Abstract
Triple-negative breast cancer (TNBC) is extremely aggressive and associated with poor prognosis. There are no known predictive or prognostic markers for TNBC. Inhibition of tumor protein P53 (TP53) has been demonstrated to increase the levels of cluster of differentiation 117 (CD117) in human colorectal cancer cells. However, the function of TP53 in the regulation of CD117 in TNBC has, to the best of our knowledge, not been reported. In the present study, the association between the expression of CD117 protein and TP53 mutations was investigated, and their prognostic value in patients with TNBC was assessed. A total of 58 TNBC and 48 non-TNBC breast cancer tissue samples were assessed for the expression of CD117, p53 and TP53 mutations. The marker of proliferation Ki-67 (MKI67) proliferation index and vascular invasion index (obtained by measuring D2-40 and CD34) was investigated via immunohistochemistry, and mutations in exons 4–8 of TP53 were measured using direct sequencing. Associations between CD117 and p53 levels or TP53 mutations and clinical parameters were statistically evaluated. The rates of CD117 or MKI67 positivity, CD117+/TP53 missense mutation+, TP53 missense mutations or recurrence were significantly higher in patients with TNBC than in patients with non-TNBC. In TNBC tissues, the presence of CD117 was associated with TP53 missense mutations (P=0.031), vascular invasion, recurrence and MKI67. CD117+/TP53 missense mutation+ also associated with vascular invasion, recurrence and MKI67. Under univariate analysis, MKI67, vascular invasion, CD117, CD117+/TP53 missense mutation+ and TP53 missense mutations were associated with the overall survival of patients with TNBC. Multivariate analysis revealed that vascular invasion and CD117+/TP53 missense mutation+ in primary tumors were independent prognostic factors in patients with TNBC. In conclusion, CD117+/TP53 missense mutation+ was associated with MKI67, vascular invasion and tumor recurrence in TNBC. The presence of CD117 and TP53 missense mutations together in the primary tumors was an independent prognostic factor for survival of patients with TNBC.
Keywords: triple-negative breast cancer, cluster of differentiation 117, tumor protein P53 missense mutations, vascular invasion, prognosis
Introduction
Triple-negative breast cancer (TNBC) is characterized by the absence of estrogen, progesterone and human epidermal growth factor receptor 2 (HER2) receptors. TNBC makes up between 15 and 20% of all types of breast cancer, are aggressive, and are associated with poorer survival rates compared with non-TNBC (1,2). Although patients with TNBC respond to chemotherapy, they are more likely to suffer early relapse than patients with other breast cancer subtypes (3). Therefore, there is a requirement to identify prognostic and predictive markers in TNBC.
Cluster of differentiation 117 (CD117; encoded by KIT) regulates cellular differentiation, proliferation, adhesion and apoptosis (4). CD117 is involved in the development of several malignant tumor types including gastrointestinal stromal cell tumors, small-cell lung, ovarian and breast cancer (4–7). Immunohistochemical staining has revealed that CD117 protein is overexpressed in primary malignant tumors, including operable esophageal squamous cell carcinoma and vulvar melanoma, and may be a valuable prognostic marker in esophageal squamous cell carcinoma (8). However, the results of studies concerning the prognostic significance of CD117 in patients with breast cancer or TNBC are conflicting: Kashiwagi et al have suggested that CD117 protein is associated with poor prognosis (9), whereas others identified no significant association between CD117 and prognosis in breast cancer or TNBC (10,11).
The tumor protein P53 (TP53) gene encodes the tumor suppressor protein p53, and is activated by various stresses including genotoxic damage, hypoxia and heat shock (12,13). TP53 mutations are most often located at exons 5–8, inhibiting the tumor suppressor activity of p53 (14). The association between TP53 mutations, p53 expression and the prognosis of patients with breast cancer or TNBC is inconsistent (15–17); a group of patients with TP53 missense mutations experienced the shortest survival and a reduction in relapse-free survival rates (15,16); however, there was no association between TP53 mutations and clinical outcome in another study (17).
A previous study revealed that mutations to TP53 upregulated CD117 expression in colon cancer cells and promoted the invasion of tumor cells (18). TP53 mutations have been demonstrated to inactivate the microRNA miR-34, and to promote CD117 expression in human colorectal cancer cells (18); however, there are no reports regarding the association between TP53 mutations and CD117 expression in TNBC.
In the present study, it was hypothesized that CD117 expression was associated with TP53 mutations in patients with TNBC. The expression levels of CD117 and p53 were measured and TP53 mutations were assessed in 58 TNBC tumor specimens and 48 control specimens, and their association with the clinicopathological characteristics of patients with TNBC was analyzed.
Materials and methods
Patients
The Ethics Committee for Human Studies at Shanghai Jiao Tong University (Shanghai, China) approved the present study, which was conducted in accordance with The Declaration of Helsinki. Participants were fully informed of the procedures, and written informed consent was obtained from each.
Between January 2008 and August 2012 at Shanghai Jiao Tong University Affiliated Sixth People's Hospital, 58 TNBC tissues and 48 non-TNBC samples were collected during surgery. All patients were women, with a mean age of 55.30±9.95 years (range, 32–80 years). Patients with non-TNBC had a mean age of 55.27±8.75 years (range, 43–77 years); patients with TNBC had a mean age of 55.33±10.92 (range, 32–80 years). Stages of breast cancer were determined using Nottingham staging, recommended by the American Joint Committee on Cancer (19). A total of 39 specimens (20 TNBC and 19 non-TNBC cases) were determined to be stage I, and 67 specimens (38 TNBC and 29 non-TNBC cases) as stage II–III. All patients received modified radical mastectomy without preoperative chemotherapy or radiotherapy. Following surgery, routine neoadjuvant chemotherapy was administered.
The end of follow-up was June 2014. The 5-year survival rate for the entire cohort of 106 patients was 49%. The recurrence rate was 35.8%. During the follow-up period, recurrence occurred in 38 cases (30 patients succumbed to metastasis, and 8 patients succumbed whilst receiving chemotherapy). The median follow-up period was 50.6 months. The overall survival time of patients was calculated from the date of surgery to the date of mortality or the date of last contact.
