Dear Editor,
Mycobacterium smegmatis is commonly used as a laboratory surrogate in studying the physiology and pathogenesis of disease-causing mycobacteria, including Mycobacterium tuberculosis, which causes tuberculosis (TB) in human. Nearly half of the existing antibiotics target cellular protein biosynthesis to kill or inhibit the growth of bacteria (Wilson, 2014). Owing to its great potential as a therapeutic target for antimicrobial drugs, the structures of the ribosomes from pathogenic microbes and model organisms have been of major interest for several decades [for reviews, see (Ramakrishnan, 2014; Yusupova and Yusupov, 2017)]. So far, only low-resolution cryo-EM structures were available for the mycobacterial ribosomes; M. smegmatis 70S, 12 Å (FSC = 0.5 criterion) (Shasmal and Sengupta, 2012) and the M. tuberculosis 50S, 8.5 Å (FSC = 0.143 criterion) (Li et al., 2015). These structures have revealed a few interesting differences from other bacterial ribosomes, particularly the length variation of certain rRNA helices (Shasmal and Sengupta, 2012; Li et al., 2015). However, they are not in sufficient resolution to depict the essential structural differences that might contribute to the designing of the mycobacterial ribosome specific anti-TB drugs.
To gain atomic details of mycobacterial ribosomes, we first purified the 70S ribosome from M. smegmatis (mc2155 strain) (MS70S) (Fig. S1). An in vitro reconstituted translation system based on M. smegmatis components was developed to check the activity of the ribosomes in peptide bond formation and tRNA translocation. The activity of the purified MS70S, determined from the dipeptide (fMet-Leu) synthesis assay, was 50% (Fig. S2A), likely due to the fact the buffer was optimized for E. coli translation assays. The MS70S ribosomes also formed tripeptide (fMet-Leu-Leu) with MS EF-G, with a rate k obs = 1.36 ± 0.12 s−1, comparable to E. coli ribosomes under similar conditions (k obs = 2.8 ± 0.2 s−1) (Fig. S2B). Next, cryo-EM single-particle analysis was employed to determine its structure. After a cascade of 2D and 3D classification (Fig. S3), we were able to obtain a near atomic structure for the MS70S. Mask-based refinement resulted in two improved density maps for the MS30S and MS50S at the resolution of 3.45 Å and 3.08 Å, respectively (Fig. S4). Most rRNA residues and protein side chains in the MS50S and MS30S are clearly separated (Fig. S5).
With the MS30S map, we modeled 19 proteins and more than 90% of 16S rRNA residues (Fig. 1A, 1B and Table S1). Compared to the E. coli 70S ribosome (EC70S), h10 and h17 are distinctively shorter in the MS70S (Figs. 1C and S6C), while h9 is about 17-nt longer (Figs. 1C and S7B). Notably, there is no density present at the expected position of bS21 in the 30S map and no bS21 sequence could be found in the M. smegmatis protein database. In addition, we did not observe strong density for bS1 in the MS70S as well. Protein bS1 is a flexible component of the 30S subunit at the mRNA exit site and plays a pivot role in translation initiation on canonical mRNAs by interacting with the 5′-UTR to facilitate unfolding of secondary structures (Byrgazov et al., 2015). The absence of bS21 and weak association of bS1 in the MS70S seem to correlate with the fact that non-canonical translation with leaderless mRNA (without 5′-UTR and Shine Dalgarno sequence) contributes to a distinctly large portion (~25%) of mycobacterial proteome (Shell et al., 2015).
Figure 1.
