Abstract
Long non-coding RNA HOXA11 antisense RNA (HOXA11-AS) has been previously reported to be involved in the tumorigenesis and progression of ovarian cancer and glioma. However, the function of HOXA11-AS in lung cancer remains unclear. Following the knockdown of HOXA11-AS in A549 cells, a microarray analysis was performed in order to detect the differences in microRNA (miRNA/miR) profiles. Subsequently, miR-642b-3p was selected for further analysis. Four miRNA target prediction algorithms were used to identify potential target genes of miR-642b-3p. Bioinformatics analyses, including Gene Ontology (GO), Kyoto Encyclopaedia of Genes and Genomes, protein-protein interactions (PPIs) and network analysis, were performed to investigate the potential functions, pathways and networks of the target genes. Furthermore, the differential expression of miR-642b-3p and its target genes between normal lung and non-small cell lung cancer (NSCLC) tissues was verified using The Cancer Genome Atlas (TCGA) database. Six target genes [zinc finger protein 350, heterogeneous nuclear ribonucleoprotein U, high mobility group box 1, phosphodiesterase 4D (PDE4D), synaptotagmin binding cytoplasmic RNA interacting protein and basic helix-loop-helix family member B9] of miR-642b-3p were predicted using all 4 algorithms. It was revealed that miR-642b-3p was overexpressed in adenocarcinoma and squamous cell carcinoma tissues compared with non-cancerous lung tissues based on the TCGA database. From the 6 target genes, PDE4D was downregulated in lung adenocarcinoma and squamous cell carcinoma tissues, and a weak negative correlation between HOXA11-AS and PDE4D was identified. The area under the curve of PDE4D was 0.905 [95% confidence interval (CI), 0.879–0.931] for patients with lung adenocarcinoma and 0.665 (95% CI, 0.606–0.725) for patients with squamous cell carcinoma. Additionally, GO analysis of the target genes revealed that miR-642b-3p was specifically involved in complex cellular pathways. The target gene RAN binding protein 2 possessed the highest degree of interactions in the PPI network (degree=40). It was hypothesized that HOXA11-AS may have a function in NSCLC by regulating the expression of miR-642b-3p and PDE4D, which laid the foundation for the further elucidation of the potential molecular mechanisms of NSCLC.
Keywords: HOXA11 antisense RNA, non-small cell lung cancer, micro RNA-642b-3p, gene ontology, Kyoto Encyclopaedia of Genes and Genomes, protein-protein analyses
Introduction
Lung cancer has a substantial mortality rate and the incidence of lung cancer has been increasing gradually (1–5). Based on histological type, lung cancer may be divided into two categories: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). From all types of lung cancer, 80–85% are classified as NSCLC (6,7). Additionally, in >70% of newly diagnosed NSCLC cases, the disease is at an advanced stage, and the 5-year survival rate of patients with NSCLC is only 16% (8). Hence, it is imperative to identify potential molecular mechanisms of NSCLC tumorigenesis and progression.