Immunohistochemistry
Immunohistochemistry was performed on 4-µm-thick sections from 10% formalin-fixed paraffin-embedded (FFPE) tissue specimens as previously described (20). Sections were deparaffinized with 100% xylene (25°C) and rehydrated through a graded ethanol series (100, 95 and 70% ethanol) and were subjected to microwave antigen retrieval by incubating sections at 98 to 100°C in 0.01 M citrate buffer (pH 6.0) for 15 min. Antibodies targeting D2-40 (cat. no. sc-59347; 1:100) were purchased from Santa Clara Biotechnology, Inc. (Dallas, TX, USA). Other antibodies were purchased from Dako (Agilent Technologies, Inc.), directed against the following: Estrogen receptor (ER; cat. no. IS657; 1:80), progesterone receptor (PR; cat. no. IS068; 1:50), human epidermal growth factor receptor 2 (HER2; cat. no. 20027850; 1:200), CD117 (cat. no. 10109679; 1:200), CD34 (cat. no. GA632; 1:50), marker of proliferation Ki-67 (MKI67; cat. no. IR626; 1:200), and p53 (cat. no. IS616; 1:100), which reacts with an epitope between amino acids 19–26 and recognizes wild-type and mutant p53 protein. The sections were incubated with the primary antibodies for 1 h at 25°C, followed by an incubation for 30 min at 37°C with a horseradish peroxidase-conjugated secondary antibody [Dako EnVision+ System-HRP (DAB); Dako, Agilent Technologies, Inc.]. The slides were assessed in an auto immunostainer Link 48 (Agilent Technologies, Inc.; Dako) according to the manufacturer's protocols. Slides were counterstained with Mayer's hematoxylin. H&E staining of other slides from the same samples was also performed in order to assess the presence/absence of vascular invasion of tumors. Slides were stained with hematoxylin (Harris Formula, Surgipath Medical Industries, Inc., Richmond, IL, USA) at 25°C for 1 min, rinsed in running distilled water (10 min), and then stained with an eosin solution (Surgipath Medical Industries, Inc.) for 1 min and washed with distilled water again, dehydrated with 100% ethanol (25°C) and mounted.
All the results of immunohistohchemistry were blindly accessed by 2 independent pathologists with long-standing experience in the Department of Pathology, Shanghai Jiao Tong University Affiliated Sixth People's Hospital. Nuclear staining >10% in tumor cells was considered to indicate p53- or MKI67-positivity. Nuclear staining in >1% of tumor cell nuclei was considered a positive indication of the presence of ER and PR. Immunostaining for HER2 and CD117 (11,21) was graded according to the percentage of cells: 0, none; 1+, <10% staining weak and incomplete; 2+, ≥10%, staining weak to moderate; and 3+, ≥10% strong staining. HER2 immunostaining was considered positive if graded 3+, or if gene amplification was confirmed by fluorescence in situ hybridization (FISH) in patients exhibiting 2+ immunostaining. Breast cancer samples with ≥10% positive cells and with scores ≥2 were considered to indicate CD117 positivity. Vascular invasion was identified by staining for CD34 (endothelial cell marker) or D2-40 (podoplanin, a membrane protein specific for lymphatic endothelium), in accordance with a previous study (22).
FISH
HER2 status was detected using FISH. A PathVysion dual-color fluorescence probes kit (HER2/CEP17) was purchased from Vysis, Inc. (Abbott Pharmaceutical Co. Ltd., Lake Bluff, IL, USA). The assay was performed according to the manufacturer's protocol. All samples were analyzed using an Olympus BX51 fluorescence microscope (Olympus Corporation, Tokyo, Japan, original magnification, ×1,000) equipped with a set of the appropriate filters (Vysis Inc.; Abbott Pharmaceutical Co. Ltd.). The hybridization results were evaluated by 2 independent pathologists in the Department of Pathology, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, blinded to the clinical outcomes. Categories of FISH were classified as previously described (23).
TP53 mutation analysis
For each case, 10 5-µm-thick slices were collected in a 1.5 ml tube, these FFPE specimen tissue sections were first deparaffinized with 100% xylene at 25°C and 100% ethanol at 25°C. Subsequently, DNA was extracted from FFPE tissue specimens using a QIAamp DNA extraction kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. The samples were incubated in 20 µl proteinase K solution at 56°C for 1 h, and then incubated in FTB buffer at 90°C for 1 h, then wash buffers (500 µl AW1 and 500 µl AW2) washing for 1 min at 25°C. Finally, the DNA was eluted in DTE buffer (all Qiagen GmbH).
Mutations in the TP53 gene were identified via direct sequencing. Polymerase chain reaction (PCR) amplification and direct sequencing of the TP53 gene (exon 4–8) were performed in 106 breast cancer samples. The primers were designed to amplify exons 4–8 of TP53 (Table I). PCR was performed using a Mastercycler gradient PCR machine (Eppendorf, Hamburg, Germany) under the following amplification conditions: 94°C for 10 min; 40 cycles of 94°C for 45 sec, 61°C for 45 sec, and 72°C for 45 sec; with a final extension at 72°C for 7 min. The PCR products were purified using a QIAquick Gel Extraction kit (Qiagen GmbH) and were prepared for sequencing via a 3500Dx genetic analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions were: 94°C for 1 min; 24 cycles of 94°C for 10 sec, 50°C for 5 sec, and 60°C for 1 min; and final extension at 72°C for 5 min. The sequences were analyzed using Chromas Lite software version 2.01 (Technelysium Pty Ltd., South Brisbane, Australia).
Table I.
Polymerase chain reaction primers for the tumor protein P53 gene.
| Exon | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|---|---|
| 4I | GCTCTTTTCACCCATCTACAG | GAAGGGACAGAAGATGACAG |
| 4II | CTGCACCAGCAGCTCCTA | GAAGTCTCATGGAAGCCAG |
| 5 | TCACTTGTGCCCTGACTTTCA | TCTCCAGCCCCAGCTGCT |
| 6 | TTCCTCACTGATTGCTCTTAG | GACCCCAGTTGCAAACCAG |
| 7 | GCGCACTGGCCTCATCTTG | CACAGCAGGCCAGTGTGCA |
| 8 | AGGACCTGATTTCCTTACTGC | GAATCTGAGGCATAACTGCAC |
Statistical analysis
The significance of association between TNBC and the various clinical factors or potential prognostic markers was determined using the χ2 test. Survival analysis was performed using a Kaplan-Meier plot analysis and a log-rank test. Hazard ratios with 95% confidence intervals were calculated using the Cox proportional hazards model, which was used to compute univariate and multivariate hazard ratios for the study parameters. P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA).