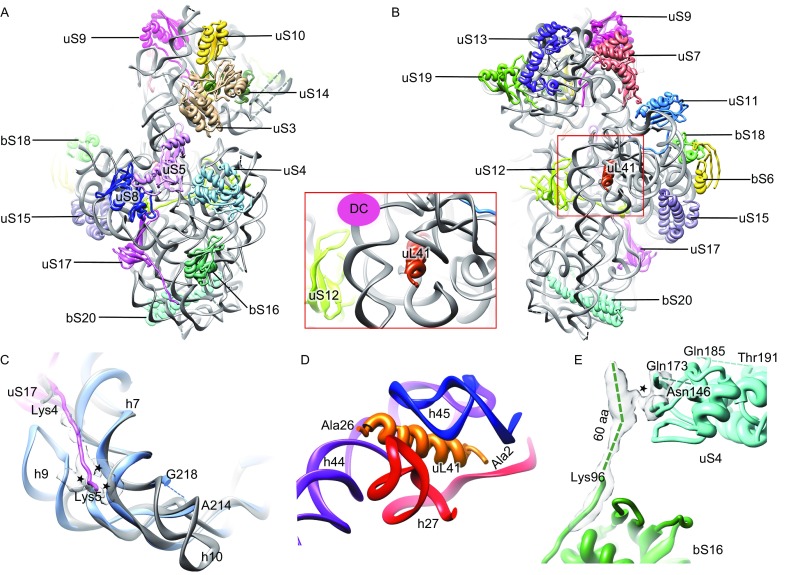
Overall atomic model and unique features of the MS 30S. (A and B) Atomic model of the MS30S, viewed from the solvent surface (A) and intersubunit surface (B). Inset, zoom-in view of uL41 close to the decoding center (DC). (C) The extended N-terminus of uS17 interacts with h7 and h9. Counterparts in the EC70S are colored gray. The coordinates of the E. coli components are from a crystallography study (PDB 4KIY) (Pulk and Cate, 2013). (D) Ribosomal protein uL41 is surrounded by h27, h44, and h45. (E) The extended C-terminus of bS16 interacts with globular domain of uS4. The interaction sites are indicated by asterisks
Interestingly, a rod-like density, which is absent from other known prokaryotic ribosomes such as the EC70S (Pulk and Cate, 2013), T. thermophilus 70S (TT70S) (Selmer et al., 2006), and Staphylococcus aureus 70S (SA70S) (Khusainov et al., 2016), locates in a pocket formed by h27, h44, and h45 close to the decoding center (DC) in the 30S subunit (Figs. 1B, 1D and S5E). This position is reminiscent of protein eL41 in eukaryotic ribosomes, which creates a central inter-subunit bridge (Ben-Shem et al., 2011). A survey of mycobacterial protein database indeed located the identity and sequence of this unannotated protein (UniProt # A0QR10 for M. smegmatis and UniProt # P9WKT5 for M. tuberculosis). The flexible C-terminus points to the 50S subunit. We propose to change the name of protein eL41 to uL41, because it is no longer a eukaryote-specific ribosomal protein. The close contact of uL41 with decoding helices (Fig. 1D), such as h44 and h27 (Fig. S8), may have a profound effect on mycobacterial translation, particularly in the decoding steps.
Almost all of the 30S proteins we have modeled have conserved globular domain, and more than half of them have variable N- or C-terminus compared with other bacterial species (Table S1). Particularly, many of them have extended terminal sequences in M. smegmatis, and these extensions are even longer in M. tuberculosis. For example, protein uS17 bears an extended N-terminus (~13 residues) (Fig. S6A), which establishes a species-specific interaction with h7 and h9 of the 16S rRNA (Fig. 1C). Notably, this N-terminus is even longer in M. tuberculosis (Fig. S6A). Another example is the mycobacterial bS16 protein, which possesses a very long C-terminus (about 60 residues longer than E. coli bS16). The extension is not resolved in atomic resolution, but apparently it forms species-specific interaction with the globular domain of uS4 (Figs. 1E and S6B). Besides 30S body proteins (bS16 and uS17), a few proteins in the neck region of the 30S have extended N- and/or C-terminus, including uS3 and uS5 (at the mRNA entry site), and uS11 (at the mRNA exit site) (Table S1). As these N- and C-termini are quite flexible, we were not able to build any of them (Table S1). Nevertheless, one of them, the N-terminal extension of uS5 is expected to be in the proximity of the mRNA entry site. Altogether, the compositional difference of the MS30S and unique extensions of mycobacterial ribosomal proteins might reflect the physiological difference of mycobacteria with regard to their widespread non-canonical translation on leaderless mRNA (Cortes et al., 2013; Shell et al., 2015).
