Abstract
Nisin, an important bacteriocin from Lactococcus lactis subsp., is primarily active against various Gram-positive bacteria. Leucocin C, produced by Leuconostoc carnosum 4010, is a class IIa bacteriocin used to inhibit the growth of Listeria monocytogenes. Because two bacteriocins have different modes of action, the combined use of them could be a potential strategy for effective inhibition of foodborne pathogens. In this study, L. lactis N8-r-lecCI (N8 harboring lecCI gene) coexpressing nisin–leucocin C was constructed based on the food-grade carrier L. lactis N8. Production of both bacteriocins was stably maintained. Antimicrobial measurements showed that the recombinant strain is effectively against Listeria monocytogenes and Staphylococcus aureus and moderately against Salmonella enterica serovar Enteritidis and Escherichia coli because of its stronger antibacterial activity than the parental strain, this result first demonstrated that the co-expression of nisin and leucocin C results in highly efficient antimicrobial activity. The checkerboard assay showed that the antibacterial activity of L. lactis N8-r-lecCI supernatant was enhanced in the presence of low concentration of EDTA. Analysis of the scanning electron microscope image showed the biggest cellular morphology change in L. monocytogenes treated with a mixture of EDTA and L. lactis N8-r-lecCI supernatant. The practical effect was verified in pasteurized milk through time-kill assay. The L. lactis N8-r-lecCI strain expressing both nisin and leucocin C has a promising application prospect in pasteurized milk processing and preservation because of its strong antibacterial activity.
Keywords: foodborne pathogens, Listeria monocytogenes, nisin, leucocin C, Lactococcus lactis, antimicrobial activity
Introduction
Pathogen control is a significant issue in the food industry. Foodborne diseases can spread further and more rapidly than before because of the increasing transnational trade, travel, and migration (Kaferstein et al., 1997). Although various measures have been implemented to decrease infections caused by foodborne pathogens (Gandhi and Chikindas, 2007), foodborne disease outbreaks from microbial contamination, chemicals, and toxins remain prevalent worldwide (Velusamy et al., 2010). For example, in the outbreak of listeriosis in the United States in September 2011, a total of 146 invasive illnesses, 30 deaths, and 1 miscarriage were recorded (Laksanalamai et al., 2012). The foodborne pathogen Listeria, the causative agent of listeriosis (Yousef and Lou, 1999), can grow at refrigeration temperatures and adapt to low pH and high salt concentration (Cole et al., 1990). As such, this pathogen can overcome various bacteriostatic measures, increasing the difficulty of anti-listeria treatments.
Researchers have developed various methods for controlling foodborne pathogens to reduce their potential risks to human health (Gandhi and Chikindas, 2007). Such methods include the addition of bacteriocins and bacteriocin-producing lactic acid bacteria (LAB) in food (Roberts and Zottola, 1993) as effective alternatives to chemical preservatives (Lucera et al., 2012). Lactococcus, Lactobacillus, Leuconostoc, and other LAB can secrete bacteriocins (Barefoot and Klaenhammer, 1983; Klaenhammer, 1993; Wan et al., 2013, 2015), which are ribosomally synthesized and extracellularly released peptides with antimicrobial activity. LAB bacteriocin has been widely characterized because of their “food-grade quality” and industrial importance (Barbosa et al., 2017). Nisin, an important bacteriocin from LAB, is primarily active against Gram-positive bacteria (Delves-Brougthon, 1990). Nisin is used as a food protection agent in more than 60 countries and regions worldwide because of its low toxicity and high efficiency. In addition to nisin, one of the available commercial antibacterial agents (Barbosa et al., 2017), other bacteriocins exhibit promising perspectives. Class IIa bacteriocin piscicolin P126 displays antilisterial activity in milk and Camembert cheese (Wan et al., 1997). Enterocin AS-48 is active against Alicyclobacillus acidoterrestris, which is a spoilage-causing bacterium in fruit juices (Grande et al., 2005). Leucocin C kills the foodborne pathogen Listeria monocytogenes, and leucocin C-producing strain Leuconostoc carnosum 4010 is used as protective culture in meat products (Wan et al., 2013).
Bacteriocins confer protection against foodborne pathogens; however, single application of bacteriocins is hindered by their limited activity spectrum, high concentration demand and the prevalence of antimicrobial resistance. Several studies have demonstrated that some bacteriocins show additive or synergistic effects when used in combination with other antimicrobial agents; in this regard, combining bacteriocins is a good alternative to the use of single bacteriocin for effective pathogen inhibition. For example, the combination of nisin and its variants with carvacrol and/or citric acid inhibited Gram-negative foodborne pathogens (Campion et al., 2017). The activity of lysozyme and nisin against L. monocytogenes and Escherichia coli was enhanced with the addition of EDTA (Branen and Davidson, 2004). The efficacy of the antibiotics polymyxin and colistin for preventing the biofilm formation of Pseudomonas aeruginosa increased when they were combined with nisin treatment (Field et al., 2016).
In this study, a strain of L. lactis stably coexpressing nisin–leucocin C was constructed to control a wide range of foodborne pathogens, including L. monocytogenes. LAB are generally recognized as safe (GRAS) microorganisms and widely used in dairy and fermented food (Roberts and Zottola, 1993; O’Sullivan et al., 2002; Swetwiwathana and Visessanguan, 2015). Application of bacteriocin-producing LAB strains may be more effective than direct addition of preservatives because of the continuous in situ production of bacteriocins (Langa et al., 2017). However, the use of genetically modified lactic acid bacteria (GM-LAB) was not widely applied and accepted in many Countries (Sybesma et al., 2006). Risk assessment and expected benefits will determine the future use of GM-LAB in the domains of food technology and health (Renault, 2002; LeBlanc et al., 2010; Capozzi et al., 2012). We aimed to construct a L. lactis strain, based on food-grade biotechnological strategy, with improved antimicrobial activity and broad antimicrobial spectrum without using genes from non-GRAS bacteria and antibiotics selection pressure for maintaining genetic stability.
Materials and Methods
Plasmids, Strains, and Growth Conditions
Lactococcus lactis N8 (N8) (Qiao et al., 1996), a wild-type (WT) nisin Z producer, and L. lactis NZ9000 (NZ9000) (Kuipers et al., 1998), which is usually used as cloning host, were cultured without agitation at 30°C in SGM17 broth [M17 broth supplemented with 0.5% (w/v) glucose and 0.55% (w/v) sucrose]. When needed the broth was supplemented with 10 ng/mL nisin for induction expression. Antibiotic susceptibility screening was conducted using 150 μg/mL erythromycin (Em) and 15 μg/mL chloramphenicol (Cm) for E. coli DH5α and 5 μg/mL Em and 5 μg/mL Cm for L. lactis.