Results
Clinical characteristics of TNBC patients
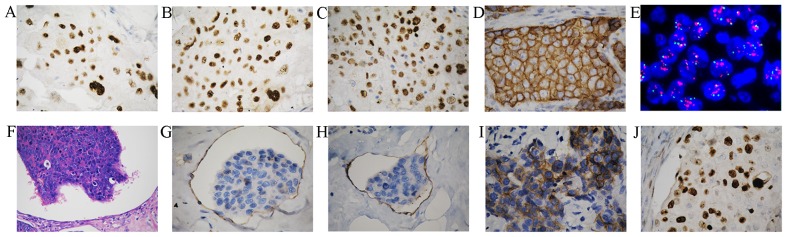
A total of 58 patients with TNBC and 48 patients with non-TNBC were included in the study. Positive stains of ER, PR, and MKI67 were primarily located in the nuclei (Fig. 1A-C). Immunohistochemistry revealed that HER2 protein was located at the cell membrane, and gene amplification of HER2 was detected using FISH analysis (Fig. 1D and E). Vascular invasion was assessed by hematoxylin and eosin staining (Fig. 1F) and by immunohistochemistry using anti-CD34 (recognizing endothelial cells) and anti-D2-40 (recognizing lymphatic endothelial cells) antibodies (Fig. 1G and H). Compared with patients with non-TNBC, those with TNBC had a higher rate of vascular invasion (P=0.043), lymph node metastasis (P=0.017) and tumor recurrence (P=0.012; Table II), and a higher proliferation index (MKI67; P=0.025; Table III).
Figure 1.
Immunohistochemical analysis of breast cancer tissues. Examples of positive immunohistochemical staining for the (A) estrogen receptor, (B) progesterone receptor, (C) MKI67, and (D) HER2 in tumor cells (original magnification, ×400). (E) Representative image of HER2 gene amplification in tumor tissue was observed using FISH in non-TNBC tumor cells (original magnification, ×1,000). (F) Tumor thrombus was observed in vessels (original magnification, ×200). (G) Blood vessel invasion was indicated by CD34 staining (original magnification, ×400). (H) Lymphatic invasion indicated by D2-40 staining (original magnification, ×400). Representative examples of (I) positive CD117 protein staining (J) nuclear localization of p53 and were observed in breast cancer (original magnification, ×400). HER2, human epidermal growth factor receptor 2; CD, cluster of differentiation; FISH, fluorescence in situ hybridization; TNBC, triple negative breast cancer; MKI67, marker of proliferation Ki-67.
Table II.
Summary of clinical and pathological features of 106 patients with breast cancer.
| Variable | Non-TNBC, n (%) | TNBC, n (%) | Total, n (%) | P-value |
|---|---|---|---|---|
| Subjects | 48 | 58 | 106 | |
| Age at operation, years | 0.245 | |||
| ≤50 | 21 (43.8) | 19 (32.8) | 40 (37.7) | |
| >50 | 27 (56.3) | 39 (67.2) | 66 (62.3) | |
| Tumor grade | 0.588 | |||
| I | 19 (39.6) | 20 (34.5) | 39 (36.8) | |
| II/III | 29 (60.4) | 38 (65.5) | 67 (63.2) | |
| Tumor, cm | 0.302 | |||
| ≤2 | 28 (58.3) | 28 (48.3) | 56 (52.8) | |
| >2 | 20 (41.7) | 30 (51.7) | 50 (47.2) | |
| Lymph node status | 0.017 | |||
| Negative | 31 (64.6) | 24 (41.4) | 55 (51.9) | |
| Positive | 17 (35.4) | 34 (58.6) | 51 (48.1) | |
| Vascular invasion | 0.043 | |||
| No | 43 (89.6) | 43 (74.1) | 85 (80.2) | |
| Yes | 5 (10.4) | 15 (25.9) | 21 (19.8) | |
| Nerve invasion | 0.726 | |||
| No | 37 (77.1) | 43 (74.1) | 80 (75.5) | |
| Yes | 11 (22.9) | 15 (25.9) | 26 (24.5) | |
| Recurrence | 0.012 | |||
| No | 37 (77.1) | 31 (53.4) | 68 (64.2) | |
| Yes | 11 (22.9) | 27 (46.6) | 38 (35.8) |
TNCB, triple negative breast cancer.
Table III.
TP53 and CD117 expression in patients with breast cancer.
| Variable | Non-TNBC, n (%) | TNBC, n (%) | P-value |
|---|---|---|---|
| CD117 | 0.045a | ||
| Negative | 34 (70.8) | 30 (51.7) | |
| Positive | 14 (29.2) | 28 (48.3) | |
| CD117/TP53MIS | 0.009a | ||
| Negative | 42 (87.5) | 38 (65.5) | |
| Positive | 6 (12.5) | 20 (34.5) | |
| TP53 MIS | 0.027a | ||
| Negative | 31 (64.6) | 25 (43.1) | |
| Positive | 17 (35.4) | 33 (56.9) | |
| MKI67 | 0.025a | ||
| Negative | 27 (56.3) | 20 (34.5) | |
| Positive | 21 (43.8) | 38 (65.5) | |
| p53 | 0.530 | ||
| Negative | 31 (64.6) | 34 (58.6) | |
| Positive | 17 (35.4) | 24 (41.4) | |
| TP53 mutation | 0.055 | ||
| Negative | 16 (33.3) | 10 (17.2) | |
| Positive | 32 (66.7) | 48 (82.8) | |
| Codon 72 | 0.530 | ||
| Negative | 31 (64.6) | 34 (58.6) | |
| Positive | 17 (35.4) | 24 (41.4) |
P<0.05. TNCB, triple negative breast cancer; CD, cluster of differentiation; TP53, tumor protein p53; MKI67, marker of proliferation Ki-67.
