium for 10 min. Cells were then washed in cold PBS supplemented with protease inhibitor cocktail and scraped from the plate gently. Cell lysis and chromatin immunoprecipitation were performed using a ChIP Assay Kit (Millipore, USA). To fragment chromatin, cells were sonicated in 30 s bursts with 1 min cooling on ice for a total time of 4 min. For immunoprecipitations, Sp1 antibody (1:100) was used to capture protein-DNA complexes, and isotype-matched IgG was served as negative control. All resulting precipitated DNA samples were quantified by real-time PCR and expressed as the percentage of input DNA. The primers for three Sp1 binding sites on tert promoter are: Sp1-site1 forward, 5′-CACCAGCATTGTGACCATCA-3′ and reverse, 5′-GCGGACCAAGCGTTGTAG-3′; Sp1-site2 forward, 5′-TTCCGCTACAACGCTTGG-3′ and reverse, 5′-ACTGAGAGTCCACGACGAA-3′; Sp1-site3 forward, 5′-GCTTGGTCCGCCTGAAT-3′ and reverse, 5′-GACCCAGGCCACTGAGA-3′.
Statistics
For normal data, Student’s t-test was conducted to compare the differences between the means of two groups, while one-way analysis of variance analysis was used when comparing three or more means. For non-parametric data, the Mann-Whitney U-test was used for comparisons between groups. Analysis was performed with the SPSS 13.0 Software. P value less than 0.05 was considered significant.
RESULTS
Altered oral microbiota in mice treated with antibiotics
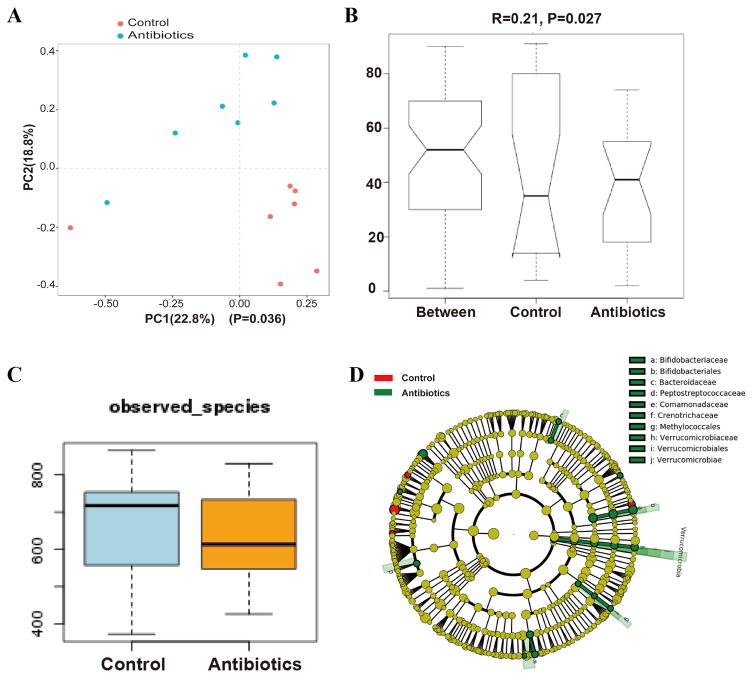
Oral microbiota from oral mucosa tissue samples was collected after antibiotics treatment to analysize the differences in oral bacterial communities in mice treated with antibiotics and control mice. 16S rRNA gene sequencing analysis revealed that in spite of individual variations in microbiota, a general discrete clustering pattern of bacterial taxa was depicted between mice treated with antibiotics and the control group, based on principal component analysis (PCA) (Figure 1A). ANOISM analysis further demonstrated that the oral microbiota between control and antibiotics-treated mice was significantly different (Figure 1B). We then examined changes in commensal microbiota diversity between the two groups. Although there is no statistical difference between the two groups, the microbial communities in antibiotics treatment group were found to have fewer OTUs (Figure 1C). The significant differences in microbial community composition were due mainly to the enrichment of antibiotics-treated mice in Verrucomicrobiace, Bacteroidaceae and Methylococcales, which mostly belong to gram-negative bacteria, while the predominant bacterial taxa in control mice were not defined (Figure 1D).
Figure 1.
Altered oral microbiota in mice treated with antibiotics. (A): A principal coordinate analysis (PCA) based on the weighted UniFrac distance values. The microbial communities in the oral cavity of mice treated with antibiotics (blue dot) cluster differently from the microbial communities in the oral cavity of control mice (red dot) (p=0.036). (B): ANOISM analysis showed that the oral microbiota between the two groups were significantly different (R=0.21, p=0.027). (C): Diversity of the microbiota (Observed OTUs) in control and antibiotics-treated mice. The microbial communities in antibiotics treatment group had fewer OTUs, but there is no statistical difference between the two groups (p>0.05). (D): The microbial composition variation was compared using the LEfSe online tool. The difference in the microbial community at the genus level was mainly due to Verrucomicrobiace, Bacteroidaceae and Methylococcales.