Escherichia coli O157:H7, L. monocytogenes WSLC 1018, and Micrococcus luteus A1CNCIMB861666 were obtained from our laboratory. E. coli ATCC8739, Staphylococcus aureus ATCC29213, Salmonella Enteritidis ATCC14128, S. aureus ATCC6538, and L. monocytogenes ATCC 19115 were obtained from the Guangdong Culture Collection Center. Except for nisin-sensitive indicator strain M. luteus A1CNCIMB861666 and leucocin C-sensitive indicator strain L. monocytogenes WSLC 1018, all these strains are common foodborne pathogens and used as indicator in this study. These strains were cultured in LB broth (Hope Bio, Qingdao, China), except for L. monocytogenes, which was cultured in TSB-YE broth (Hope Bio, Qingdao, China).
Plasmids used in this study are listed in Table 1. pLEB124 was used to construct the expression vector. pNZ5319 was used to express lecCI (Leucocin C and its immunity gene) in the chromosome of N8 through Cre–loxP gene recombination system.
Table 1.
Plasmids used in this study.
| Plasmids | Relevant properties | Reference |
|---|---|---|
| pLEB690 | L. lactis secretion vector harboring lactococcal promoters P45 and PnisZ, and signal sequence SSusp45 | Li et al., 2011 |
| pLEB729 | Leucocin C and the immunity gene lecCI fused to SSusp45 in pLEB690 | Wan et al., 2013 |
| pLEB124 | L. lactis secretion vector harboring lactococcal promoter P45 | Qiao et al., 1995 |
| pLEB124-lecCI | fragment (PnisZ+SSusp45+lecCI) obtained from pLEB729 cloned into pLEB124 | This study |
| pNZ5319 | vector for gene replacements in Gram-positive bacteria | Lambert et al., 2007 |
| pNZ5319-up-down | Upstream and downstream sequences of the N8GL37-38 genes cloned into pNZ5319 | This study |
| pNZ5319-up-down-lecCI | Fragment (P45+PnisZ+SSusp45+lecCI) obtained from pLEB124-lecCI replaced the cat gene in pNZ5319-up-down | This study |
Construction of Leucocin C Expression Strains
The plasmid pLEB729 (Wan et al., 2013) was digested with HindIII and BamHI (Takara, Dalian, China) to obtain the expression fragment PnisZ+SSusp45+lecCI. The fragment was purified with DNA gel extraction kit (Solarbio, Beijing, China) and cloned into HindIII-BamHI digested pLEB124 to produce pLEB124-lecCI. pLEB124-lecCI was then electroporated into L. lactis NZ9000 and L. lactis N8 to obtain NZ9000-p-lecCI and N8-p-lecCI (plasmid expression method) expression strains, respectively.
The non-necessary genes N8GL37-38 were replaced with lecCI to stably express lecCI in the genome of N8 without antibiotic pressure. In brief, 633-bp upstream and 528-bp downstream sequences of the N8GL37-38 genes were amplified with the N8 genome as template by using LA Taq (Takara, Dalian, China) and cloned into the XhoI-SwaI and SacI–BglII restriction sites of pNZ5319, respectively, to obtain the pNZ5319–up-down vector. P45+PnisZ+SSusp45+lecCI was amplified with the pLEB124-lecCI as template by PCR using Phusion High-Fidelity DNA polymerase (Thermo Fisher, China) with primer pairs LecC F/R. PCRs were performed using the primers listed in Table 2. The PCR products and pNZ5319-up-down were digested with BglII and XhoI and then cloned into the BglII-XhoI sites of pNZ5319-up-down to replace the cat gene and obtain the recombination vector pNZ5319-up-down-lecCI, which was introduced into N8. The genomes of transformants and N8 were used as templates to amplify the target genes with the primers Ery-F/R, LecCI-F/R, and Out-F/R to identify the single or double cross-over recombinants. The PCR products were analyzed on 1% agarose gels. The potential nisin and leucocin C co-expression strain N8-r-lecCI (homologous recombination expression method) was identified.
Table 2.
Primers used in this study.
| Primer | Sequence (5′–3′) | Annealing temperature (°C) |
|---|---|---|
| For the construction of leucocinC expression vector | ||
| (pNZ5319-up-down-lecCI) | ||
| UP-F | CCGCTCGAG AGATGAGGGTAATCTGAAGGA | 52 |
| UP-R | CCCATTTAAAT ACCTTATCTTAAATGCCTATCTT | |
| Down-F | CCCGAGCTC GAAAGGGGGATTAGTGATCA | 54 |
| Down-R | GAAGATCT ACTCGCTACCGCCTGAAA | |
| LecCI-F | CGTTAGGGGCTTGAACAAG | 56 |
| LecCI-R | CTAATATCCATAACGAATACTAGAT | |
| For the identification of leucocinC expression strain (N8-r-lecCI) | ||
| Ery-F | CGATACCGTTTACGAAATTGG | 52 |
| Ery-R | CTTGCTCATAAGTAACGGTAC | |
| Out-F | ACGAACAAAACAGTTGCG | 52.5 |
| Out-R | TTCTCTGCCATACTGGCTC | |
Growth Profile of Wild and Constructed Strains
N8, N8-p-lecCI, and N8-r-lecCI were cultured to OD600 of 0.8 in the SGM17 broth and diluted to an optical density of OD600 of 0.2. Afterward, 200 μl of the cultures were inoculated into 20 mL of SGM17 medium in a 50 mL triangular flask. Growth profiles were monitored by measuring OD600 for 12 h at 30°C. All tests were performed in triplicate.
Anti-bacterial Activity
Agar diffusion assay was performed in triplicate to detect leucocin C and nisin activities of the culture supernatants of N8, N8-p-lecCI, N8-r-lecCI, and NZ9000-p-lecCI. In brief, 20 mL of soft agar and 80 μl of the corresponding indicator strain (5 × 108 CFU/mL) were mixed and poured onto a plate with Oxford cups. The holes of the dried agar plates were added with 150 μl of the pasteurized supernatant (75°C, 10 min) from the overnight cultures of L. lactis strains. The plates were incubated overnight at corresponding temperature.