Expression of CD117 is increased in TNBC patients
CD117 expression was identified in 28/58 of TNBC samples (48.3%), and in 14/48 of non-TNBC samples (29.2%). Positive staining for CD117 was observed in the cytoplasm or at the cell membrane (Fig. 1I). The TNBC tissues exhibited a higher rate of positive staining for CD117 compared with non-TNBC tissues (P=0.045; Table III). The presence of p53 protein, localized in the nuclei (Fig. 1J), was similar in TNBC and non-TNBC patients (P=0.530; Table III). Furthermore, in TNBC, positive expression of CD117 was associated with vascular invasion (P=0.024), proliferation index (P=0.01) and tumor recurrence (P=0.001; Table IV). No association was identified between CD117 expression and tumor size, tumor grade, nerve invasion, lymph node metastasis, p53 protein or TP53 mutation in TNBC (Table IV).
Table IV.
Association of CD117 with clinicopathological features of 58 patients with TNBC.
| CD117 | |||
|---|---|---|---|
| Variable | Negative, n (%) | Positive, n (%) | P-value |
| p53 | 0.825 | ||
| Negative | 18 (60.0) | 16 (57.1) | |
| Positive | 12 (40.0) | 12 (42.9) | |
| TP53 MIS | 0.031 | ||
| Negative | 17 (56.7) | 8 (28.6) | |
| Positive | 13 (43.3) | 20 (48.3) | |
| TP53 mutation | 0.380 | ||
| Negative | 7 (23.3) | 4 (14.3) | |
| Positive | 23 (76.7) | 24 (85.7) | |
| Codon 72 | 0.754 | ||
| Negative | 17 (56.7) | 17 (60.7) | |
| Positive | 13 (43.3) | 11 (39.3) | |
| MKI67 | 0.010 | ||
| Negative | 15 (50.0) | 5 (17.9) | |
| Positive | 15 (50.0) | 23 (82.1) | |
| Age, years | 0.512 | ||
| ≤50 | 11 (36.7) | 8 (28.6) | |
| >50 | 19 (63.3) | 20 (71.4) | |
| Tumor grade | 0.849 | ||
| I | 10 (33.3) | 10 (35.7) | |
| II and III | 20 (66.7) | 18 (64.3) | |
| Tumor, cm | 0.786 | ||
| ≤2 | 15 (50.0) | 13 (46.4) | |
| >2 | 15 (50.0) | 15 (53.6) | |
| Lymph node status | 0.451 | ||
| Negative | 11 (36.7) | 13 (46.4) | |
| Positive | 19 (63.3) | 15 (53.6) | |
| Vascular invasion | 0.024 | ||
| No | 26 (86.7) | 17 (60.7) | |
| Yes | 4 (13.3) | 11 (39.9) | |
| Recurrence | 0.001 | ||
| No | 23 (76.7) | 9 (32.1) | |
| Yes | 7 (23.3) | 19 (67.9) | |
| Nerve invasion | 0.649 | ||
| No | 23 (76.7) | 20 (71.4) | |
| Yes | 7 (23.3) | 8 (28.6) | |
TNCB, triple negative breast cancer; CD, cluster of differentiation; p53/TP53, tumor protein p53; MKI67, marker of proliferation Ki-67.
TP53 missense mutations occur at a high frequency in TNBC
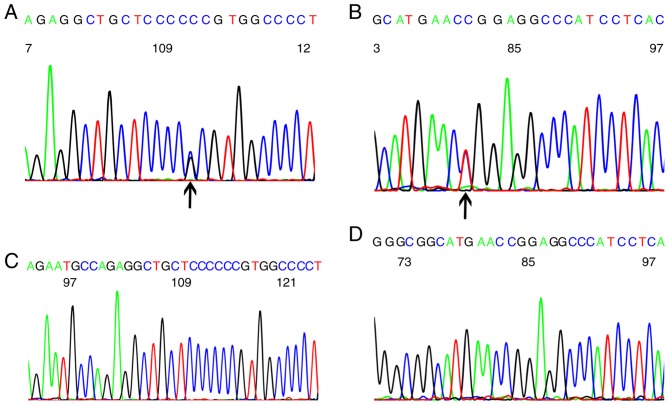
Missense mutations were the most common type of TP53 mutations (40/106, 37.7%) in breast cancer. Frame-shift mutations were identified in 7 patients (6.6%), nonsense mutations were observed in 6 patients (5.67%) and silent mutations in 3 (1.89%). A polymorphism of codon 72 in exon 4 was present in 38.68% (41/106) of patients. A codon 72 polymorphism (CCG to CCC) in exon 4 (Fig. 2A), and codon 248 mutations (CGG to TGG) in exon 7 (Fig. 2B) were the most common mutations. Wild-type codons 72, 61.3% (65/106) of breast cancer patients, and codon 248, 82.1% (87/106) of breast cancer patients were observed (Fig. 2C and D).
Figure 2.
TP53 gene mutations in breast cancer. (A) Codon 72 polymorphism identified in breast cancer (black arrow). (B) Codon 248 point mutations marked by black arrow were observed in breast cancer. (C) Codon 72 with no mutation. (D) Codon 248 with no mutation. TP53, tumor protein P53.
TP53 missense mutations in exons 4–8 were all functional mutations, with the exception of codon 72 mutations. The rate of missense mutations in TNBC tissues (33/58, 56.9%) was significantly higher than in non-TNBC tissues (17/48, 35.4%; P=0.027); however, rates of total TP53 mutations (82.8 vs. 66.7%, respectively) and polymorphisms at codon 72 (41.4 vs. 35.4%, respectively) were similar in TNBC vs. non-TNBC groups (Table III). The presence of p53 protein was similar in the TNBC and non-TNBC patients (P=0.530; Table III). No association was identified between TP53 mutations and the positive expression of p53 protein (Table V). TP53 mutations (P=0.001) were associated with tumor recurrence, whereas expression of p53 protein was not associated with tumor recurrence (Table V). TP53 missense mutations were associated with tumor recurrence (P=0.006), but not tumor grade, size, vascular invasion or lymph node metastasis (Table V).
Table V.