Non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs) and microRNAs (miRNA/miR), have been demonstrated to function differently in different types of cancer. Different ncRNAs in the same cancer may have different underlying functions (9,10). Previous studies demonstrated that ncRNAs are extensively involved in the tumorigenesis and progression of NSCLC (11,12). Various lncRNAs have been confirmed to have significant functions in epigenetic gene regulation, transcriptional regulation or disease development (13–16). It is of note that lncRNAs may regulate the transcription of corresponding genes by combining with polymerases or transcription factors due to their pervasive distribution in the nucleus (15,17). Previous studies have revealed that lncRNAs may regulate miRNA expression in lung cancer. With regards to the interplay between lncRNAs and miRNAs during tumorigenesis and progression, the competing endogenous RNA (ceRNA) hypothesis has attracted attention (18,19). The ceRNA hypothesis posits that lncRNAs function as molecular sponges for miRNAs and functionally liberate the targeted mRNAs regulated by the aforementioned miRNAs (20–22). For example, the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) may function as a ceRNA for miR-377-3p, and NEAT1 promotes NSCLC progression by regulating the expression of miR-377-3p (23). The lncRNA colon cancer-associated transcript 1 (non-protein coding) reduced miR-218 levels via the use of BMI1 proto-oncogene, polycomb ring finger to promote cell cycle transition in cigarette smoke extract-induced lung carcinogenesis (24). The lncRNA urothelial cancer associated one performed oncogenic functions in NSCLC by targeting miR-193a-3p (25). Furthermore, lncRNAs may co-express with microRNA in diseases. For example, Keniry et al (26) revealed that lncRNA H19, imprinted maternally expressed transcript (non-protein coding) may function as the pre-miRNA of miR-675, and they may interplay to suppress growth. Furthermore, miRNAs regulate the expression of their target transcripts by binding to the 3′-untranslated region (3′-UTR) (27–32). For example, Huang et al (33) revealed that miRNA-186 may suppress the cell proliferation and metastasis of NSCLC by targeting mitogen-activated protein kinase kinase kinase 2. Lu et al (34) revealed that miR-541-3p may reverse cancer progression by directly targeting TGFB induced factor homeobox 2 in NSCLC. Hence, the further investigation of miRNA profiling associated with lncRNAs in NSCLC is useful in order to identify novel targeted therapeutic strategies.
In the present study, a microarray analysis was performed to verify the differential expression of miRNAs between the RNA interference (RNAi) and control samples according to the fold change filtering (fold change ≥1.5 or ≤1) and the P-value (P<0.05). The five most downregulated miRNAs were selected (miR-1264, miR-337-3p, miR-302c-5p, miR-642b-3p and miR-3621). Then, based on original data from The Cancer Genome Atlas (TCGA), miR-642b-3p was selected for further analysis. Four miRNA target prediction algorithms were used to identify the potential target genes of miR-642b-3p. Bioinformatics analysis, including Gene Ontology (GO), the Kyoto Encyclopaedia of Genes and Genomes (KEGG), protein-protein interactions (PPIs) and network analysis were performed in order to investigate the potential functions, pathways and networks of the target genes (35–38). Additionally, the expression of miR-642b-3p and its targeted genes [zinc finger protein 350 (ZNF350), heterogeneous nuclear ribonucleoprotein U (HNRNPU), high mobility group box 1 (HMGB1), phosphodiesterase 4D (PDE4D), synaptotagmin binding cytoplasmic RNA interacting protein (SYNCRIP), basic helix-loop-helix family member B9 (BHLHB9)] in NSCLC was analysed based on original data from TCGA database.
Materials and methods
Microarray analysis and the knockdown of HOXA11 antisense RNA (HOXA11-AS) in the NSCLC cell line A549
The A549 human NSCLC cell line was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultivated in a humidified atmosphere of 5% CO2 at 37°C with 10% heat-inactivated foetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The A549 NSCLC cell line was transfected with HOXA11-AS small interfering RNA and the subsequent experiments were performed at least 72 h following transfection. Lipofectamine™ 2000 (cat no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection, according to the manufacturer's protocol. A microarray analysis was performed in order to verify the differential expression of miR-642b-3p between the RNAi and control samples according to fold-change filtering (fold-change ≥1.5 or ≤1) and P-value (P<0.05).
miRNA target prediction
Four miRNA target prediction algorithms were utilized to predict the potential target genes of miR-642b-3p. The four corresponding prediction algorithms were miRDB (version 4.0, http://www.mirdb.org/) (39,40), mirTarBase (http://mirtarbase.mbc.nctu.edu.tw/) (41), TargetScan (version 6.2; http://www.targetscan.org/) (42) and DIANA-microT (version v4.0; http://www.microrna.gr/microT) (43,44). Candidate genes were identified and compared with Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/).