SDS-PAGE and Identification of the Leucocin C Activity Band
Total protein was obtained from 100 mL of the cell-free overnight culture supernatants of NZ9000 (blank control) and NZ9000-p-lecCI (leucocin C) and precipitated by 50% ammonium sulfate at 4°C. The precipitate was centrifuged at 14,000 × g and 4°C for 30 min and released in 1 mL of sterile water. The concentrated supernatant samples were analyzed by tricine-SDS-PAGE (Schägger and von Jagow, 1987). Gels were prepared using 16% acrylamide resolving gel and 1 cm of 4% acrylamide spacer gel. After electrophoresis (Bio-Rad), the gel was divided vertically into two parts; the first part was stained with Coomassie brilliant blue G-250 (Takara, Dalian, China), and the second part was used for testing antimicrobial activity according to the method described earlier (Bhunia et al., 1987). The gel was washed for 30 min by sterile water and placed in a plate. The plate was poured with soft agar containing L. monocytogenes ATCC19115 and incubated at 37°C overnight.
Comparison of Genetic Stability
The expression strains N8-p-lecCI (plasmid expression method) and N8-r-lecCI (homologous recombination expression method) were inoculated into 1.5 mL tube containing SMG17 broth (non-selective conditions). The strains were cultivated at 30°C for 8 h, transferred into a second tube, and cultivated under the same conditions. Serial transferring was carried out 70 times, and each transfer was referred to as 10 generation times (results not shown). A single colony was harvested to inspect the stability of the strain by calculating the percentage of leucocin C expressing cells in the population.
Determination of the Minimum Inhibitory Concentration
The minimum inhibitory concentrations (MICs) of EDTA and N8-r-lecCI supernatant against L. monocytogenes WSLC1018, L. monocytogenes ATCC 19115, Staphylococcus aureus ATCC6538, Staphylococcus aureus ATCC29213, E. coli ATCC25922, E. coli O157:H7, Salmonella Enteritidis ATCC 14128 and M. luteus A1CNCIMB 861666 were determined using a broth micro-dilution assay (Clinical and Laboratory Standards Institute [CLSI], 2014). These strains were adjusted to a final concentration of 105 CFU/mL in the corresponding broth, and 100 μl of the prepared suspension was added to the twofold serial dilutions of antibacterial agents (100 μl). After 24 h of incubation at 37°C, MIC was defined as the lowest antimicrobial concentration that did not result in visible growth. All tests were performed in triplicate.
Checkerboard Assay
Checkerboard method (Martinez-Irujo et al., 1996) is usually used to evaluate the interaction of different drugs. In this study, each combination (A × B) of compound A (50 μl of EDTA) and B (50 μl of N8-r-lecCI supernatant) was diluted by twofold along the ordinate and abscissa axes in a 96-well microtiter plate. The final concentrations of the compounds ranged from 1/32–2 times the MIC for EDTA and 1/64–2 times the MIC for the N8-r-lecCI supernatant. The pathogenic bacteria were adjusted to the final concentration of 105 CFU/mL in the corresponding broth (100 μl) and added to the combinations of the compounds. The plate was incubated at 37°C for 24 h. All tests were performed in triplicate.
Time-Kill Assay in Pasteurized Milk
Listeria monocytogenes, as a common food contaminating bacterium poses significant threat to public health (Jemmi and Stephan, 2006). According to preliminary checkerboard assays, the positive interaction between EDTA and the N8-r-lecCI supernatant against L. monocytogenes ATCC 19115 was chosen for time–kill assay in pasteurized milk. The growth curve-based time–kill assay was determined according to a previous method with minor modification (Garcia et al., 2010; Shi et al., 2017). The experimental tubes containing pasteurized milk initially inoculated with 105 CFU/mL pathogens were added with either single antibacterial agent or the combination of antibacterial agents. The tube with pathogen was used as control. The samples were cultured at 4°C (refrigeration temperature) and 25°C (room temperature) for 24 h. Bacterial counts were determined by spreading appropriate dilutions on TSB-YE agar every 4 h. The plates were incubated at 37°C overnight, and bacterial colonies were counted. In these experiments, the concentration of EDTA was consistent with 1/2 of the MIC, and the concentration of the N8-r-lecCI supernatant was consistent with the MIC. All tests and measurements were performed in triplicate.
Scanning Electron Microscopy (SEM)
Logarithmic-phase bacteria (L. monocytogenes ATCC 19115) were collected at 4000 g for 5 min and washed twice with PBS (pH 7.4). The cells were suspended in PBS (OD600 = 0.2) and treated with EDTA (0.125 mg/mL)/N8-r-lecCI supernatant (MIC) or their combination [EDTA (0.125 mg/mL) and N8-r-lecCI supernatant (MIC)] for 1 h. The bacterial cells without antimicrobial treatment were similarly processed and used as controls. After incubation, the cells were washed with PBS and fixed overnight at 4°C with 2.5% glutaraldehyde. The cells were washed in the same buffer and dehydrated using gradient ethanol concentrations ranging from 50 to 100%, with each gradient for about 15 min. Ethanol was replaced with tertiary butyl alcohol. The samples were drop on the plate and prepared for SEM (QUANTA 200) observation.
Results
Identification of Activity and Protein Band of Leucocin C
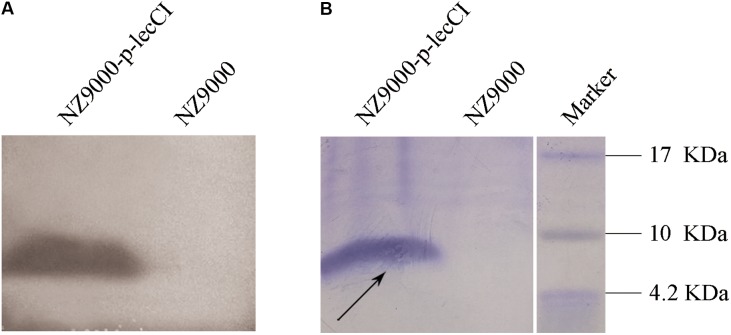
Bacteriocin production was determined by agar diffusion assay on a plate. Leucocin C was effectively secreted by recombinant L. lactis strains and shown as inhibition zones on L. monocytogenes indicator plates (Figure 1). All constructed strains showed excellent anti-listeriosis activity compared with the wild strain N8 (slight inhibition). The inhibitory effect of N8-r-lecCI was more distinct than the plasmid expression strains N8-p-lecCI and NZ9000-p-lecCI, indicating the superior antibacterial activity of N8-r-lecCI. The proteins from the culture supernatants were concentrated and analyzed with SDS-PAGE to confirm whether or not leucocin C inhibits the growth of L. monocytogenes. Results of Coomassie brilliant Blue G-250 staining indicated the migration of the putative leucocin C band between 4.2 and 10 kDa (Figure 2B). An inhibition zone was found around the potential leucocin C band in the second part of the gel which was used for testing antimicrobial activity against L. monocytogenes, whereas no antimicrobial activity was detected in the remaining parts of the gel (Figure 2A). This finding suggested that the protein band in SDS-PAGE is leucocin C. All these results indicated that the constructed strains can successfully secrete leucocin C.