Association between clinicopathological features and TP53 missense mutations.
| TP53 missense mutations | |||
|---|---|---|---|
| Variable | Negative | Positive | P-value |
| MKI67 | 0.442 | ||
| Negative | 10 (40.0) | 10 (30.3) | |
| Positive | 15 (60.0) | 23 (69.7) | |
| p53 | 0.853 | ||
| Negative | 15 (60.0) | 19 (57.6) | |
| Positive | 10 (40.0) | 14 (42.4) | |
| Age, years | 0.502 | ||
| ≤50 | 7 (28.0) | 12 (36.4) | |
| >50 | 18 (72.0) | 21 (63.6) | |
| Tumor grade | 0.059 | ||
| I | 12 (48.0) | 8 (24.2) | |
| II and III | 13 (52.0) | 25 (75.8) | |
| Tumor, cm | 0.971 | ||
| ≤2 | 12 (48.0) | 16 (48.5) | |
| >2 | 13 (52.0) | 17 (51.5) | |
| Lymph node status | 0. 853 | ||
| Negative | 10 (40.0) | 14 (42.4) | |
| Positive | 15 (60.0) | 19 (57.6) | |
| Vascular invasion | 0.135 | ||
| No | 21 (84.0) | 22 (66.7) | |
| Yes | 4 (16.0) | 11 (33.3) | |
| Recurrence | 0.006 | ||
| No | 19 (76.0) | 13 (39.4) | |
| Yes | 6 (24.0) | 20 (60.6) | |
| Nerve invasion | 0.746 | ||
| No | 18 (72.0) | 25 (75.8) | |
| Yes | 7 (28.0) | 8 (24.2) | |
TP53/p53, tumor protein p53; MKI67, marker of proliferation Ki-67.
Expression of CD117 is associated with TP53 missense mutations in TNBC
The TNBC tissues exhibit a higher rate of CD117 positive/TP53 missense mutations (20/58, 34.5%) than non-TNBC tissues (6/48, 12.5%; P=0.009; Table III). The presence of CD117 was associated with TP53 missense mutations (P=0.031) in TNBC (Table IV). The CD117 positive/TP53 missense mutations were associated with vascular invasion (P=0.016), proliferation index (P=0.004) and particularly tumor recurrence (P<0.001). There was no association identified between CD117 positive/TP53 missense mutations and age, grade, tumor size, node status and nerve invasion (Table VI).
Table VI.
Association of CD117+/TP53 missense mutation+ with clinicopathological features of 58 patients with TNBC.
| CD117/TP53 missense | |||
|---|---|---|---|
| Variable | Negative | Positive | P-value |
| MKI67 | 0.004 | ||
| Negative | 18 (47.4) | 2 (10.0) | |
| Positive | 20 (52.6) | 18 (90.0) | |
| Age, years | 0.792 | ||
| ≤50 | 12 (31.6) | 7 (35.0) | |
| >50 | 26 (68.4) | 13 (65.0) | |
| Tumor grade | 0.270 | ||
| I | 15 (39.5) | 5 (25.0) | |
| II and III | 23 (60.5) | 15 (75.0) | |
| Size, cm | 0.360 | ||
| ≤2 | 20 (52.6) | 8 (40.0) | |
| >2 | 18 (47.4) | 12 (60.0) | |
| Lymph node status | 0.877 | ||
| Negative | 16 (42.1) | 8 (40.0) | |
| Positive | 22 (57.9) | 12 (60.0) | |
| Vascular invasion | 0.016 | ||
| No | 32 (84.2) | 11 (55.0) | |
| Yes | 6 (15.8) | 9 (45.0) | |
| Recurrence | <0.001 | ||
| No | 28 (73.7) | 4 (20.0) | |
| Yes | 10 (26.3) | 16 (80.0) | |
| Nerve invasion | 0.913 | ||
| No | 28 (73.7) | 15 (75.0) | |
| Yes | 10 (26.3) | 5 (25.0) | |
TNBC, triple negative breast cancer; CD, cluster of differentiation; MKI67, marker of proliferation Ki-67.
CD117+/TP53 missense mutation+ and vascular invasion are independent prognostic factors in TNBC
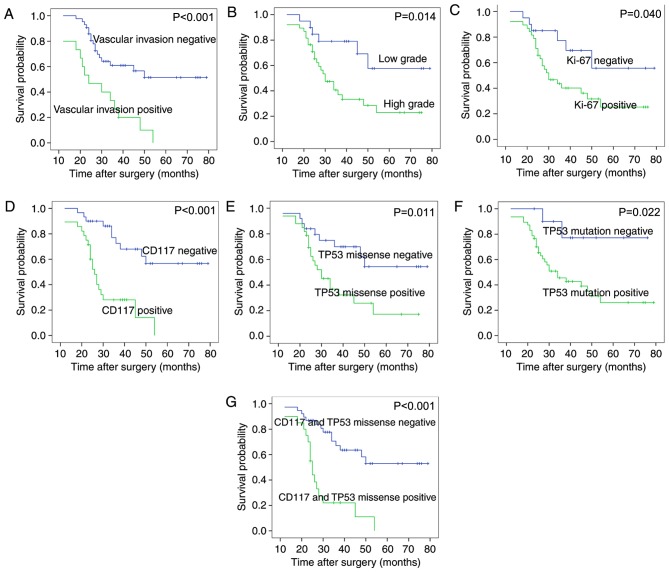
Survival analysis of the 58 patients with TNBC revealed that tumor grade, vascular invasion, expression of CD117 and MKI67 protein, TP53 missense mutations, TP53 mutations or the CD117+/TP53 missense mutation+ (CD117 and TP53 missense mutations together), were associated with overall survival (Fig. 3 and Table VII). The following features were not associated with the overall survival rate of patients with TNBC: Age, tumor size, lymph node status, nerve invasion, codon 72 mutations or expression of p53 protein. The Cox proportional hazards model revealed that CD117+/TP53 missense mutation+ [positive vs. negative group, P=0.004, RR=3.153; 95% confidence interval (CI), 1.430–6.952] and vascular invasion (present vs. not present, P=0.043, RR=2.234; 95% CI, 1.026–4.863) were independent prognostic factors in patients with TNBC (Fig. 3; Table VII).
Figure 3.
Survival analysis in patients with TNBC. Kaplan-Meier plots of overall survival in patients with TNBC and with (A) positive and negative vascular invasion, (B) high and low grade tumors, (C) positive and negative MKI67 protein expression, (D) CD117 protein expression, (E) TP53 missense mutations, (F) TP53 mutations, and (G) CD117 with TP53 missense mutations. TNBC, triple negative breast cancer; MKI67, marker of proliferation Ki-67; TP53, tumor protein P53.
Table VII.