For the MS50S, we modeled 29 proteins and about 84% of 23S rRNA residues (Fig. 2A, 2B and Table S2). An unexpected finding is the presence of a piece of additional density in the 50S map. After rounds of “trial and error” attempts with small ORFs in the M. Smegmatis databse, a protein of 24 amino acids (UniProt # A0QTP4) was found to match very well with the density (Figs. S5A and S9). This protein, consisting of an α-helical N-terminus (~13 residues) and a C-terminal loop, is encircled by a pocket formed by H39, H40, joint of H41 and H42, H72 and H89 (Fig. 2C), while in other prokaryotic ribosomes (EC70S, TT70S, and SA70S), there is no protein in this pocket. Located deep inside an rRNA pocket, it is a previously unidentified and uncharacterized ribosomal protein; we propose to name it as bL37. Bioinformatic analysis shows that this bL37 only exists in high G + C, actinobacterial species, including mycobacteria (Fig. S10B and S10C). Evolutionary relationship based 16S-like rRNA sequences confirms that bL37-containing species are in a same bacterial branch that is separated from a few well-studied species, such as E. coli, T. thermophilus and B. subtilis (Fig. S11). Notably, bL37 strongly interacts with the buried side of a helix H89 at the peptidyl transferase center (Figs. 2, inset and S7C). Therefore, bL37 might be able to affect the peptidyl transferase function of the 50S subunit, likely through modulating H89 conformation.
Figure 2.
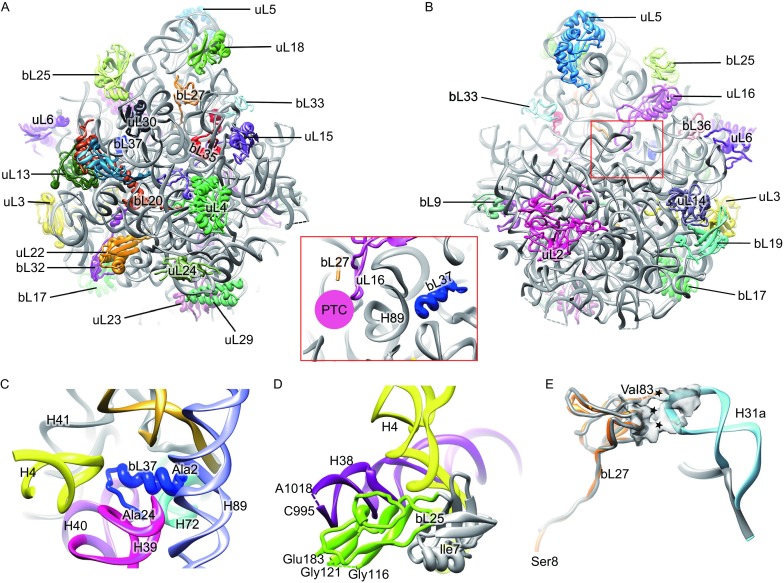
Overall atomic model and unique features in the MS 50S. (A and B) Atomic model of the MS50S, viewed from solvent surface (A) and inter-subunit surface (B). Inset, relative orientation of bL27 with respect to the peptidyl transferase center (PTC). (C) Ribosomal protein bL37 is encircled by H4, H39, H40, base of H41 and H42, H72 and H89. (D) The extra C-terminal domain of bL25 is in proximity to H38 (purple). Conserved N-terminal domain of bL25 is colored gray while the extra C-terminal domain is colored green. (E) The extended H31 interacts with bL27, N-terminus of which extends to the peptidyl transfer center. The interaction sites are indicated by asterisks. Protein bL27 and H31a in the MS70S are colored with orange and cyan, respectively, while their counterparts in the EC70S are in gray. The coordinates of the E. coli components are from a crystallography study (PDB 4KIX) (Pulk and Cate, 2013)
Among all modelled 50S proteins, about 40% of them have variable N- or C-terminus in the MS50S (Table S2). For example, protein bL25 of M. smegmatis, similar as those of Bacillus subtilis, T. thermophilus and S. aureus, has two domains while it only has one domain in E. coli. The extra C-terminal domain of mycobacterial bL25, locates near H38 in the MS50S (Fig. 2D). This domain is involved in tRNA proofreading in T. thermophilus (Jenner et al., 2010). While in B. subtilis, bL25 is only expressed and bind to ribosome under stress conditions (Schmalisch et al., 2002).
As to the 23S rRNA of the MS50S, some variable helices from the solvent surface, mainly clustered in the L1 stalk region, are remarkably extended. Consistent with previous low-resolution structure (Shasmal and Sengupta, 2012), the most distinct one is H54a. This helix stretches all the way from the bottom of the L1 stalk base to a region near bS6 in the MS30S (open state) (Fig. S7) (Shasmal and Sengupta, 2012). Strikingly, in the structure of the M. tuberculosis 50S, this helix is pointing to a different direction: it locates on the inter-subunit side of the 50S subunit, with its tip occupying the exit site of tRNA (E-site) on the 50S subunit (close state) (Li et al., 2015). Thus, this helix has to be in the open state before the 50S subunit can bind to the 30S subunit, suggesting the presence of additional steps on translation initiation by regulating the structural dynamics of H54a in mycobacteria.