GO and pathway analysis and construction of a PPI network
To elucidate the potential functions of the target genes, GO and KEGG pathway analyses were conducted at the biological process (BP), cellular component (CC) and molecular function (MF) levels, as previously described (45). The enrichment of potential target genes was further analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.7, http://david.abcc.ncifcrf.gov/) (46,47) or Search Tool for the Retrieval of Interacting Genes (STRING; version 9.0; http://string-db.org) (48). Genes with a false discovery rate of ≤0.05 and P<0.05 were identified as enriched in the target genes. Then, a functional network of GO analysis was constructed using Cytoscape (version 2.8; http://cytoscape.org) (49).
The interaction pairs of target genes were additionally analysed using STRING version 9.0, and the interaction data was downloaded and analysed as described (50). A combined score >0.4 was selected as a threshold in order to construct the PPI network.
Additional analysis of miR-642b-3p and target genes in NSCLC from TCGA
TCGA (http://cancergenome.nih.gov/) is a collection of RNA sequencing, miRNA sequencing, exome sequencing, single nucleotide polymorphism array and DNA methylation data (51). TCGA may also be used to analyse clinical parameters and complicated cancer genomics (52,53). In the present study, raw data from RNASeq version 2 (54,55) in lung adenocarcinoma and squamous cell carcinoma for miR-642b and the 6 target genes (ZNF350, HNRNPU, HMGB1, PDE4D, SYNCRIP and BHLHB9) were extracted from TCGA. The expression of miR-642b and the target genes in each case was subsequently calculated according to the distribution of exon reads.
Statistical analysis
SPSS version 20.0 (IBM Corporation, Armonk, NY, USA) was used for statistical analysis. An unpaired Student's t-test was used for comparing the expression of miR-642b-3p and its target genes in lung adenocarcinoma and squamous cell carcinoma. The association between HOXA11-AS, miR-642b-3p and target genes were evaluated using Spearman's test. The association between gene expression and clinical diagnostic values was analysed using receiver operating characteristic curves. A Kaplan-Meier survival analysis was used to produce survival curves. P<0.05 was considered to indicate a statistically significant difference (two-tailed).
Results
miR-642b-3p profiling was associated with lncRNA HOXA11-AS
The transfection efficiency was ~100%, and the knockdown efficiency of HOXA11-AS in NSCLC cell lines was >75%, as determined by a reverse transcription-quantitative polymerase chain reaction (RT-qPCR; data not presented). Following the knockdown of HOXA11-AS, miR-642b-3p was significantly downregulated in A549 NSCLC cells (fold-change=0.355 and P=0.008).
miRNA target prediction
In the present study, four miRNA target prediction algorithms (miRDB, mirTarBase, TargetScan and DIANA-microT) were used to identify the target genes of miR-642b-3p. Venn diagrams were generated in order to analyse and compare the candidate genes. Amongst these target genes, 705 genes (including ZNF350, MAPK8, ASTN2 and YOD1) that were predicted by >2 algorithms were selected and these genes were used for GO and pathway analyses.
GO and pathway analysis and construction of a PPI network
The GO analysis was performed at the BP, CC and MF levels. The top 5 GO functional annotations for the target genes of miR-642b are presented in Table I. GO analysis indicated the significant functional groups, including multicellular organism development, intracellular part and molecular function. To further elucidate the relevant functions of the target genes, a functional network was constructed based on the GO analysis using Cytoscape (Fig. 1).
Table I.
The top 5 GO functional annotation for the target genes of miR-642b-3p.