Univariate and multivariate survival analysis of 58 patients with TNBC.
| P-value | ||||
|---|---|---|---|---|
| Variable | Univariate | Multivariate | RR | 95% CI |
| Age, years (≤50 vs. >50) | 0.738 | |||
| Tumor size, cm (≤2 vs. >2) | 0.389 | |||
| Lymph node status (+ vs. −) | 0.601 | |||
| Histological grade (high vs. low) | 0.014 | |||
| Vascular invasion (yes vs. no) | <0.001a | 0.043a | 2.234 | 1.026–4.863 |
| Nerve invasion (yes vs. no) | 0.898 | |||
| CD117 (+ vs. −) | <0.001 | |||
| MKI67 (+ vs. −) | 0.040 | |||
| P53 (+ vs. −) | 0.651 | |||
| TP53 MIS (+ vs. −) | 0.011 | |||
| TP53 mutation (+ vs. −) | 0.022 | |||
| Codon 72 (+ vs. −) | 0.837 | |||
| CD117/TP53MIS (+ vs. −) | <0.001a | 0.004a | 3.153 | 1.430–6.952 |
Statistically significant. CI, confidence interval; RR, risk ratio; CD, cluster of differentiation; MKI67, marker of proliferation Ki67; TP53, tumor protein P53.
Discussion
In the present study, patients with TNBC had a higher rate of CD117 expression, TP53 missense mutations and CD117+/TP53 missense mutation+ than patients with non-TNBC. CD117, TP53 missense mutations and CD117+/TP53 missense mutation+ were associated with poor prognosis and tumor recurrence in patients with TNBC. It was confirmed that CD117+/TP53 missense mutation+ was an independent prognostic factor in patients with TNBC. CD117+/TP53 missense mutation+ was associated with positive expression of MKI67 and vascular invasion. It was identified that tumor grade and the proliferation index (as indicated by MKI67 levels) were poor prognostic factors, associated with a high risk of relapse in patients with TNBC, consistent with previous reports (24–26). Vascular invasion, an independent prognostic factor, was associated with tumor recurrence in patients with TNBC in the present study. The data generated in the present study were consistent with that of a previous study (27), which reported that the 5-year metastasis-free survival rate in patients with TNBC and vascular invasion was lower than in TNBC patients without vascular invasion (P=0.003) (27).
Simon et al (28) demonstrated that none of these tumors contained mutated KIT; however, in the present study a higher rate of CD117 expression was identified in patients with TNBC than in patients with non-TNBC, consistent with previous studies (11,29). It was also confirmed that the presence of CD117 was associated with the expression of MKI67 and vascular invasion in patients with TNBC, consistent with previous observations (6,29).
There are conflicting reports regarding the prognostic value of CD117 in TNBC; Several reports have demonstrated that CD117 may not influence the survival of patients with TNBC (11,30), whereas Kashiwagi et al have confirmed a poorer outcome for CD117-positive TNBC (9). Overexpression of CD117 protein was associated with poor prognosis and recurrence in TNBC, but was not identified as an independent prognostic factor in the present study. A possible explanation for these opposing results is differences in threshold for categorizing samples as KIT-positive; Thike et al (6) used ≥1% of tumor cells as positive criteria, whereas Kashiwagi et al (9) set their threshold as ≥10%. In the present study, the threshold was set at 10% of stained cells. Another explanation for these opposing results is differences in criteria of HER2 positive expression for the determination of TNBC. Nielsen et al (30) classified the HER2 expression according to a three-point scoring system, in which patients were divided into three groups (negative, weakly-positive and positive groups); overexpression of CD117 protein was considered not to be associated the poor prognosis of TNBC patients. However, in the present study, HER2 immunostaining was considered positive (graded 3+, or gene amplification confirmed by FISH), since it is a standard threshold for TNBC assessment.
A number of previous studies have investigated TP53 gene mutations in TNBC (31). In the present study, the frequency of TP53 missense mutations was increased in TNBC compared with non-TNBC tissues, consistent with previous observations (32). A positive association was not observed between the expression of p53 protein and TP53 missense mutations in TNBC in the present study using direct sequencing, consistent with previous reports (33). However, Kim et al (34) reported that expression levels of p53 were also influenced by TP53 mutation status and mRNA level of TP53. The authors detected TP53 mutations using next-generation sequencing (34), while direct sequencing in our study. Taylor et al (35) found that results of p53 by IHC could identifying TP53 missense mutations in exons 4–8 in breast cancer, because moderate/average nuclear staining intensity for TP53 as a positive criterion, rather than the >10% positive tumor cells that was used in the present study. Furthermore, a previous study reported the Arg72Pro polymorphism in the TNBC NIPBC-2 cell line (36). In the present study, the Arg72Pro polymorphism was identified in 41.4% of patients with TNBC and in 35.4% of the non-TNBC patients; the positive rate in patients of TNBC compared with in the non-TNBC patients was similar (P=0.530).
It has been reported that p53 expression is associated with the prognosis of patients with TNBC (15). The present study revealed that TP53 missense mutations were associated with poor patient prognosis and a high risk of relapse in patients with TNBC, which was consistent with a previous report (37). Silwal-Pandit et al identified that TP53 mutations in TNBC are not associated with an unfavorable prognosis (38). TP53 mutations were also detected in exon2-8 (38), whereas the present study analyzed TP53 missense mutations in exon4-8.
To the best of our knowledge, the present study reported an association between CD117 expression and TP53 missense mutations for the first time, but not the association between CD117 expression and codon 72 polymorphisms, or synonymous mutations in patients with TNBC. CD117+/TP53 missense mutation+ was associated with positive MKI67 expression, vascular invasion and tumor recurrence. On the basis of these results, it may be hypothesized that CD117+/TP53 missense mutation+ is associated with the malignant biological behavior of patients with TNBC. p53 has been reported to regulate numerous genes, including MYC proto-oncogene and KIT, affecting the aggressive biological behavior of cancer (39–41). p53 may also directly regulate microRNA-34 (miR-34), with inactivation of p53 reducing miR-34 expression, promoting the invasion of colon cancer cells (18). Furthermore, the present study demonstrated that the expression of CD117 protein in TNBC tissues was an independent prognostic factor in patients with TNBC with TP53 missense mutations.
It was not possible to perform sub-categorization of patients with TNBC according to biological characteristics such as EGFR amplification, which may be one limitation of the present study. CD117+/TP53 missense mutation+ was not analyzed in patients with TNBC of different genotypes. The present study also did not detect the level of miR-34 or analyze the association between miR-34 and CD117+/TP53 missense mutation+. Therefore, further study into the effect of CD117+/TP53 missense mutation+ in patients with TNBC with different genotypes and the association between miR-34 and CD117+/TP53 missense mutation+ is required.