Microbiome imbalance impairs wound-healing process
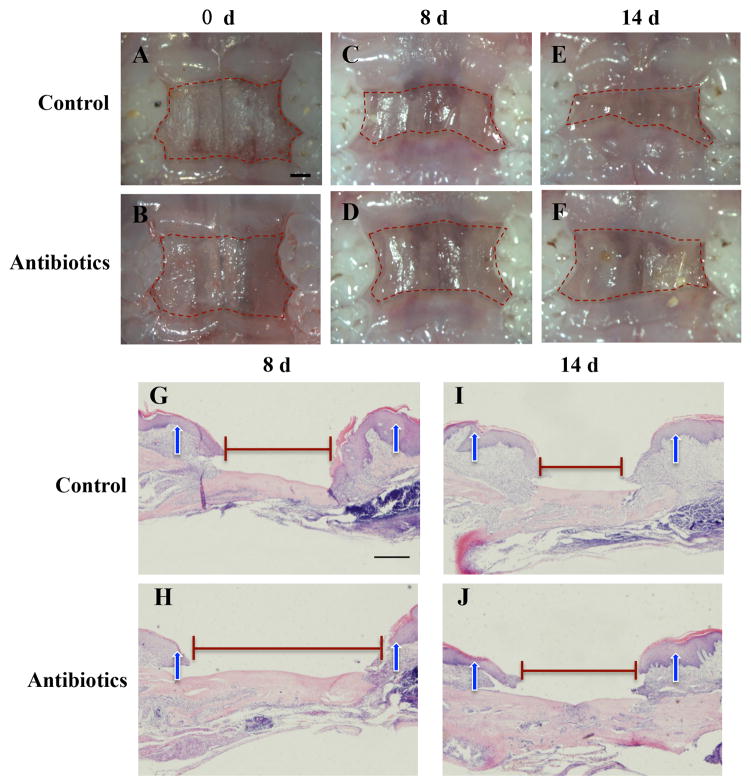
To eliminate the direct impact of antibiotics on wound healing, palatal wounds were made 1 day after the end time of antibiotics treatment when the drugs were completely metabolized in the body. Wound healing was assessed at 8 and 14 days after surgery. Macroscopic observation showed that antibiotics-treated mice exhibited a decrease in percentage of wound closure after day 8 and day 14 post wounding compared with control group (Figure 2A–2F, Table 1). These macroscopic findings were confirmed by histological assessment at 8 and 14 days after surgery. HE staining showed large residual defect of the palatal wound was observed in mice treated with antibiotics compared with that in control mice (Figure 2G–2J).
Figure 2.
Microbiome imbalance impairs wound-healing process. (A–F): Macroscopic observation showed that antibiotics-treated mice exhibited a decrease in percentage of wound closure after day 8 and 14 post wounding compared with control group. Scale bar = 1 mm. (G–J): Representative photomicrographies of H&E-stained sections evidencing time-course of wound closure. Scale bar = 100 μm. Arrowheads show the original wound sites.
Table 1.
Unhealing area/original area
| Control | Antibiotics | P value | |
|---|---|---|---|
| 8 day | 0.569 (0.480–0.734) | 0.860 (0.829–0.983) | 0.038 |
| 14 day | 0.419 (0.347–0.462) | 0.730 (0.675–0.765) | 0.007 |
The data are expressed as median (25%–75% interquartile range).
Microbiome imbalance decreases gingival MSCs proliferation capacity via downregulation of TERT
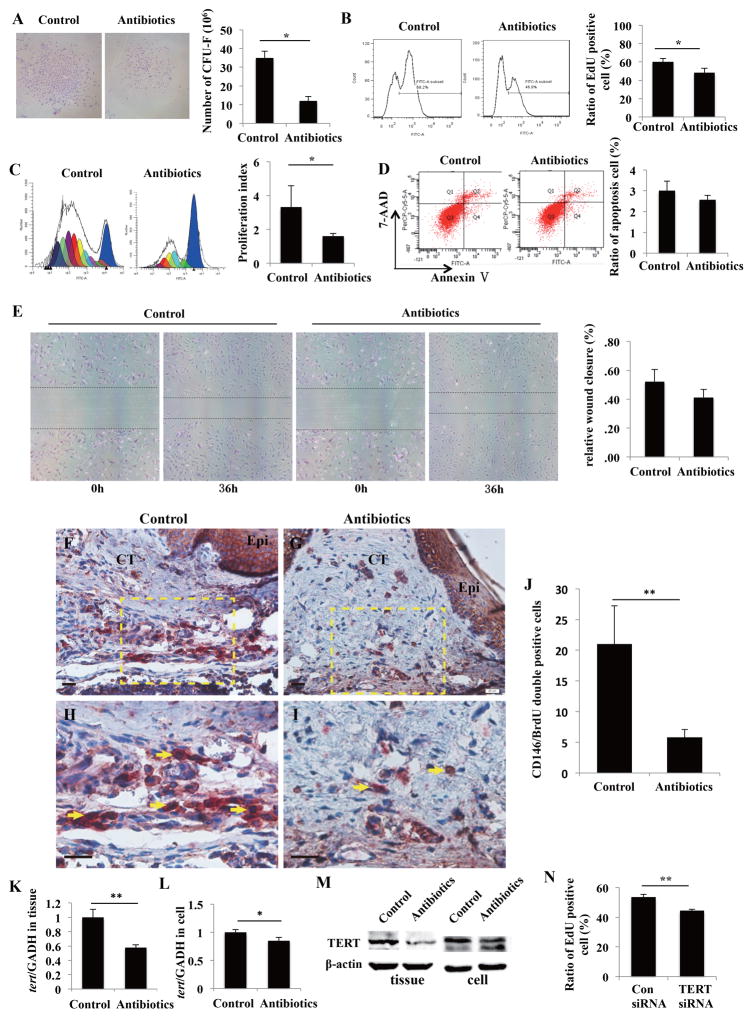
Because the involvement of MSCs in wound-healing process is critical [15], we examined the biological properties of oral MSCs derived from the mice in the two groups. Since attached gingival and hard palatal mucosa both belong to masticatory mucosa and MSCs from hard palatal mucosa is difficult to culture, we used gingival MSCs as the substitution in subsequent experiments. The phenotype characteristics of gingival MSCs were shown in Figure S1A–1C. We first found that gingival MSCs from antibiotics-treated mice formed fewer single-colony clusters than control cells (Figure 3A). EdU assay and CFSE analysis showed gingival MSCs from antibiotics-treated mice had a decreased proliferation capacity when compared with those from control mice, as suggested by the lower percentage of EdU positive cells and decreased proliferation index (Figure 3B and 3C). Apoptosis assay and in vitro wound healing assay indicated there was no significant difference in apoptotic rate and migration ability between gingival MSCs from the two groups (Figure 3D and 3E). To further elucidate the effects of microbiome imbalance on MSCs proliferation in vivo, mice were injected with BrdU at 3 days after wounding for the next 7 days, and animals were sacrificed at 10 days after surgery. Anti-CD146 antibody and anti-BrdU antibody were used to detect MSCs and proliferative cells, respectively. Immunostaining revealed that fewer CD146/BrdU double positive mesenchymal cells were observed in palatal connective tissues of antibiotics-treated mice compared with that in control mice (Fig. 3F–3J). In order to investigate whether antibiotics treatment impairs gingival MSCs proliferation directly by the effects of drug or indirectly by shifting oral microbiota, we treated gingival MSCs with the combinations of various doses of metronidazole and levofloxacin. We found that normal blood concentration, even 10-fold blood concentration of antibiotics had no effects on gingival MSCs proliferation and apoptosis (Supplementary information, Figure S2A and 2B).