| A, Biological processes | ||||
|---|---|---|---|---|
| GO ID | GO term | Count in network | FDR | Gene symbol (top 30 genes) |
| GO.0007275 | Multicellular organism development | 184 | 0.000122 | ABCA12, ABI1, ADAR, ADRBK1, AGPAT5, AHR, AK4, AMBN, ANKRD1, ANKRD17, ARL6, ASB2, ATP8B1, ATXN3, BBS4, BCL11B, BCOR, BHLHB9, BMP3, CACNA1G, CALM1, CAMSAP1, CASQ1, CCK, CCNA2, CD109, CDH22, CDK5R2, CDKN1A |
| GO.0048731 | System development | 165 | 0.000122 | ABCA12, ABI1, ADAR, ADRBK1, AGPAT5, AHR, AK4, AMBN, AMHR2, ANKRD1, ANKRD17, ARL6, ASB2, ATP8B1, ATXN3, BBS4, BCL11B, BCOR, BHLHB9, BMP3, CACNA1G, CALM1, CAMSAP1, CASQ1, CCK, CCNA2, CD109, CDH22, CDK5R2, CDKN1A |
| GO.0008152 | Metabolic process | 353 | 0.000356 | ABCA12, ABHD10, ABHD2, ABI1, ABRA, ADAM22, ADAR, ADORA2B, ADRBK1, AFF4, AGPAT5, AHR, AK4, ALDH4A1, AMHR2, ANGPT1, ANKRD1, ARSD, ASB2, ASCL1, ATAD1, ATG10, ATG12, ATP9A, B4GALT4, BAZ1A, BBS4, BCL11B, BCLAF1, BCOR |
| GO.0009893 | Positive regulation of metabolic process | 159 | 0.000356 | ABI1, ACAP2, ADAR, ADRBK1, AFF1, ANKRD1, ANKRD6, APP, ARHGAP18, ARHGAP31, ARHGAP6, ARHGEF37, ASB2, ASCL1, ATAD1, ATG10, ATXN3, BCLAF1, CALM1, CAND2, CARD8, CASP10, CCK, CCNA2, CCPG1, CDC25B, CDK5R2, CDKN1A, CENPE, CHRNA7 |
| GO.0042325 | Regulation of phosphorylation | 75 | 0.000356 | ABI1, ADAR, ADORA2B, ANKRD6, APP, BMP3, CALM1, CCK, CCNA2, CD109, CDC25B, CDKN1A, CENPE, CHRNA7, CISH, CREBL2, DNAJC27, EIF2AK2, EPGN, EPHA7, EPHB1, FAM129A, FZD1, FZD4, GAB1, GDF6, GMFB, GRM1, HIPK3, IBTK |
| B, Cellular components | ||||
| GO ID | GO term | Count in network | FDR | Gene symbol (top 30 genes) |
| GO.0005622 | Intracellular | 470 | 3.88E-05 | ABCA12, ABHD10, ABI1, ABRA, ACAP2, ADAR, ADORA2B, ADRBK1, AFF1, AFF4, AGPAT5, AHR, AK4, ALDH4A1, ANKRD1, ANKRD17, ANKRD6, ANKS1B, ANO5, ANTXR2, APP, ARHGAP18, ARHGAP31, ARHGAP6, ARHGEF37, ARSD, ASB2, ASCL1, ASTN2, ATG10 |
| GO.0043227 | Membrane-bound organelle | 423 | 0.000126 | ABCA12, ABHD10, ABI1, ACAP2, ADAR, AFF1, AFF4, AGPAT5, AHR, AK4, AKAP4, ALDH4A1, AMOTL2, ANGPT1, ANKRD1, ANKRD17, ANKRD6, ANKS1B, ANO5, ANTXR2, AP1S3, APP, ARL6, ARSD, ASCL1, ASTN2, ATG12, ATP8B1, ATP9A, ATXN3 |
| GO.0044424 | Intracellular part | 455 | 0.000126 | ABCA12, ABHD10, ABI1, ABRA, ACAP2, ADAR, ADRBK1, AFF1, AFF4, AGPAT5, AHR, AK4, ALDH4A1, ANKRD1, ANKRD17, ANKRD6, ANKS1B, ANO5, ANTXR2, APP, ARHGAP18, ARHGAP31, ARHGAP6, ARHGEF37, ARSD, ASB2, ASCL1, ASTN2, ATG10 |