In conclusion, the present study confirmed that CD117+/TP53 missense mutation+ was associated with MKI67 expression, vascular invasion and tumor recurrence in patients with TNBC. CD117 expression was indicative of poor prognosis in patients with TNBC with TP53 missense mutations.
Acknowledgements
The authors would like to thank thank Keyang Sun, Liang Liu and Jie Chen for preparing the paraffin-embedded slides, and Miss Liang Liu and Mr Jie Chen as technicians (Department of Pathology, Shanghai No. 6 People's Hospital, Shanghai, China) for their help with immunohistochemical staining was.
Funding
This project was supported by grants from the National Natural Science Foundation (grant no. 81100223) and from Shanghai Municipal Health Bureau Program (grant no. 20134052).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author, on reasonable request.
Authors' contributions
Preparation of the DNA, sequencing, IHC and analysis of data were performed by YLL. Statistical analysis was also performed by YLL. The slides were analyzed by HZZ and WTH. GL participated in analysis of experimental data and conducted the PCR experiment. The manuscript was written by YLL and reviewed by HZZ and GL.
Ethics approval and consent to participate
The Ethics Committee for Human Studies at Shanghai Jiao Tong University (Shanghai, China) approved the present study, which was conducted in accordance with The Declaration of Helsinki. Participants were fully informed of the procedures, and written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Changavi AA, Shashikala A, Ramji AS. Epidermal growth factor receptor expression in triple negative and nontriple negative breast carcinomas. J Lab Physicians. 2015;7:79–83. doi: 10.4103/0974-2727.163129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA, Jr, Ellis P, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 4.Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245–266. doi: 10.1111/j.1365-2559.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida C, Tsuji AB, Sudo H, Sugyo A, Kikuchi T, Koizumi M, Arano Y, Saga T. Therapeutic efficacy of c-kit-targeted radioimmunotherapy using 90Y-labeled anti-c-kit antibodies in a mouse model of small cell lung cancer. PLoS One. 2013;8:e59248. doi: 10.1371/journal.pone.0059248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thike AA, Iqbal J, Cheok PY, Chong AP, Tse GM, Tan B, Tan P, Wong NS, Tan PH. Triple negative breast cancer: Outcome correlation with immunohistochemical detection of basal markers. Am J Surg Pathol. 2010;34:956–964. doi: 10.1097/PAS.0b013e3181e02f45. [DOI] [PubMed] [Google Scholar]
- 7.Blassl C, Kuhlmann JD, Webers A, Wimberger P, Fehm T, Neubauer H. Gene expression profiling of single circulating tumor cells in ovarian cancer-Establishment of a multi-marker gene panel. Mol Oncol. 2016;10:1030–1042. doi: 10.1016/j.molonc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan H, Yuan Y, Wang J, Zhou F, Zhang M, Giercksky KE, Nesland JM, Suo Z. CD117 expression in operable oesophageal squamous cell carcinomas predicts worse clinical outcome. Histopathology. 2013;62:1028–1037. doi: 10.1111/his.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashiwagi S, Yashiro M, Takashima T, Aomatsu N, Kawajiri H, Ogawa Y, Onoda N, Ishikawa T, Wakasa K, Hirakawa K. c-Kit expression as a prognostic molecular marker in patients with basal-like breast cancer. Br J Surg. 2013;100:490–496. doi: 10.1002/bjs.9021. [DOI] [PubMed] [Google Scholar]
- 10.Medinger M, Kleinschmidt M, Mross K, Wehmeyer B, Unger C, Schaefer HE, Weber R, Azemar M. c-kit (CD117) expression in human tumors and its prognostic value: An immunohistochemical analysis. Pathol Oncol Res. 2010;16:295–301. doi: 10.1007/s12253-010-9247-9. [DOI] [PubMed] [Google Scholar]
- 11.Jansson S, Bendah PO, Grabau DA, Falck AK, Fernö M, Aaltonen K, Rydén L. The three receptor tyrosine kinases c-KIT, VEGFR2 and PDGFRα, closely spaced at 4q12, show increased protein expression in triple-negative breast cancer. PLoS One. 2014;9:e102176. doi: 10.1371/journal.pone.0102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HW, Lee HM, Hwang SH, Ahn SG, Lee KA, Jeong J. Patterns and biologic features of p53 mutation types in Korean breast cancer patients. J Breast Cancer. 2014;17:1–7. doi: 10.4048/jbc.2014.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 14.Fernández-Cuesta L, Oakman C, Falagan-Lotsch P, Smoth KS, Quinaux E, Buyse M, Dolci MS, Azambuja ED, Hainaut P, Dell'orto P, et al. Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: Results from the BIG 02–98 phase III trial. Breast Cancer Res. 2012;14:R70. doi: 10.1186/bcr3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chae BJ, Bae JS, Lee A, Park WC, Seo YJ, Song BJ, Kim JS, Jung SS. p53 as a specific prognostic factor in triple-negative breast cancer. Jpn J Clin Oncol. 2009;39:217–224. doi: 10.1093/jjco/hyp007. [DOI] [PubMed] [Google Scholar]
- 16.Olivier M, Langerød A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bièche I, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 17.Foedermayr M, Sebesta M, Rudas M, Berghoff AS, Promberger R, Preusser M, Dubsky P, Fitzal F, Gnant M, Steger GG, et al. BRCA-1 methylation and TP53 mutation in triple-negative breast cancer patients without pathological complete response to taxane-based neoadjuvant chemotherapy. Cancer Chemother Pharmacol. 2014;73:771–778. doi: 10.1007/s00280-014-2404-1. [DOI] [PubMed] [Google Scholar]
- 18.Siemens H, Jackstadt R, Kaller M, Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittendorf EA, Ballman KV, McCall LM, Yi M, Sahin AA, Bedrosian I, Hansen N, Gabram S, Hurd T, Giuliano AE, Hunt KK. Evaluation of the stage IB designation of the American Joint Committee on Cancer staging system in breast cancer. J Clin Oncol. 2015;33:1119–1127. doi: 10.1200/JCO.2014.57.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penault-Llorca F, André F, Sagan C, Lacroix-Triki M, Denoux Y, Verriele V, Jacquemier J, Baranzelli MC, Bibeau F, Antoine M, et al. Ki67 expression and docetaxel efficacy in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009;27:2809–2815. doi: 10.1200/JCO.2008.18.2808. [DOI] [PubMed] [Google Scholar]