Figure 3.
Microbiome imbalance decreases gingival MSCs proliferation capacity via downregulation of TERT. (A): CFU-F assay revealed that gingival MSCs from antibiotics-treated mice formed fewer single-colony clusters than control cell (*p < 0.05). (B): EdU assay showed gingival MSCs from antibiotics-treated mice had a lower percentage of EdU positive cells when compared with those from control mice (*p < 0.05). (C): CFSE analysis verified that gingival MSCs from antibiotics-treated mice had decreased proliferation index compared to cells from control mice (*p < 0.05). (D, E): Apoptosis assay (D) and in vitro wound healing assay (E) indicated there was no significant difference in apoptotic rate and migration ability between gingival MSCs from the two groups. (F–J): Mice were injected with BrdU at 3 days after wounding for 7 days, and then sacrificed. Immunochemistry staining revealed that fewer CD146/BrdU double positive mesenchymal cells were observed in palatal connective tissues of antibiotics-treated mice compared with that in control mice. Epi, epithelium; CT, connective tissue. Scale bars = 20 μm. (H) is the higher magnification of the dotted box in F. (I) is the higher magnification of the dotted box in G. Scale bars = 20 μm. Red arrows showed BrdU positive cells. Arrows showed CD146/BrdU double positive cells. (K–M): TERT mRNA (K, L) and protein (M) levels were reduced in both gingival tissues and MSCs from antibiotics-treated mice compared with those from control mice (*p < 0.05, **p < 0.01). (N): EdU assay revealed that TERT siRNA impaired gingival MSCs proliferation (**p < 0.01).
Since TERT is critical for controlling cell proliferation and tissue homeostasis by maintaining telomere length [16], we next investigated whether microbiome imbalance affects gingival MSCs proliferation via regulating TERT. We first found that the expression of TERT was reduced in both gingival tissues and MSCs from antibiotics-treated mice compared with those from control mice (Figure 3K–3M). EdU assay further revealed that knockdown of TERT impaired gingival MSCs proliferation (Figure 3N). These data indicated the involvement of TERT in microbiome-imbalance-mediated cell proliferation impairment.
Gingival MSCs proliferation is inhibited by increased LPS level in mice gingival tissues caused by antibiotics treatment
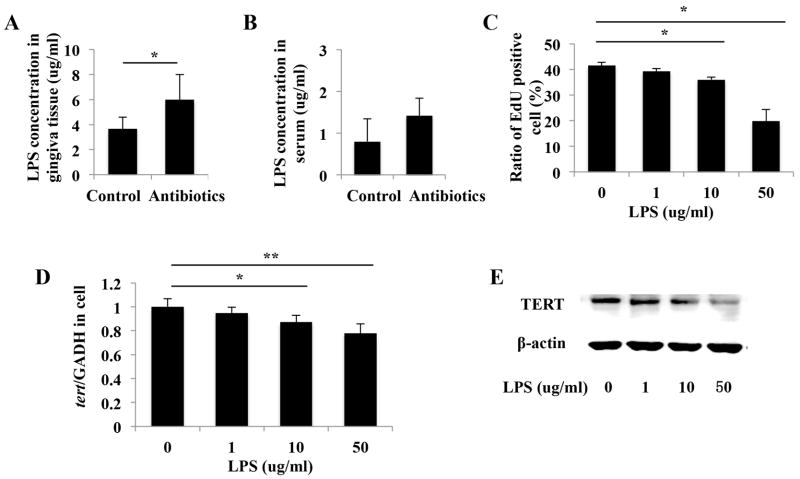
Since microbiota analysis revealed that the dominant bacterial taxa in mice treated with mainly belong to gram-negative bacteria, we investigated the major component of the outer membrane of gram-negative bacteria, LPS, in gingival tissues and serum. Increased levels of LPS were found in gingival tissues of antibiotics-treated mice compared with control mice (Figure 4A). Although no significant difference was detected, there was an increasing trend in serum LPS levels in mice treated with antibiotics (Figure 4B). We next treated gingival MSCs with Pg-LPS to examine the effect of exogeneous LPS on cell proliferation. EdU assay showed Pg-LPS inhibited cell proliferation in a dose-dependent manner (Figure 4C). We further found that when treated with Pg-LPS, gingival MSCs showed decreased TERT expression, as evidenced by results from real time RT-PCR and western blot (Figure 4D and 4E).