| GO.0043226 | Organelle | 439 | 0.00018 | ABCA12, ABHD10, ABI1, ABRA, ACAP2, ADAR, ADRBK1, AFF1, AFF4, AGPAT5, AHR, AK4, ALDH4A1, AMOTL2, ANGPT1, ANKRD1, ANKRD17, ANKRD6, ANKS1B, ANTXR2, AP1S3, APP, ARHGAP6, ARL6, ARSD, ASCL1, ASTN2, ATG12, ATP8B1, ATP9A |
| GO.0043229 | Intracellular organelle | 411 | 0.000197 | ABCA12, ABHD10, ABI1, ABRA, A CAP2, ADAR, AFF1, AFF4, AGPAT5, AHR, AK4, ALDH4A1, AMOTL2, ANKRD1, ANKRD17, ANKRD6, ANKS1B, ANO5, ANTXR2, AP1S3, APP, ARHGAP6, ARL6, ARSD, ASCL1, ASTN2, ATP8B1, ATP9A, ATXN3, B3GALT1 |
| C, Molecular functions | ||||
| GO ID | GO term | Count in network | FDR | Gene symbol (top 30 genes) |
| GO.0005488 | Binding | 392 | 0.00069 | ABCA12, ABI1, ABRA, ACAP2, ADAR, ADRBK1, AHR, AK4, AKAP4, ALDH4A1, AMBN, AMHR2, ANGPT1, ANKMY1, ANKRD1, ANKRD17, ANKS1B, ANTXR2, APP, ARHGAP6, ARL6, ARSB, ARSD, ATAD1, ATP8B1, ATP9A, ATXN3, B4GALT4, BAIAP3, BAZ1A |
| GO.0003674 | Molecular function | 456 | 0.000857 | ABCA12, ABHD10, ABHD2, ACAP2, ADAR, ADORA2B, ADRBK1, AFF1, AFF4, AGPAT5, AHR, AK4, AKAP4, ALDH4A1, AMBN, ANGPT1, ANKMY1, ANKRD1, ANKRD17, ANKS1B, ANO5, ANTXR2, AP1S3, APP, ARHGAP18, ARHGAP31, ARHGAP6, ARHGEF37, ARL6, ARSB |
| GO.0005515 | Protein binding | 200 | 0.000857 | ABCA12, ABI1, ABRA, ADAM22, ADAMTS8, ADRBK1, AHR, AKAP4, ALDH4A1, AMBN, ANGPT1, ANKRD1, ANKS1B, APP, ARHGAP6, ASCL1, ATXN3, BAIAP3, BAZ2A, BBS4, BCOR, BHLHB9, BICD2, BMP3, C18orf42, CACNA1B, CACNA1G, CALM1, CAMSAP1, CARD8 |
| GO.0004705 | JUN kinase activity | 3 | 0.0274 | MAPK10, MAPK8, MAPK9 |
| GO.0061578 | Lys63-specific deubiquitinase activity | 3 | 0.0274 | ATXN3, CYLD, YOD1 |
GO, Gene Ontology; FDR, false discovery rate.
Figure 1.
A functional network of Gene Ontology terms for the target genes of microRNA-642b-3p in non-small cell lung cancer. The network for terms for (A) biological process, (B) cellular component (C) and molecular function.
Using the online tools DAVID or STRING, the KEGG analysis demonstrated that the target genes may be involved in different pathways, including the avian erythroblastosis oncogene B2 signalling pathway, RIG-I-like receptor signalling pathway or mitogen-activated protein kinases (MAPK) signalling pathway (data not shown), with the false discovery rate value >0.05. Thus, perform further analyses on these pathways from KEGG were not performed.