FIGURE 1.
Results of agar-diffusion experiment with leucocin C-producing L. lactis and the wild type N8 (indicator strain: L. monocytogenes WSLC 1018). Three independent experiments were conducted, and the image represents the results of one of the experiments (unit: mm). (A) L. lactis N8; (B) L. lactis N8-p-lecCI; (C) L. lactis N8-r-lecCI; and (D) L. lactis NZ9000-p-lecCI. The difference between two groups was compared by t-test, the diameter of inhibitions zone of N8-p-lecCI, N8-r-lecCI and NZ9000-p-lecCI were significantly different (P < 0.05) compared with N8.
FIGURE 2.
SDS-PAGE of supernatant concentrations from overnight cultures of L. lactis NZ9000 (control) and leucocin C-producing strain L. lactis NZ9000-p-lecCI. (A) The gel was covered with soft agar containing L. monocytogenes WSLC1018. (B) Coomassie blue G-250-stained gel. A putative leucocin C band (black arrow) in (B) and the corresponding inhibition zone in (A) appeared between 4.2 and 10 kDa. The actual size of leucocin C is about 4.6 kDa.
Growth Profiles of Wild and Constructed Strains
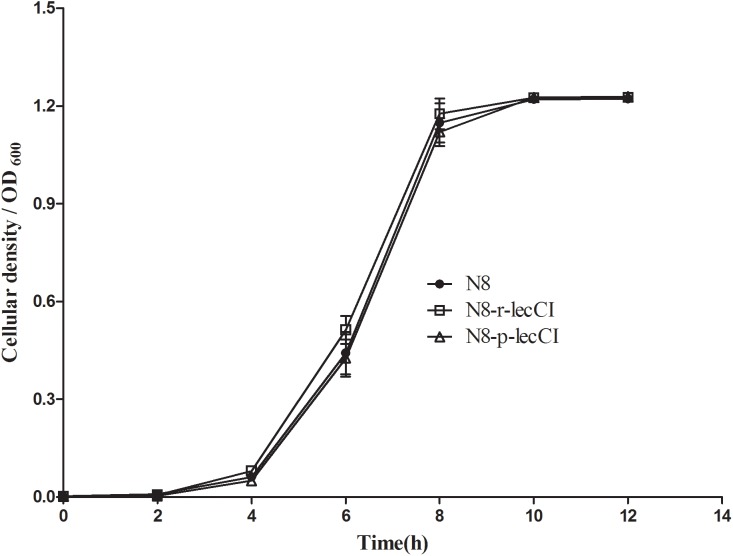
The growth curves of N8, N8-p-lecCI, and N8-r-lecCI in SMG17 (no-selective pressure) are shown in Figure 3. The growth profiles were similar among the wild-type strain N8, N8-p-lecCI, and N8-r-lecCI. Hence, the growth of N8 was not affected by the expression of leucocin C.
FIGURE 3.
Growth profiles of N8, N8-p-lecCI and N8-r-lecCI in SMG17 (non-selective pressure). The cellular density of the cultures was monitored once every 2 h by measuring OD600 for 12 h. All growth curve experiments were carried out in triplicate, and each point represents the average of separate experiments with standard deviation indicated by vertical bars. The difference between two groups was compared by t-test, the growth profiles of different strains exhibited no significant difference.
Determination of Genetic Stability
The genetic stability of different lecCI expression methods was compared. The genetic stability of N8-r-lecCI and N8-p-lecCI was tested by continuously reculturing in SGM17 broth without selective pressure for 500 and 700 generation times. At the 500th generation time, the percentage of leucocin C expression cells of population of N8-p-lecCI reduced to 99.33 ± 0.47% and this figure dropped to 98.33 ± 0.47% after 700 generations. By contrast, N8-r-lecCI maintained excellent genetic stability (the percentage of leucocin C expression cells is 100%) after 700 generation times. Hence, homologous recombination (lecC-R expression method) was superior to lecC-P expression method because the plasmid of N8-p-lecCI can be lost in the absence of antibiotic pressure.
Determination of Antimicrobial Spectrum and Antimicrobial Activity
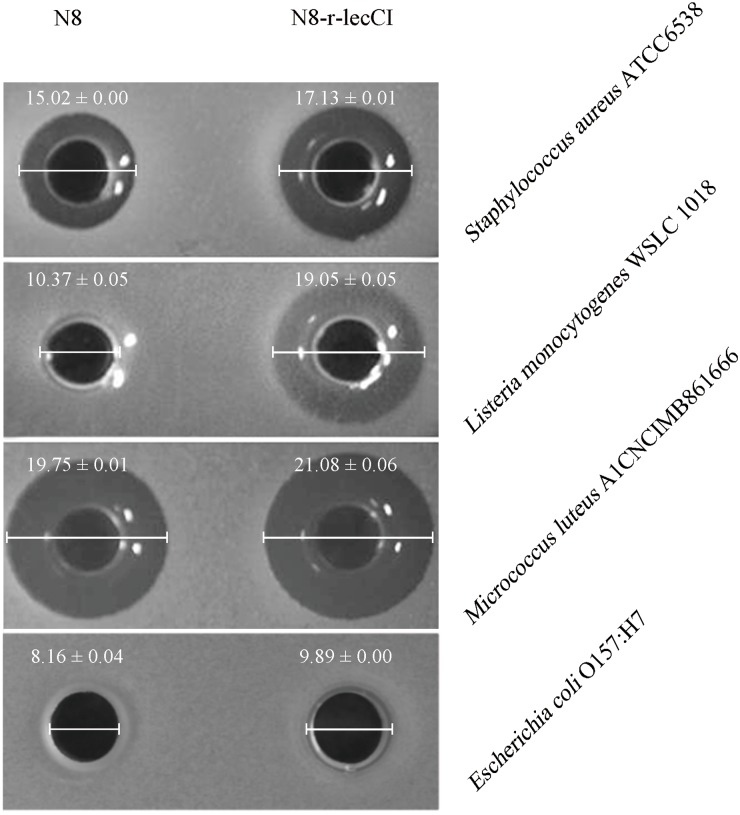
To verify the advantages of the co-expression strain N8-r-lecCI in terms of scope and activity of bacteriostasis, we compared it with the wild-type strain N8. The results of antimicrobial spectrum analysis are shown in Table 3. The pasteurized supernatant (75°C for 10 min) from N8-r-lecCI inhibited the growth of the eight strains tested, and the wild-type strain N8 inhibited only four strains. The antimicrobial spectrum of the recombinant strain N8-r-lecCI evidently increased. As shown in Figure 4, the co-expression strain N8-r-lecCI was superior to the parental strain N8 in terms of bacteriostatic activity against S. aureus ATCC6538, L. monocytogenes WSLC 1018, M. luteus A1CNCIMB861666, and E. coli O157:H7.