Figure 4.
Gingival MSCs proliferation is inhibited by increased LPS level in mice gingival tissues caused by antibiotics treatment. (A): ELISA assay showed increased levels of LPS in gingival tissues of antibiotics-treated mice compared with control mice (*p < 0.05). (B): There was no significant difference in serum LPS levels between antibiotics-treated mice and control mice. (C): EdU assay showed Pg-LPS inhibited cell proliferation in a dose-dependent manner (*p < 0.05, **p < 0.01). (D, E): Real time RT-PCR (D) and western blot (E) showed that the expression of TERT mRNA and protein were decreased in response to LPS stimulation (*p < 0.05, **p < 0.01).
LPS inhibits gingival MSCs proliferation and decreases TERT expression via miR-21
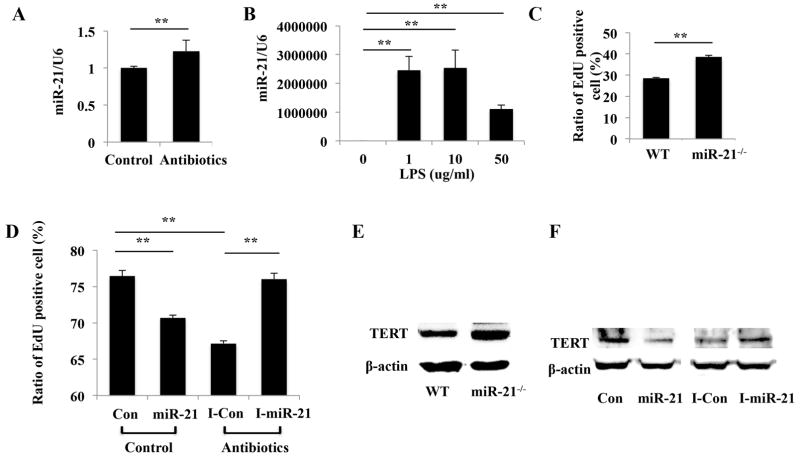
Recent studies pointed to the significant role of miR-21 as regulatory signals for maintaining the stemness and for determining the fate of mesenchymal stem cells [17–19]. Moreover, emerging evidence indicates that miR-21 regulate LPS-induced inflammatory response [20, 21]. We first found that miR-21 was expressed at higher level in gingival MSCs from antibiotics-treated mice compared with those from control mice (Figure 5A). Then we tested if miR-21 involved in LPS-mediated gingival MSCs impairment. Real time RT-PCR results revealed that following LPS treatment, the expression of miR-21 was dramatically increased (Figure 5B). We next demonstrated higher proliferative potential of gingival MSCs from miR-21 knockout (miR-21−/−) mice than those from wild type (WT) mice (Figure 5C). To further strengthen the conclusion that overexpression of miR-21 affects gingival MSCs proliferation, we utilized miR-21 mimic and inhibitor to transfect into gingival MSCs derived from mice treated without or without antibiotics, respectively. EdU assay revealed that overexpression of miR-21 significantly inhibited cell proliferation in gingival MSCs from control mice, whereas miR-21 inhibition enhanced cell proliferation in gingival MSCs from antibiotics-treated mice (Figure 5D). We further used gingival MSCs from WT and miR-21−/− mice, as well as gingival MSCs transfected with miR-21 mimic or inhibitor to examine whether miR-21 controls TERT expression. Overexpression of miR-21 inhibits TERT mRNA and protein expression, and these effects could be reversed by knockdown of miR-21 (Figure 5E and 5F).
Figure 5.
LPS inhibits gingival MSCs proliferation and decreases TERT expression via miR-21. (A): miR-21 was expressed at higher level in gingival MSCs from antibiotics-treated mice compared with those from control mice (**p < 0.01). (B): Real time RT-PCR showed that when stimulated with LPS the expression of miR-21 was dramatically increased (**p < 0.01). (C): Gingival MSCs from miR-21 knockout (miR-21−/−) mice exhibited higher proliferative potential than those from wildtype (WT) mice (**p < 0.01). (D): EdU assay revealed that overexpression of miR-21 significantly inhibited cell proliferation, whereas miR-21 inhibition enhanced cell proliferation (**p < 0.01). (E): The expression of TERT was increased in gingival MSCs from miR-21−/− mice compared with those from WT mice. (F): miR-21 mimic inhibited Sp1 expression, whereas miR-21 inhibitor could reverse this effect. Con: control mimic; I-Con: control inhibitor; miR-21: miR-21 mimic; I-miR-21: miR-21 inhibitor.