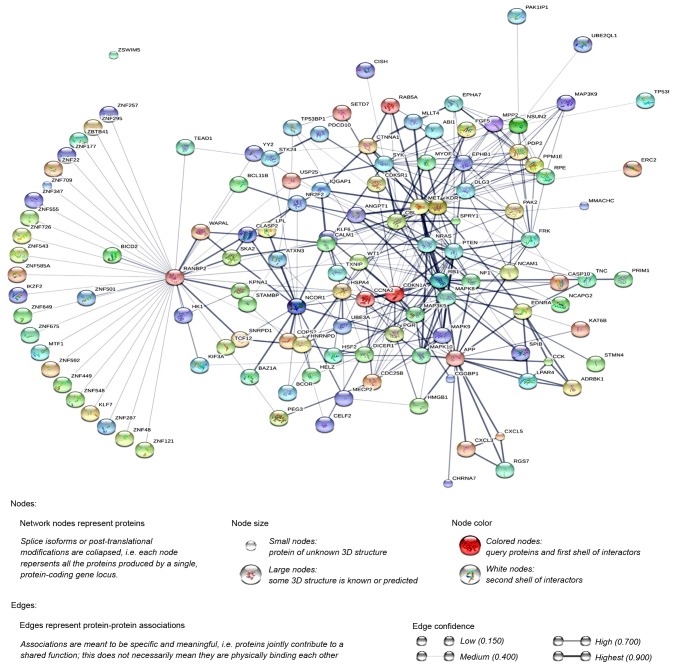
Additionally, the PPI network was constructed using STRING, and a total of 1,228 PPI pairs with a combined score of >0.4 were selected. The map of the PPI network is presented in Fig. 2. The number of nodes was 700, accounting for 99.30% of all target genes. The clustering coefficient of the PPI network was 0.626. RAN binding protein 2 (degree=40) had the highest degree of interactions in the PPI network. A sub-network of 172 PPI pairs (with degree >15) was selected for further analyses (Fig. 3).
Figure 2.
The protein-protein interaction network of differentially expressed genes.
Figure 3.
A sub-network of 172 protein-protein interaction pairs.
Supplementary information from TCGA database
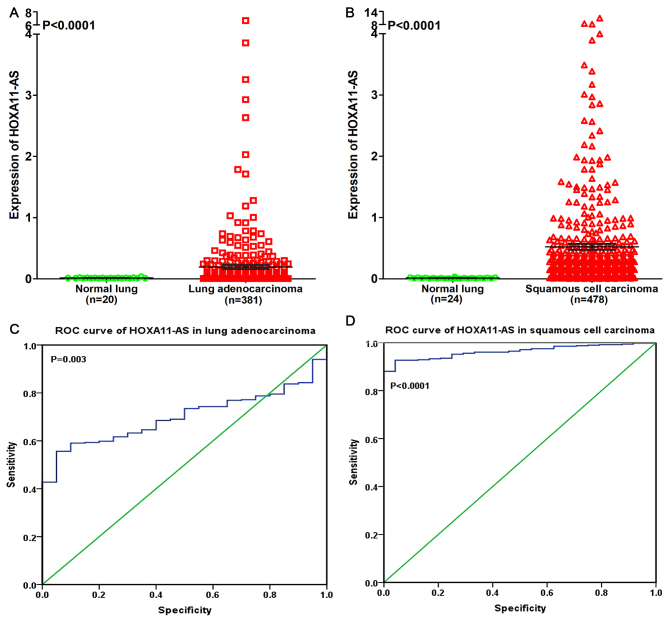
In order to reveal the association between HOXA11-AS and NSCLC, a clinical study was performed using the raw data in TCGA. It was revealed that HOXA11-AS was upregulated in lung adenocarcinoma and squamous cell carcinoma compared with non-cancerous lung tissues (P<0.0001; Fig. 4A and B). The ROC curve revealed that the area under curve (AUC) of HOXA11-AS was 0.700 (95% confidence interval (CI), 0.636~0.764; P=0.003) for patients with lung adenocarcinoma and 0.964 (95% CI, 0.946~0.981; P<0.0001) for patients with squamous cell carcinoma, which may gain a moderate or high diagnostic value of HOXA11-AS level in lung cancer (Fig. 4C and D).
Figure 4.