Table 3.
Antimicrobial spectrum of N8 and N8-r-lecCI.
| Indicative bacteria | Inhibitory effect |
|
|---|---|---|
| N8 | N8-r-lecCI | |
| S. aureus ATCC 6538 | +++ | ++++ |
| S. aureus ATCC 29213 | +++ | ++++ |
| L. monocytogenes WSLC 1018 | - | +++ |
| L. monocytogenes ATCC 19115 | + | +++ |
| E. coli ATCC 8739 | - | + |
| E. coli O157:H7 | - | + |
| S. Enteritidis ATCC 14128 | - | + |
| M. luteus A1CNCIMB 861666 | +++ | +++ |
(-), absence of inhibitory effect with twofold dilution of supernatant; (+), presence of inhibitory effect with twofold dilution of supernatant; (++), presence of inhibitory effect with fourfold dilution of supernatant; (+++), presence of inhibitory effect with eightfold dilution of supernatant; (++++), presence of inhibitory effect with 32-fold (greater or equal to) dilution of supernatant.
FIGURE 4.
Anti-bacterial assays comparing the antimicrobial activities of co-expression strain L. lactis N8-r-lecCI and wild-type N8 against S. aureus ATCC6538, L. monocytogenes WSLC 1018, M. luteus A1CNCIMB861666, and Escherichia coli O157:H7. The holes of the plates were added with 150 μl of the pasteurized supernatant (75°C, 10 min) obtained from overnight cultures of L. lactis strains. The diameter of the inhibition zones represents differences in antimicrobial activity. The difference between two groups was compared by t-test, the antimicrobial activity of N8-r-lecCI against four indicator strains was significantly different (P < 0.05) compared with N8 (unit: mm).
Effects of N8-r-lecCI Supernatant Alone or in Combination With EDTA
The activities of the N8-r-lecCI supernatant and EDTA against three common foodborne pathogens alone or in combination are summarized in Table 4. The N8-r-lecCI supernatant and EDTA showed different antimicrobial activities against the tested strains based on the calculated MICs. The MIC of the N8-r-lecCI supernatant decreased by 75% in the presence of 0.0625 mg/mL EDTA against L. monocytogenes ATCC 19155 and decreased by 75% in the presence of 0.0312 mg/mL EDTA against S. aureus ATCC 6538. The combination of EDTA and N8-r-lecCI supernatant exhibited bactericidal effect on E. coli O157:H7, and the MIC of the N8-r-lecCI supernatant decreased by 87.5% in the presence of 0.625 mg/mL EDTA. EDTA and the N8-r-lecCI supernatant showed weak or even no antibacterial activities when used alone at this low concentration.
Table 4.
Minimum inhibitory concentrations (MICs) of L. lactis N8-r-lecCI supernatants alone and in combination with EDTA against common foodborne strains.
| Strains | MIC (range) of compounds |
|||
|---|---|---|---|---|
| Alone |
Combination |
|||
| EDTA (mg/mL) | Supernatant | EDTA (mg/mL) | Supernatant | |
| L. monocytogenes ATCC 19115 | 0.25 | 1/8× | 0.0625 | 1/32× |
| S. aureus ATCC 6538 | 0.0625 | 1/64× | 0.03125 | 1/256× |
| E. coli O157:H7 | 2.5 | 1/2× | 0.625 | 1/16× |
(×), multiple.
Time-Kill Assay in Pasteurized Milk
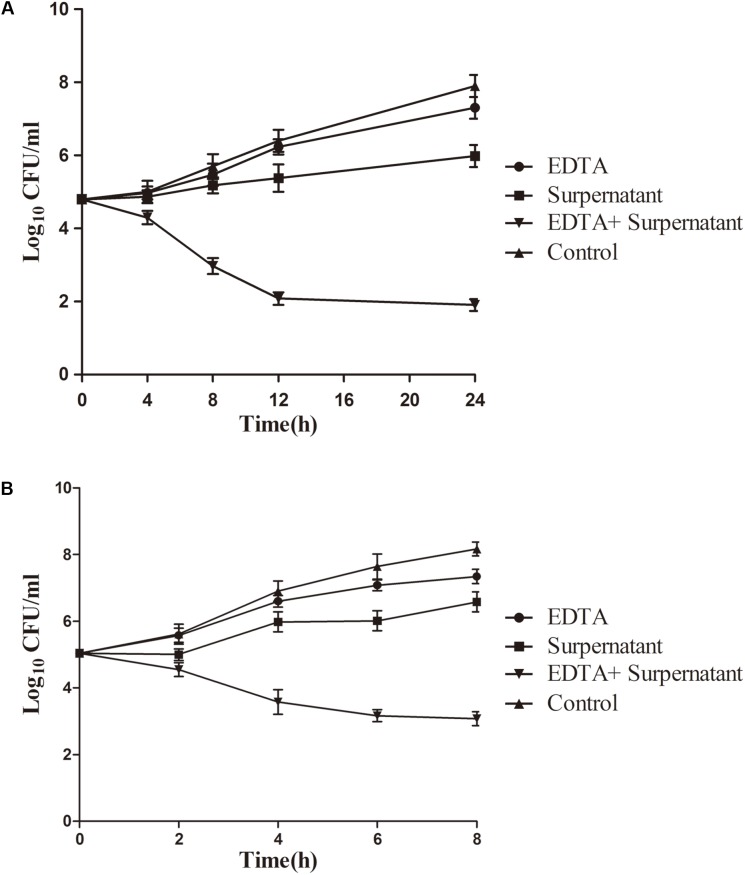
The increased antibacterial activity of the combination of EDTA and the N8-r-lecCI supernatant was further investigated in pasteurized milk to evaluate the practical application of this method. Considering the growth of L. monocytogenes at low temperature, we performed the assays at 4 and 25°C. As shown in Figure 5, bacterial growth was more effectively inhibited in the sample treated with the mixture of EDTA and N8-r-lecCI supernatant at both temperatures compared with that in the sample treated with a single antibacterial substance. The combination of EDTA and the N8-r-lecCI supernatant reduced the viable bacterial count by more than 2 log10 CFU/mL at both temperatures, indicating a positive interaction (Knezevic et al., 2013). The effect of the combination in pasteurized milk corresponded to the results presented in checkerboard assay. The results revealed that the combination of EDTA and N8-r-lecCI supernatant exerts a strong bactericidal effect in pasteurized milk against L. monocytogenes.