miR-21-mediated-TERT downregulation is regulated by Sp1
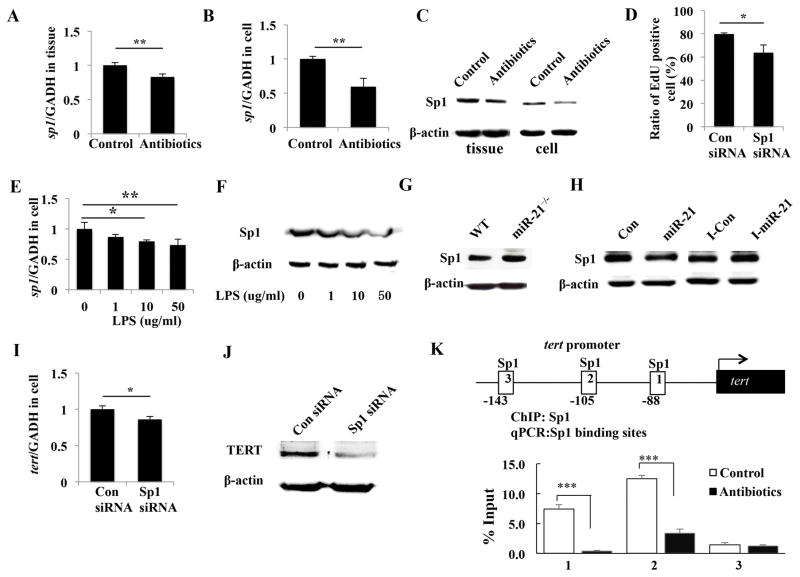
To identify the key regulator of miR-21-mediated TERT downregulation and cell proliferation inhibition, we focused on Sp1, because it is widely known as a transcriptional factor involved in cell proliferation and metastasis [22]. We first confirmed that the expression level of Sp1 decreased in both gingival tissues and MSCs from mice treated with antibiotics (Figure 6A–6C). Moreover, The knockdown of Sp1 with SP1 siRNA significantly blocked cell proliferation (Figure 6D). We then investigated whether LPS regulates Sp1 expression in gingival MSCs. As showed in Figure 6E and 6F, Sp1 mRNA and protein were downregulated by high concentration of LPS. We next used gingival MSCs from WT and miR-21−/− mice, as well as gingival MSCs transfected with miR-21 mimic or inhibitor to test whether miR-21 regulates Sp1 expression. Overexpression of miR-21 resulted in inhibition in Sp1 expression, and these effects could be reversed by knockdown of miR-21 (Figure 6G and 6H). These data indicate that LPS and miR-21 negatively regulates Sp1 expression and Sp1 involves in gingival MSCs proliferation.
Figure 6.
miR-21-mediated-TERT downregulation is regulated by Sp1. (A–C): Sp1 mRNA (A, B) and protein (C) levels were reduced in both gingival tissues and MSCs from antibiotics-treated mice compared with those from control mice (*p < 0.05, **p < 0.01). (D): EdU assay showed that knockdown of Sp1 with SP1 siRNA significantly blocked cell proliferation (*p < 0.05). (E, F): Sp1 mRNA (E) and protein (F) was downregulated by high concentration of LPS. (G): The expression of Sp1 was increased in gingival MSCs from miR-21−/− mice compared with those from WT mice. (H): miR-21 mimic inhibited Sp1, whereas miR-21 inhibitor could reverse this effect. (I, J): Sp1 knockdown reduced TERT mRNA (I) and protein (J) expression (*p < 0.05). (K): Three Sp1 binding-site are found on TERT promoter. ChIP assay showed mainly site-1 and site-2 are regulated by Sp1 binding. Gingival MSCs derived from antibiotics treated mice showed significantly decreased Sp1 binding on these sites (***p < 0.005).
Next, we investigated whether Sp1 is involved in miR-21-mediated TERT downregulation. Sp1 knockdown reduced TERT mRNA and protein expression (Figure 6I and 6J), which suggested Sp1 regulated TERT expression in gingival MSCs. ChIP assays were then performed to determine whether Sp1 modulates TERT transcription by directly binding to the promoter region of TERT. There are 3 Sp1 binding-site found on TERT promoter and ChIP assay showed mainly site-1 and site-2 are regulated by Sp1 binding. Moreover, cells derived from antibiotics treated mice showed significantly decreased Sp1 binding on these sites (Figure 6K). These fingdings suggest that Sp1 functions as a transactivator of mouse TERT gene in gingival MSCs.
DISCUSSION
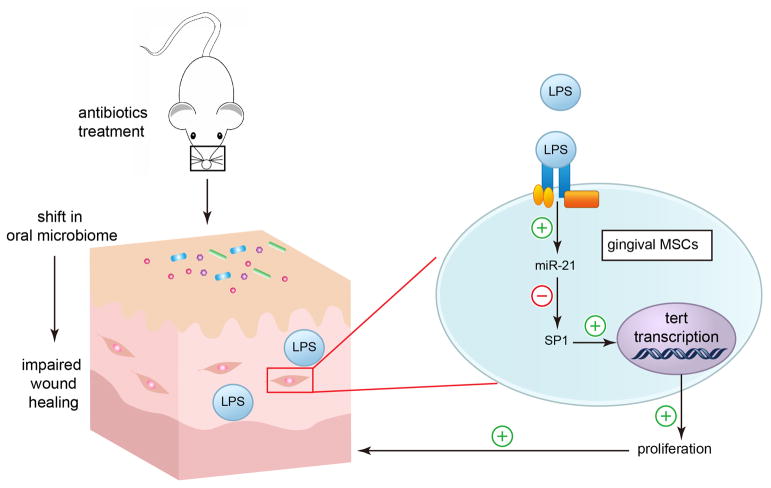
For decades, there has been numerous literature evaluating skin and gut microbial communities diversity in healthy and disease [23] and also evaluating the role of gut microbiota on intestinal stem cell activity [24], the influence of microbiota on oral wound healing and oral MSC function is largely unknown. In this study, we disclose the impact of microbiota on oral mucosa homeostasis and gingival MSCs proliferation, and revealed that microbiome imbalance impairs gingival MSCs proliferation and palatal wound healing through downregulation of Sp1 and TERT via LPS induced miR-21 expression.