In addition, two other helices H15 and H16a, which do not exist in the EC70S, are located close to the base of the L1 stalk (Fig. S7). However, both of them are rather flexible and could not be modeled at the residue precision. Nevertheless, their proximity to the L1 stalk implies that these extended helices might be involved in regulating the dynamics of the L1 stalk during translation cycle. Another lengthened component is H31a (compared to the EC70S, SA70S, TT70S), which creates a set of unique interactions with protein bL27 (Figs. S7 and 2E). Since the N-terminus of bL27 inserts into the peptidyl transfer center, the addition interactions between bL27 and H31a might affect certain kinetic steps of translation elongation on the mycobacterial ribosome.
In summary, we report the near atomic structure of the ribosome from genus mycobacterium, many of which (M. tuberculosis, M. leprae, M. avium) are high-risk pathogens for human. In particular, the unique H54a may play a role in regulating subunit association. Similarly, the two newly discovered proteins uL41 and bL37, located close to the DC and PTC respectively, may have implications in modulating the main functions of the ribosome. The structure provides a resource for studying mycobacterial translation as well as in designing anti-TB drugs.
FOOTNOTES
While this work was in the stage of manuscript submission, a similar structure was published (Hentschel et al., 2017). The major conclusions related to the structure are highly consistent to our present work.
The cryo-EM density map of the MS30S and MS50S has been deposited in the EMDB with accession number EMD-6790 and EMD-6789. The atomic model has been deposited in the PDB with accession number 5XYU and 5XYM.
We thank the Tsinghua University Cryo-EM Facility of China National Center for Protein Sciences (Beijing) for providing resources for data collection and computation. Part of the computation was performed on the Computing Platform of the Center for Life Science, Peking University. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31630087, 31422016 and 31470722 to N.G.); the Swedish Research Council (Diary No. 2013-8778, 2014-4423, 2016-06264 and 2008-6593), and the Knut and Alice Wallenberg Foundation (KAW 2011.0081) to S.S.
Zhifei Li, Xueliang Ge, Yixiao Zhang, Lvqin Zheng, Suparna Sanyal, and Ning Gao declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
Zhifei Li and Xueliang Ge have contributed equally.
Contributor Information
Suparna Sanyal, Email: Suparna.Sanyal@icm.uu.se.
Ning Gao, Email: gaon@pku.edu.cn.
References
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Byrgazov K, Grishkovskaya I, Arenz S, Coudevylle N, Temmel H, Wilson DN, Djinovic-Carugo K, Moll I. Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res. 2015;43:661–673. doi: 10.1093/nar/gku1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel J, Burnside C, Mignot I, Leibundgut M, Boehringer D, Ban N. The complete structure of the mycobacterium smegmatis 70S ribosome. Cell Rep. 2017;20:149–160. doi: 10.1016/j.celrep.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1076. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- Khusainov I, Vicens Q, Bochler A, Grosse F, Myasnikov A, Menetret JF, Chicher J, Marzi S, Romby P, Yusupova G, et al. Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucleic Acids Res. 2016;44:10491–10504. doi: 10.1093/nar/gkw933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sun Q, Jiang C, Yang K, Hung LW, Zhang J, Sacchettini JC. Structure of ribosomal silencing factor bound to mycobacterium tuberculosis ribosome. Structure. 2015;23:1858–1865. doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014;159:979–984. doi: 10.1016/j.cell.2014.10.052. [DOI] [PubMed] [Google Scholar]
- Schmalisch M, Langbein I, Stulke J. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J Mol Microbiol Biotechnol. 2002;4:495–501. [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shasmal M, Sengupta J. Structural diversity in bacterial ribosomes: mycobacterial 70S ribosome structure reveals novel features. PLoS ONE. 2012;7:e31742. doi: 10.1371/journal.pone.0031742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M (2017) Crystal structure of eukaryotic ribosome and its complexes with inhibitors. Philos Trans R Soc Lond B Biol Sci 372 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