Differential expression and ROC curve of HOXA11-AS in lung adenocarcinoma and squamous cell carcinoma based on The Cancer Genome Atlas database. The differential expression of HOXA11-AS in (A) lung adenocarcinoma and (B) squamous cell carcinoma. The ROC curve of HOXA11-AS in (C) lung adenocarcinoma and (D) squamous cell carcinoma. ROC, receiving operator characteristic; HOXA11-AS, HOXA11 antisense RNA.
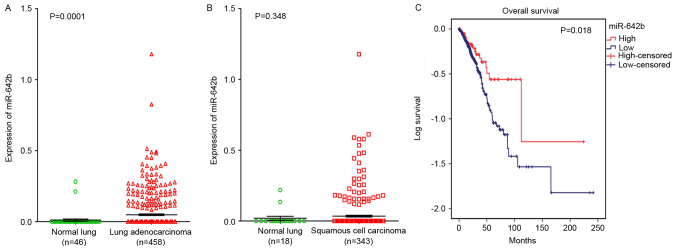
To elucidate the relationships between miR-642b and NSCLC, a clinical study was performed using the primary data in TCGA. It was revealed that miR-642b was significantly upregulated in lung adenocarcinoma (P=0.0001) and non-significantly upregulated in squamous cell carcinoma (P=0.348) compared with non-cancerous lung tissues (Fig. 5A and B). The relationship between miR-642b and the prognosis of NSCLC was also investigated, and it was revealed that the high expression of miR-642b was associated with the overall survival of patients with adenocarcinoma (108.56±25.23 vs. 79.51±8.75; P=0.018; Fig. 5C), which indicated that miR-642b may influence the prognosis of adenocarcinoma. The correlation between HOXA11-AS and miR-642b was also investigated, but no significant correlation was revealed in lung adenocarcinoma (r=−0.047, P=0.507; data not shown) and squamous