Bacteria and inflammatory processes play a pivotal role, not only in normal wound healing but also in the pathophysiology of delayed wound healing [25]. Common wound bacteria may accelerate wound healing with beneficial effects on each phase of wound healing process, including increased infiltrate of immune cells, increased granulation tissue formation and collagen formation [26]. However, if the inflammatory response is too excessive, the repair process is prolonged [27]. LPS is a biologically active bacterial endotoxin and is suggested to delay wound healing process by inhibiting epithelial cell migration [28, 29]. It is reported that antibiotic treatment could promote the release of LPS from bacterial walls by causing bacteria death or lysis, and consequently enhance inflammation [30–32]. In this study, we show that oral administration of antibiotics increases LPS release in oral mucosa which results in delayed palatal wound healing and inhibited gingival MSCs proliferation. Previous studies have found the role of LPS in gingiva derived cells proliferation, however, conflicting conclusions have been addressed [33–36]. This is correlated with the concentration of LPS, that is, LPS at high concentrations (usually > or = 10 μg/ml) generally resultes in reduced cell proliferation capacity; however, LPS at low concentration (usually < 10 μg/ml) induces enhanced cell proliferation [32–36]. These data suggest that low concentration of LPS is necessary to maintain cells viability, whereas high dose of LPS exerts toxic effect to cells. The change in LPS is probably due to the shift in the oral microbial composition.
LPS functions as a triggering factor for inflammatory response by inducing various inflammatory mediators. Increasing studies have expanded our understanding of the role of miRNAs in the response to both bacterial pathogens and commensal bacteria in host cells [37]. Some miRNAs, including miR-146, miR-155, and miR-21, are considered as an effector through which commensal bacteria impact the regulation of intestinal homeostasis [21, 38]. miR-21 has been reported to be induced in response to LPS stimulation in many cell types, including macrophages, peripheral blood mononuclear cells and mouse embryonic fibroblasts [21, 39, 40]. miR-21 is suggested to be an indicator of inflammation [21]. Here, we find that induction of miR-21 by LPS inhibites gingival MSCs proliferation. Other reports showed that miR-21 could result in the suppression of self-renewal and cell cycle arrest of embryonic and adult mesenchymal stem cells, through directly interaction with Sox2 or indirectly by the loss of expression of Oct4, Nanog, and c-Myc [19, 41]. These findings collectively indicate that miR-21 might be an important link between inflammation and stem cell function in wound healing process.
miRs exert their biological functions through targeting multiple genes expression. We report in this study that miR-21 regulates mouse Sp1 protein expression. Sp1 is a major transcription factor that mediates the expression of a large number of genes, including fibroblast growth factor and its receptors [42, 43], extracellular matrix [44, 45], and anti-inflammatory genes [46]. These gene products play vital roles in many physiological processes, in tissue repair and anti-inflammatory responses. It has been confirmed that Sp1 is a direct target of miR-21 in human smooth muscle cells and HEK293 cell lines [47, 48]. These findings collectively suggest that Sp1 may be a candidate target gene of miR-21 in mouse cells. This modulation regulates Sp1-mediated proliferation-related gene transcription.
Telomeres are specialized structures at the ends of chromosomes that are essential for maintaining the stability of the eukaryote genome [49]. Telomerase is the reverse transcriptase that maintains telomere DNA. The most important component responsible for the activity of telomerase is telomerase reverse transcriptase (TERT) [50]. TERT is regulated by a number of inducible transcription factors, including Sp family members, c-Myc and NF-κB [51–53]. In this study, we provide evidence that TERT is involved in microbiome imbalance-induced gingival MSCs proliferation impairment. Sp1 acts as positive regulator in mouse TERT gene transcription. The cellular content of TERT mRNA controlled by Sp1 decreased in parallel with antibiotics treatment, indicating that the alterations in the oral microbial profile accounts for the elimination of TERT expression.
In summary, we have shown that treatment of mice oral cavity with antibiotics leads to changes in the oral microbiome accompanied by impairment of oral MSCs function in male mice. Our study shows a previously unrecognized link between imbalances in oral microbiota and the delayed wound healing process, which implies the importance of microbiome balance in maintaining oral MSC function and oral mucosa homeostasis. However, previous studies have demonstrated that genetics, diet, sex differences and other environmental exposures are major factors in the development and composition of the intestinal microbiota of animals [54–57]. Whether these factors have impact on oral microbiota and consequently influence oral MSC function needs further investigation.
Supplementary Material
Figure 7.
An illustration of the role of oral microbiota in maintaining gingival MSCs homeostasis. Our study shows that antibiotics treatment alters the profile of oral microbiome in mice. This change impairs the wound healing process and oral MSCs proliferation. Mechanically, oral microbiota release LPS which maintains oral MSCs homeostasis via miR-21/Sp1/TERT pathway.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81470751 and 81222011 to Y.L, 81600891 to L.G, 81600829 to Y.S), Beijing Natural Science Foundation (7172087 to Y.L.), and National Institute of Dental and Craniofacial Research, National Institute of Health (K99DE025915 to C.C.).
Footnotes
AUTHOR CONTRIBUTIONS
Y.L. conceived the study; Y.S. performed the majority of experiments with the aid of C.C., J.D., and X.L.; L.G. analyzed the data; Y.S. wrote the manuscript, and Y.L. critically revised the manuscript.
DISCLOSURE OF CONFLICTS OF INTERESTS
The authors declare no competing financial interests.