FIGURE 5.
Time-kill curves for EDTA and N8-lecCI supernatant alone or in combination against L. monocytogenes ATCC 19155 in pasteurized milk at (A) refrigeration temperature (4°C) and (B) room temperature (25°C). The strains were treated with EDTA (0.125 mg/mL), N8-r-lecCI supernatant (concentration of the MIC), and EDTA (0.125 mg/mL) + N8-r-lecCI supernatant (concentration of the MIC) at a starting inoculum concentration of 105 CFU/mL. All time–kill curves were constructed in triplicate, and each point represents the average of separate experiments with standard deviation indicated by vertical bars. The difference between two groups was compared by t-test, the antimicrobial effect of EDTA + N8-r-lecCI supernatant was significantly different (P < 0.05) compared with EDTA, N8-r-lecCI supernatant and control.
Effect of Antimicrobials on Bacterial Morphology
Morphological changes in L. monocytogenes cells were observed through SEM analysis to intuitively elucidate the antibacterial effect of EDTA and the N8-r-lecCI supernatant alone or in combination. As shown in Figure 6, cell damage was observed considering the evident changes in the cell morphology. The control cells maintained a relatively intact cell morphology with a smooth surface (Figure 6A), and the cells treated with EDTA at the 1/2 MIC concentration showed slight damage (Figure 6B). Moreover, the cells treated with the N8-r-lecCI supernatant at the MIC concentration showed relatively more severe damage than those treated with EDTA alone (Figure 6C). The significant cell damage was observed in the sample treated with EDTA + N8-r-lecCI supernatant. Most of the outermost layer of the bacterial cells disappeared, and the cell inclusions leaked, leading to the death of bacteria. These results revealed that the inhibition of bacterial growth could be due to the membrane disruption of cells, that is, a stronger antibacterial activity causes more severe cell damage. This assay confirmed that the combination of EDTA and N8-r-lecCI exhibited strong antibacterial activity.
FIGURE 6.

Scanning electron microscopy (SEM) observation of L. monocytogenes ATCC 19155 treated with EDTA and N8-r-lecCI supernatant alone and in combination. (A) Control cells (no treatment), (B) cells treated with EDTA (0.125 mg/mL), (C) cells treated with the N8-r-lecCI supernatant (concentration of the MIC), and (D) cells treated with EDTA and the N8-r-lecCI supernatant.
Discussion
The food-grade strain L. lactis N8 was used as the original strain because of its potential as nisin-leucocin C co-expression carrier. The results of agar diffusion assay and SDS-PAGE verified that the constructed strains can successfully secrete leucocin C and nisin, and the secreted leucocin C exhibited distinct inhibitory effect against Listeria monocytogenes. It is also worth noting that the expression of leucocin C did not influence the growth of strains.
Accumulating evidences indicated that serial passage of plasmid-borne strains expressing a protein often result in genetic instability (O’Kennedy et al., 2000), which is an important factor that should be considered when developing recombinant strains for commercial application in industrial fermentation. Therefore, the genetic stability of different lecCI expression methods was compared. Homologous recombination method (N8-r-lecCI) was superior to plasmid expression method (N8-p-lecCI) because the plasmid of N8-p-lecCI can be lost in the absence of antibiotic pressure. In addition, the relative advantage of N8-r-lecCI compared with N8-p-lecCI is that non-food source gene was introduced into the expression host. This result verified that the recombinant strain N8-r-lecCI has excellent genetic stability and can stably coexpress nisin–leucocin C, which means it is more suitable for food fermentation and application.
The co-production strain N8-r-lecCI exhibits broad activity spectrum and highly efficient bacteriostatic ability against Gram-positive foodborne pathogens. Several studies have demonstrated that nisin show additive or synergistic effects when used in combination with other antimicrobial agents (Branen and Davidson, 2004; Field et al., 2016; Shi et al., 2017). However, to the best of our knowledge, no information regarding the antibacterial activity of nisin co-expression with leucocin C or in combination with leucocin C was available in literature. Our study first demonstrated that the co-expression of these two bacteriocins which have different modes of action results in highly efficient antimicrobial activity against Listeria monocytogenes and Staphylococcus aureus. It is a novel combination that is worthy of further investigation.
The food grade chelators EDTA and the N8-r-lecCI supernatant were combined to further enhance the antimicrobial activity of the N8-r-lecCI supernatant. Previous studies have demonstrated that EDTA could be used in the food industry due to its active antibacterial activity and its safety (Heimbach et al., 2000). Our study showed the synergistic antibacterial activity of N8-r-lecCI supernatant and EDTA against L. monocytogenes WSLC1018 and E. coli ATCC25922, and enhanced activity against S. aureus ATCC 6538. Previous reports described that the combination of nisin and EDTA can enhance antimicrobial ability against Gram-negative bacteria and L. monocytogenes (Zhang and Mustapha, 1999; Mastromatteo et al., 2010; Norhana et al., 2012). Several studies have explained the reason regarding the antibacterial efficiency of the combination of nisin and EDTA (Branen and Davidson, 2004). EDTA could release up to 50% of the lipopolysaccharide (LPS) to affect the outer membrane permeability (Leive, 1965), thereby increasing the susceptibility of cells to nisin. Hence, we hypothesized that EDTA could contribute to efficient binding of leucocin C to mannose phosphotransferase permease, which serves as target in sensitive cells (Kjos et al., 2009; Wan et al., 2013). This assumption must be further investigated. In the present study, the results revealed the positive interaction among leucocin C, nisin, and EDTA against some foodborne strains. In addition, the organic acids of the N8-r-lecCI fermentation supernatant helped to suppress the pathogens. Organic acids are widely used for food preservation (Young and Foegeding, 1993). The antimicrobial activity of nisin or other bacteriocins against L. monocytogenes F6854 can be enhanced in the presence of organic acids (Smith et al., 2016). Overall, the antibacterial activity of L. lactis N8-r-lecCI supernatant can be further enhanced in the presence of the low concentration of EDTA.
The practical antibacterial effect of this combination was verified in a food model through time-kill assay. The effect of the combination in pasteurized milk was consistent with the result presented in checkerboard assay, which shows excellent antimicrobial activity in pasteurized milk preservation. The current standards of pasteurization appear to be adequate for public health assurance of milk safety (Holsinger et al., 1997). However, outbreaks associated with pasteurized milk continue to occur (De Buyser et al., 2001; Centers for Disease Control and Prevention, 2008). The combination of bacteriocins has excellent antimicrobial potential in pasteurized milk preservation.