- 21.Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, O'Sullivan B, He Z, Peng Y, Tan AC, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaman S, Gumuskaya B, Ozkan C, Aksoy S, Guler G, Altundag K. Lymphatic and capillary invasion patterns in triple negative breast cancer. Am Surg. 2012;78:1238–1242. [PubMed] [Google Scholar]
- 23.Varga Z, Noske A, Ramach C, Padberg B, Moch H. Assessment of HER2 status in breast cancer: Overall positivity rate and accuracy by fluorescence in situ hybridization and immunohistochemistry in a single institution over 12 years: A quality control study. BMC Cancer. 2013;13:615. doi: 10.1186/1471-2407-13-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue YI, Astvatsaturyan K, Cui X, Zhang X, Fraass B, Bose S. Stratification of prognosis of triple-negative breast cancer patients using combinatorial biomarkers. PLoS One. 2016;11:e0149661. doi: 10.1371/journal.pone.0149661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricciardi GR, Adamo B, Ieni A, Licata L, Cardia R, Ferraro G, Franchina T, Tuccari G, Adamo V. Androgen Receptor (AR), E-cadherin, and Ki-67 as emerging targets and novel prognostic markers in Triple-Negative Breast Cancer (TNBC) patients. PLoS One. 2015;10:e0128368. doi: 10.1371/journal.pone.0132647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keam B, Im SA, Lee KH, Han SW, Oh DY, Kim JH, Lee SH, Han W, Kim DW, Kim TY, et al. Ki-67 can be used for further classification of triple negative breast cancer into two subtypes with different response and prognosis. Breast Cancer Res. 2011;13:R22. doi: 10.1186/bcr2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatier R, Jacquemier J, Bertucci F, Esterni B, Finetti P, Azario F, Birnbaum D, Viens P, Gonçalves A, Extra JM. Peritumoural vascular invasion: A major determinant of triple-negative breast cancer outcome. Eur J Cancer. 2011;47:1537–1545. doi: 10.1016/j.ejca.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Simon R, Panussis S, Maurer R, Spichtin H, Glatz K, Tapia C, Mirlacher M, Rufle A, Torhorst J, Sauter G. KIT (CD117)-positive breast cancers are infrequent and lack KIT gene mutations. Clin Cancer Res. 2004;10:178–183. doi: 10.1158/1078-0432.CCR-0597-3. [DOI] [PubMed] [Google Scholar]
- 29.Kanapathy Pillai SK, Tay A, Nair S, Leong CO. Triple-negative breast cancer is associated with EGFR, CK5/6 and c-KIT expression in Malaysian women. BMC Clin Pathol. 2012;12:18. doi: 10.1186/1472-6890-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immuno-histochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 31.Fountzilas G, Giannoulatou E, Alexopoulou Z, Zagouri F, Timotheadou E, Papadopoulou K, Lakis S, Bobos M, Poulios C, Sotiropoulou M, et al. TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting. Oncotarget. 2016;7:32731–32753. doi: 10.18632/oncotarget.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Kim J, Lee HD, Jeong J, Lee W, Lee KA. Spectrum of EGFR gene copy number changes and KRAS gene mutation status in korean triple negative breast cancer patients. PLoS One. 2013;8:e79014. doi: 10.1371/journal.pone.0079014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Végran F, Rebucci M, Chevrier S, Cadouot M, Boidot R, Lizard-Nacol S. Only missense mutations affecting the DNA binding domain of p53 influence outcomes in patients with breast carcinoma. PLoS One. 2013;8:e55103. doi: 10.1371/journal.pone.0055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JY, Park K, Jung HH, Lee E, Cho EY, Lee KH, Bae SY, Lee SK, Kim SW, Lee JE, et al. Association between mutation and expression of TP53 as a potential prognostic marker of triple-negative breast cancer. Cancer Res Treat. 2016;48:1338–1350. doi: 10.4143/crt.2015.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor NJ, Nikolaishvili-Feinberg N, Midkiff BR, Conway K, Millikan RC, Geradts J. Rational manual and automated scoring thresholds for the immunohistochemical detection of TP53 missense mutations in human breast carcinomas. Appl Immunohistochem Mol Morphol. 2016;24:398–404. doi: 10.1097/PAI.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandrangi SL, Raju Bagadi SA, Sinha NK, Kumar M, Dada R, Lakhanpal M, Soni A, Malvia S, Simon S, Chintamani C, et al. Establishment and characterization of two primary breast cancer cell lines from young Indian breast cancer patients: Mutation analysis. Cancer Cell Int. 2014;14:14. doi: 10.1186/1475-2867-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobes P, Podhorec J, Coufal O, Jureckova A, Petrakova K, Vojtesek B, Hrstka R. Influence of mutation type on prognostic and predictive values of TP53 status in primary breast cancer patients. Oncol Rep. 2014;32:1695–1702. doi: 10.3892/or.2014.3346. [DOI] [PubMed] [Google Scholar]
- 38.Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, Quigley DA, Kristensen VN, Aparicio S, Børresen-Dale AL, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20:3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 39.Abraham SA, Hopcroft LE, Carrick E, Drotar ME, Dunn K, Williamson AJ, Korfi K, Baquero P, Park LE, Scott MT, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yogev O, Barker K, Sikka A, Almeida GS, Hallsworth A, Smith LM, Jamin Y, Ruddle R, Koers A, Webber HT, et al. p53 loss in MYC-driven neuroblastoma leads to metabolic adaptations supporting radioresistance. Cancer Res. 2016;76:3025–3035. doi: 10.1158/0008-5472.CAN-15-1939. [DOI] [PubMed] [Google Scholar]
- 41.Lai JH, Fleming KE, Ly TY, Pasternak S, Godlewski M, Doucette S, Walsh NM. Pure versus combined Merkel cell carcinomas: Immunohistochemical evaluation of cellular proteins (p53, Bcl-2 and c-kit) reveals significant overexpression of p53 in combined tumors. Hum Pathol. 2015;46:1290–1296. doi: 10.1016/j.humpath.2015.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author, on reasonable request.