References
- 1.Wilson M. Bacteriology of Humans: An Ecological Perspective. Malden, MA: Blackwell Publishing Ltd; 2008. [Google Scholar]
- 2.Filoche S, Wong L, Sissons CH. Oral biofilms: emerging concepts in microbial ecology. J Dent Res. 2010;89:8–18. doi: 10.1177/0022034509351812. [DOI] [PubMed] [Google Scholar]
- 3.Vasconcelos RM, Sanfilippo N, Paster BJ, et al. Host-Microbiome Cross-talk in Oral Mucositis. J Dent Res. 2016;95:725–733. doi: 10.1177/0022034516641890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahla RS. Stem Cells Applications in Regenerative Medicine and Disease Therapeutics. Int J Cell Biol. 2016;2016:6940283. doi: 10.1155/2016/6940283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoltz JF, de Isla N, Li YP, Bensoussan D, Zhang L, Huselstein C, Chen Y, Decot V, Magdalou J, Li N, Reppel L, He Y. Stem Cells and Regenerative Medicine: Myth or Reality of the 21th Century. Stem Cells Int. 2015;2015:734731. doi: 10.1155/2015/734731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenette PS, Pinho S, Lucas D, Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu Rev Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Chen L, Scott PG, Tredget EE. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells. 2007;25(10):2648–59. doi: 10.1634/stemcells.2007-0226. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Menocal L, Shareef S, Salgado M, Shabbir A, Van Badiavas E. Role of whole bone marrow, whole bone marrow cultured cells, and mesenchymal stem cells in chronic wound healing. Stem Cell Res Ther. 2015;6:24. doi: 10.1186/s13287-015-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogacka AM, Salazar N, Arboleya S, Suárez M, Fernández N, Solís G, de Los Reyes-Gavilán CG, Gueimonde M. Early microbiota, antibiotics and health. Cell Mol Life Sci. 2017 Oct 7; doi: 10.1007/s00018-017-2670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanvit P, Konkel JE, Jiao X, et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun. 2015;6:8424. doi: 10.1038/ncomms9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Shi S, Liu Y, Uyanne J, Shi Y, Shi S, Le AD. Mesenchymal stem cells derived from human gingiva are capable of immunomodulatory functions and ameliorate inflammation-related tissue destruction in experimental colitis. J Immunol. 2009;183(12):7787–7798. doi: 10.4049/jimmunol.0902318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang L, Li N, Xie H, Jin Y. Characterization of mesenchymal stem cells from human normal and hyperplastic gingiva. J Cell Physiol. 2011;226(3):832–842. doi: 10.1002/jcp.22405. [DOI] [PubMed] [Google Scholar]
- 13.Zhang QZ, Nguyen AL, Yu WH, Le AD. Human oral mucosa and gingiva: a unique reservoir for mesenchymal stem cells. J Dent Res. 2012;91(11):1011–1018. doi: 10.1177/0022034512461016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su Y, Liu D, Liu Y, Zhang C, Wang J, Wang S. Physiologic Levels of Endogenous Hydrogen Sulfide Maintain the Proliferation and Differentiation Capacity of Periodontal Ligament Stem Cells. J Periodontol. 2015;86(11):1276–1286. doi: 10.1902/jop.2015.150240. [DOI] [PubMed] [Google Scholar]
- 15.Maxson S, Lopez EA, Yoo D, et al. Concise Review: Role of Mesenchymal Stem Cells in Wound Repair. Stem Cells Transl Med. 2012;1:142–149. doi: 10.5966/sctm.2011-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Cheng D, Wang S, et al. Human Specific Regulation of the Telomerase Reverse Transcriptase Gene. Genes (Basel) 2016;7 doi: 10.3390/genes7070030. Pii:E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei Y, Bian C, Li J, et al. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114:1374–1384. doi: 10.1002/jcb.24479. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Xu C, Zhang Y, et al. Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomal MicroRNAs Suppress Myofibroblast Differentiation by Inhibiting the Transforming Growth Factor-β/SMAD2 Pathway During Wound Healing. Stem Cells Transl Med. 2016 doi: 10.5966/sctm.2015-0367. pii: sctm.2015–0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trohatou O, Zagoura D, Bitsika V, et al. Sox2 suppression by miR-21 governs human mesenchymal stem cell properties. Stem Cells Transl Med. 2014;3:54–68. doi: 10.5966/sctm.2013-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das A, Ganesh K, Khanna S, et al. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol. 2014;192:1120–1129. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 22.Cook T, Gebelein B, Urrutia R. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann N Y Acad Sci. 1999;880:94–102. doi: 10.1111/j.1749-6632.1999.tb09513.x. [DOI] [PubMed] [Google Scholar]
- 23.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WJ. Bacterial-modulated host immunity and stem cell activation for gut homeostasis. Genes Dev. 2009;23:2260–2265. doi: 10.1101/gad.1858709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones SG, Edwards R, Thomas DW. Inflammation and wound healing: the role of bacteria in the immuno-regulation of wound healing. Int J Low Extrem Wounds. 2004;3:201–208. doi: 10.1177/1534734604271810. [DOI] [PubMed] [Google Scholar]
- 26.Scales BS, Huffnagle GB. The microbiome in wound repair and tissue fibrosis. J Pathol. 2013;229:323–331. doi: 10.1002/path.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis. 2004;17:91–96. doi: 10.1097/00001432-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Brothers KM, Stella NA, Hunt KM, et al. Putting on the brakes: Bacterial impediment of wound healing. Sci Rep. 2015;5:14003. doi: 10.1038/srep14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loryman C, Mansbridge J. Inhibition of keratinocyte migration by lipopolysaccharide. Wound Repair Regen. 2008;16:45–51. doi: 10.1111/j.1524-475X.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- 30.Lepper PM, Held TK, Schneider EM, Bölke E, Gerlach H, Trautmann M. Clinical implications of antibiotic-induced endotoxin release in septic shock. Intensive Care Med. 2002;28(7):824–833. doi: 10.1007/s00134-002-1330-6. [DOI] [PubMed] [Google Scholar]