Conclusion
Lactococcus lactis N8-r-lecCI co-expressing nisin and leucocin C is characterized by high genetic stability and antibacterial efficiency, and exhibits high potential in food preservation and processing.
Author Contributions
DZ, YF, and MQ designed the experiments. YF, XW, and WQ performed all the major experiments. DM, FL, and MQ analyzed the experimental results. DM and PS provided the additional experimental assistance. YF and DZ wrote the manuscript. OK and MQ helped to revise the manuscript. MQ and HX provided the laboratory equipment and place.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- EDTA
ethylenediaminetetraacetic acid
- lecCI
bacteriocin leucocin C and its immunity gene.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31770102), the Sino-Swiss Science and Technology Cooperation Project supported by the Ministry of Science and Technology of the People’s Republic of China (2015DFG32140).
References
- Barbosa A. A., Mantovani H. C., Jain S. (2017). Bacteriocins from lactic acid bacteria and their potential in the preservation of fruit products. Crit. Rev. Biotechnol. 37 1–13. 10.1080/07388551.2016.1262323 [DOI] [PubMed] [Google Scholar]
- Barefoot S. F., Klaenhammer T. R. (1983). Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45 1808–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia A. K., Johnson M., Ray B. (1987). Direct detection of an antimicrobial peptide of Pediococcus acidilactici in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Ind. Microbiol. Biotechnol. 2 319–322. 10.1007/BF01569434 [DOI] [Google Scholar]
- Branen J. K., Davidson P. M. (2004). Enhancement of nisin, lysozyme, and monolaurin antimicrobial activities by ethylenediaminetetraacetic acid and lactoferrin. Int. J. Food Microbiol. 90 63–74. 10.1016/s0168-1605(03)00172-7 [DOI] [PubMed] [Google Scholar]
- Campion A., Morrissey R., Field D., Cotter P. D., Hill C., Ross R. P. (2017). Use of enhanced nisin derivatives in combination with food-grade oils or citric acid to control Cronobacter sakazakii and Escherichia coli O157:H7. Food Microbiol. 65 254–263. 10.1016/j.fm.2017.01.020 [DOI] [PubMed] [Google Scholar]
- Capozzi V., Russo P., Dueñas M., López P., Spano G. (2012). Lactic acid bacteria producing B-group vitamins: a great potential for functional cereals products. Appl. Microbiol. Biotechnol. 96 1383–1394. 10.1007/s00253-012-4440-2 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2008). Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy–Massachusetts, 2007. MMWR Morb. Mortal. Wkly. Rep. 57 1097–1100. [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement, M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cole M. B., Jones M. V., Holyoak C. (1990). The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol. 69 63–72. 10.1111/j.1365-2672.1990.tb02912.x [DOI] [PubMed] [Google Scholar]
- De Buyser M.-L., Dufour B., Maire M., Lafarge V. (2001). Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67 1–17. 10.1016/S0168-1605(01)00443-3 [DOI] [PubMed] [Google Scholar]
- Delves-Brougthon J. (1990). Nisin and its uses as a food preservative. Food Technol. 44 100–117. [Google Scholar]
- Field D., Seisling N., Cotter P. D., Ross R. P., Hill C. (2016). Synergistic nisin-polymyxin combinations for the control of Pseudomonas biofilm formation. Front. Microbiol. 7:1713. 10.3389/fmicb.2016.01713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Chikindas M. L. (2007). Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113 1–15. 10.1016/j.ijfoodmicro.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Garcia P., Martinez B., Rodriguez L., Rodriguez A. (2010). Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int. J. Food Microbiol. 141 151–155. 10.1016/j.ijfoodmicro.2010.04.029 [DOI] [PubMed] [Google Scholar]
- Grande M. J., Lucas R., Abriouel H., Omar N. B., Maqueda M., Martinez-Bueno M., et al. (2005). Control of Alicyclobacillus acidoterrestris in fruit juices by enterocin AS-48. Int. J. Food Microbiol. 104 289–297. 10.1016/j.ijfoodmicro.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Heimbach J., Rieth S., Mohamedshah F., Slesinski R., Samuel-Fernando P., Sheehan T., et al. (2000). Safety assessment of iron EDTA [sodium iron (Fe3+) ethylenediaminetetraacetic acid]: summary of toxicological, fortification and exposure data. Food Chem. Toxicol. 38 99–111. 10.1016/S0278-6915(99)00125-8 [DOI] [PubMed] [Google Scholar]
- Holsinger V. H., Rajkowski K. T., Stabel J. R. (1997). Milk pasteurisation and safety: a brief history and update. Rev. Sci. Tech. 16 441–451. 10.20506/rst.16.2.1037 [DOI] [PubMed] [Google Scholar]
- Jemmi T., Stephan R. (2006). Listeria monocytogenes: food-borne pathogen and hygiene indicator. Rev. Sci. Tech. 25 571–580. 10.20506/rst.25.2.1681 [DOI] [PubMed] [Google Scholar]
- Kaferstein F. K., Motarjemi Y., Bettcher D. W. (1997). Foodborne disease control: a transnational challenge. Emerg. Infect. Dis. 3 503–510. 10.3201/eid0304.970414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M., Nes I. F., Diep D. B. (2009). Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 155 2949–2961. 10.1099/mic.0.030015-0 [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R. (1993). Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12 39–85. 10.1111/j.1574-6976.1993.tb00012.x [DOI] [PubMed] [Google Scholar]
- Knezevic P., Curcin S., Aleksic V., Petrusic M., Vlaski L. (2013). Phage-antibiotic synergism: a possible approach to combatting Pseudomonas aeruginosa. Res. Microbiol. 164 55–60. 10.1016/j.resmic.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Kuipers O. P., de Ruyter P. G., Kleerebezem M., de Vos W. M. (1998). Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64 15–21. 10.1016/S0168-1656(98)00100-X [DOI] [Google Scholar]
- Laksanalamai P., Joseph L. A., Silk B. J., Burall L. S., Tarr L. C., Gerner-Smidt P., et al. (2012). Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One 7:e42448. 10.1371/journal.pone.0042448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. M., Bongers R. S., Kleerebezem M. (2007). Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73 1126–1135. 10.1128/AEM.01473-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langa S., Arques J. L., Medina M., Landete J. M. (2017). Coproduction of colicin V and lactic acid bacteria bacteriocins in lactococci and enterococci strains of biotechnological interest. J. Appl. Microbiol. 122 1159–1167. 10.1111/jam.13439 [DOI] [PubMed] [Google Scholar]
- LeBlanc J. G., Van Sinderen D., Hugenholtz J., Piard J.-C., Sesma F., de Giori G. S. (2010). Risk assessment of genetically modified lactic acid bacteria using the concept of substantial equivalence. Curr. Microbiol. 61 590–595. 10.1007/s00284-010-9657-7 [DOI] [PubMed] [Google Scholar]
- Leive L. (1965). Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem. Biophys. Res. Commun. 21 290–296. 10.1016/0006-291X(65)90191-9 [DOI] [PubMed] [Google Scholar]
- Li R., Takala T. M., Qiao M., Xu H., Saris P. E. (2011). Nisin-selectable food-grade secretion vector for Lactococcus lactis. Biotechnol. Lett. 33 797–803. 10.1007/s10529-010-0503-6 [DOI] [PubMed] [Google Scholar]
- Lucera A., Costa C., Conte A., Del Nobile M. A. (2012). Food applications of natural antimicrobial compounds. Front. Microbiol. 3:287 10.3389/fmicb.2012.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Irujo J. J., Villahermosa M. L., Alberdi E., Santiago E. (1996). A checkerboard method to evaluate interactions between drugs. Biochem. Pharmacol. 51 635–644. 10.1016/S0006-2952(95)02230-9 [DOI] [PubMed] [Google Scholar]
- Mastromatteo M., Lucera A., Sinigaglia M., Corbo M. R. (2010). Synergic antimicrobial activity of lysozyme, nisin, and EDTA against Listeria Monocytogenes in ostrich meat patties. J. Food Sci. 75 M422–M429. 10.1111/j.1750-3841.2010.01732.x [DOI] [PubMed] [Google Scholar]
- Norhana M. N. W., Poole S. E., Deeth H. C., Dykes G. A. (2012). Effects of nisin, EDTA and salts of organic acids on Listeria monocytogenes, Salmonella and native microflora on fresh vacuum packaged shrimps stored at 4°C. Food Microbiol. 31 43–50. 10.1016/j.fm.2012.01.007 [DOI] [PubMed] [Google Scholar]
- O’Kennedy R. D., Baldwin C., Keshavarz-Moore E. (2000). Effects of growth medium selection on plasmid DNA production and initial processing steps. J. Biotechnol. 76 175–183. 10.1016/S0168-1656(99)00187-X [DOI] [PubMed] [Google Scholar]
- O’Sullivan L., Ross R. P., Hill C. (2002). Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie 84 593–604. 10.1016/S0300-9084(02)01457-8 [DOI] [PubMed] [Google Scholar]
- Qiao M., Immonen T., Koponen O., Saris P. E. (1995). The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131 75–80. 10.1016/0378-1097(95)00238-Z [DOI] [PubMed] [Google Scholar]
- Qiao M., Ye S., Koponen O., Ra R., Usabiaga M., Immonen T., et al. (1996). Regulation of the nisin operons in Lactococcus lactis N8. J. Appl. Microbiol. 80 626–634. 10.1111/j.1365-2672.1996.tb03267.x [DOI] [PubMed] [Google Scholar]
- Renault P. (2002). Genetically modified lactic acid bacteria: applications to food or health and risk assessment. Biochimie 84 1073–1087. 10.1016/s0300-9084(02)00029-9 [DOI] [PubMed] [Google Scholar]
- Roberts R., Zottola E. (1993). Shelf-life of pasteurized process cheese spreads made from cheddar cheese manufactured with a nisin-producing starter culture1. J. Dairy Sci. 76 1829–1836. 10.3168/jds.S0022-0302(93)77515-3 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- Shi C., Zhang X., Zhao X., Meng R., Liu Z., Chen X., et al. (2017). Synergistic interactions of nisin in combination with cinnamaldehyde against Staphylococcus aureus in pasteurized milk. Food Control 71 10–16. 10.1016/j.foodcont.2016.06.020 [DOI] [Google Scholar]
- Smith M. K., Draper L. A., Hazelhoff P. J., Cotter P. D., Ross R. P., Hill C. (2016). A bioengineered nisin derivative, M21A, in combination with food grade additives eradicates biofilms of Listeria monocytogenes. Front. Microbiol. 7:1939. 10.3389/fmicb.2016.01939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swetwiwathana A., Visessanguan W. (2015). Potential of bacteriocin-producing lactic acid bacteria for safety improvements of traditional Thai fermented meat and human health. Meat Sci. 109 101–105. 10.1016/j.meatsci.2015.05.030 [DOI] [PubMed] [Google Scholar]
- Sybesma W., Hugenholtz J., De Vos W. M., Smid E. J. (2006). Safe use of genetically modified lactic acid bacteria in food. Bridging the gap between consumers, green groups, and industry. Electron. J. Biotechnol. 9 424–448. 10.2225/vol9-issue4-fulltext-12 [DOI] [Google Scholar]
- Velusamy V., Arshak K., Korostynska O., Oliwa K., Adley C. (2010). An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol. Adv. 28 232–254. 10.1016/j.biotechadv.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Wan J., Harmark K., Davidson B. E., Hillier A. J., Gordon J. B., Wilcock A., et al. (1997). Inhibition of Listeria monocytogenes by piscicolin 126 in milk and Camembert cheese manufactured with a thermophilic starter. J. Appl. Microbiol. 82 273–280. 10.1046/j.1365-2672.1997.00349.x [DOI] [PubMed] [Google Scholar]
- Wan X., Li R., Saris P. E., Takala T. M. (2013). Genetic characterisation and heterologous expression of leucocin C, a class IIa bacteriocin from Leuconostoc carnosum 4010. Appl. Microbiol. Biotechnol. 97 3509–3518. 10.1007/s00253-012-4406-4 [DOI] [PubMed] [Google Scholar]
- Wan X., Saris P. E., Takala T. M. (2015). Genetic characterization and expression of leucocin B, a class IId bacteriocin from Leuconostoc carnosum 4010. Res. Microbiol. 166 494–503. 10.1016/j.resmic.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Young K., Foegeding P. (1993). Acetic, lactic and citric acids and pH inhibition of Listeria monocytogenes Scott A and the effect on intracellular pH. J. Appl. Bacteriol. 74 515–520. [PubMed] [Google Scholar]
- Yousef A. E., Lou Y. (1999). “Characteristics of Listeria monocytogenes important to food processors,” in Listeria, Listeriosis, and Food Safety 2nd Edn, eds Ryser E. T., Marth E. H. (New York, NY: Marcel Dekker, Inc.), 131–224. [Google Scholar]
- Zhang S. S., Mustapha A. (1999). Reduction of Listeria monocytogenes and Escherichia coli O157:H7 numbers on vacuum-packaged fresh beef treated with nisin or nisin combined with EDTA. J. Food Prot. 62 1123–1127. 10.4315/0362-028X-62.10.1123 [DOI] [PubMed] [Google Scholar]