- 31.Walters SM, Dubey VS, Jeffrey NR, Dixon DR. Antibiotic-induced Porphyromonas gingivalis LPS release and inhibition of LPS-stimulated cytokines by antimicrobial peptides. Peptides. 2010;31(9):1649–1653. doi: 10.1016/j.peptides.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Buijs J, Dofferhoff AS, Mouton JW, van der Meer JW. Continuous administration of PBP-2- and PBP-3-specific beta-lactams causes higher cytokine responses in murine Pseudomonas aeruginosa and Escherichia coli sepsis. J Antimicrob Chemother. 2007;59(5):926–933. doi: 10.1093/jac/dkm073. [DOI] [PubMed] [Google Scholar]
- 33.Bozkurt SB, Hakki SS, Hakki EE, et al. Porphyromonas gingivalis Lipopolysaccharide Induces a Pro-inflammatory Human Gingival Fibroblast Phenotype. Inflammation. 2017;40:144–153. doi: 10.1007/s10753-016-0463-7. [DOI] [PubMed] [Google Scholar]
- 34.Hill SJ, Ebersole JL. The effect of lipopolysaccharide on growth factor-induced mitogenesis in human gingival fibroblasts. J Periodontol. 1996;67:1274–1280. doi: 10.1902/jop.1996.67.12.1274. [DOI] [PubMed] [Google Scholar]
- 35.Basso FG, Soares DG, Pansani TN, et al. Effect of LPS treatment on the viability and chemokine synthesis by epithelial cells and gingival fibroblasts. Arch Oral Biol. 2015;60:1117–1121. doi: 10.1016/j.archoralbio.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Tang J, Wu T, Xiong J, et al. Porphyromonas gingivalis lipopolysaccharides regulate functions of bone marrow mesenchymal stem cells. Cell Prolif. 2015;48:239–248. doi: 10.1111/cpr.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staedel D, Darfeuille F. MicroRNAs and bacterial infection. Cell Microbiol. 2013;15:1496–1507. doi: 10.1111/cmi.12159. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 39.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asirvatham AJ, Magner WJ, Tomasi TB. miRNA regulation of cytokine genes. Cytokine. 2009;45:58–69. doi: 10.1016/j.cyto.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh SK, Kagalwala MN, Parker-Thornburg J, et al. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luster TA, Johnson LR, Nowling TK, et al. Effects of three Sp1 motifs on the transcription of the FGF-4 gene. Mol Reprod Dev. 2000;57:4–15. doi: 10.1002/1098-2795(200009)57:1<4::AID-MRD3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 43.Parakati R, DiMario JX. Sp1- and Sp3-mediated transcriptional regulation of the fibroblast growth factor receptor 1 gene in chicken skeletal muscle cells. J Biol Chem. 2002;277:9278–9285. doi: 10.1074/jbc.M108411200. [DOI] [PubMed] [Google Scholar]
- 44.Verrecchia F, Rossert J, Mauviel A. Blocking sp1 transcription factor broadly inhibits extracellular matrix gene expression in vitro and in vivo: implications for the treatment of tissue fibrosis. J Invest Dermatol. 2001;116:755–763. doi: 10.1046/j.1523-1747.2001.01326.x. [DOI] [PubMed] [Google Scholar]
- 45.Magee C, Nurminskaya M, Faverman L, et al. SP3/SP1 transcription activity regulates specific expression of collagen type X in hypertrophic chondrocytes. J Biol Chem. 2005;280:25331–25338. doi: 10.1074/jbc.M412549200. [DOI] [PubMed] [Google Scholar]
- 46.Tone M, Powell MJ, Tone Y, et al. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 47.Ye X, Liu H, Gong YS, et al. LPS Down-Regulates Specificity Protein 1 Activity by Activating NF-κB Pathway in Endotoxemic Mice. PLoS One. 2015;10:e0130317. doi: 10.1371/journal.pone.0130317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Shi YQ, Yan CC, et al. Up-regulation of miR-21 and miR-23a Contributes to As2O3-induced hERG Channel Deficiency. Basic Clin Pharmacol Toxicol. 2015;116:516–523. doi: 10.1111/bcpt.12348. [DOI] [PubMed] [Google Scholar]
- 49.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 50.Martín-Rivera L, Herrera E, Albar JP, et al. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci U S A. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nozawa K, Maehara K, Isobe K. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J Biol Chem. 2001;276:22016–22023. doi: 10.1074/jbc.M011181200. [DOI] [PubMed] [Google Scholar]
- 52.Takakura M, Kyo S, Kanaya T, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 53.Yin L, Hubbard AK, Giardina C. NF-kappa B regulates transcription of the mouse telomerase catalytic subunit. J Biol Chem. 2000;275:36671–36675. doi: 10.1074/jbc.M007378200. [DOI] [PubMed] [Google Scholar]
- 54.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, Lusis AJ. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7(4):313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoy YE, Bik EM, Lawley TD, Holmes SP, Monack DM, Theriot JA, Relman DA. Variation in Taxonomic Composition of the Fecal Microbiota in an Inbred Mouse Strain across Individuals and Time. PLoS One. 2015;10(11):e0142825. doi: 10.1371/journal.pone.0142825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friswell MK1, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, McBain AJ. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One. 2010;5(1):e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng B, Li G, Yuan J, Li W, Tang H, Wei H. Effects of age and strain on the microbiota colonization in an infant human flora-associated mouse model. Curr Microbiol. 2013;67(3):313–321. doi: 10.1007/s00284-013-0360-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.