cell carcinoma (r=0.123, P=0.148; data not shown) partly due to the fact that only 203 lung patients with adenocarcinoma and 140 patients with squamous cell carcinoma were included in the data from TCGA.
Figure 5.
Differential expression of miR-642b in lung adenocarcinoma and squamous cell carcinoma based on the Cancer Genome Atlas database. The expression of miR-642b between normal lung and (A) lung adenocarcinoma cells or (B) lung squamous cell carcinoma cells. (C) Kaplan-Meier curves for miR-642b expression in lung adenocarcinoma. miR, microRNA.
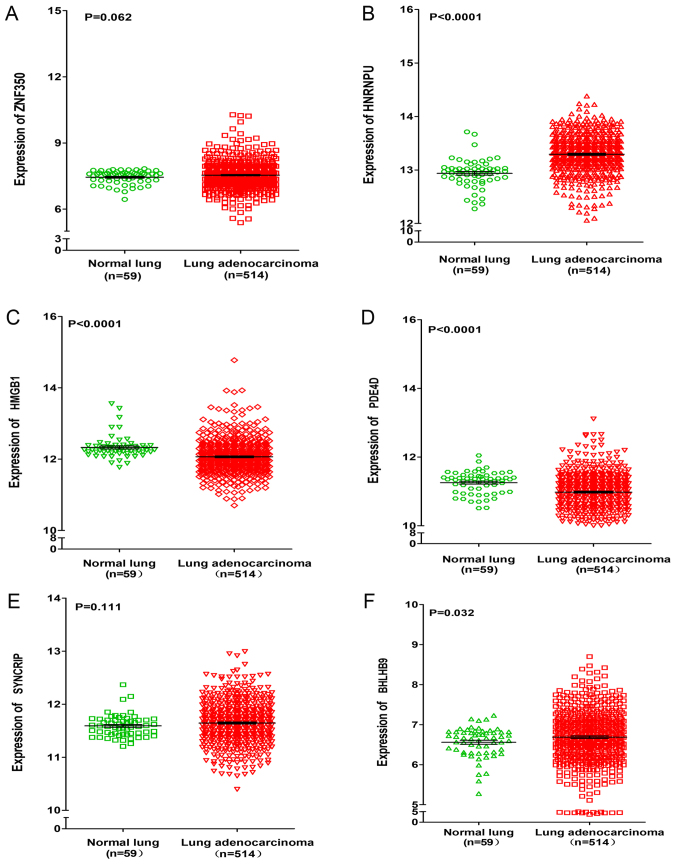
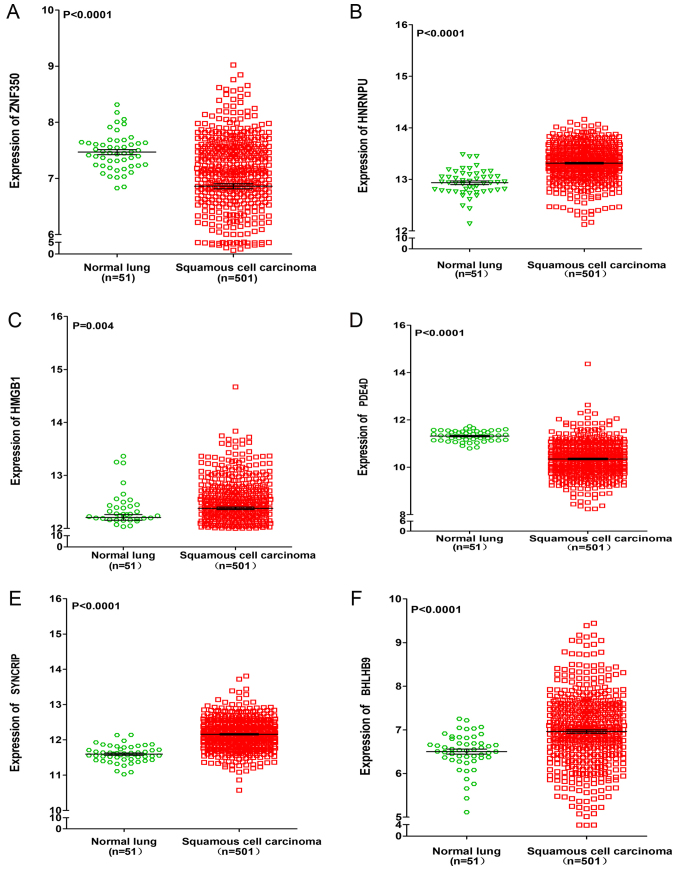
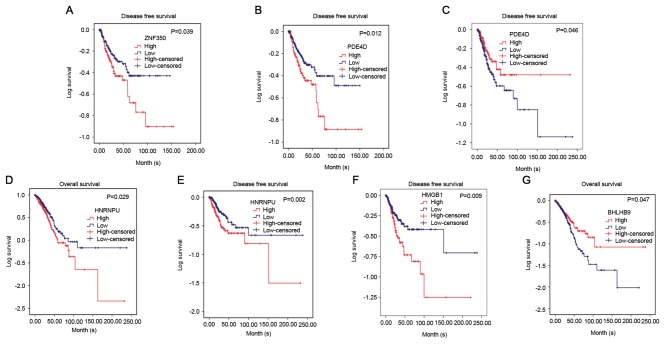
For target gene prediction, six target genes (ZNF350, HNRNPU, HMGB1, PDE4D, SYNCRIP and BHLHB9) of miR-642b-3p were identified by all four prediction algorithms. Then, further analyses were performed in order to determine whether the expressions of these genes were associated with NSCLC, based on the data from TCGA. It was revealed that the expressions of HNRNPU, SYNCRIP and BHLHB9 were upregulated in lung adenocarcinoma and squamous cell carcinoma, whereas PDE4D was significantly downregulated in lung adenocarcinoma and squamous cell carcinoma (P<0.001). For the remaining two genes (ZNF350 and HMGB1), it was revealed that ZNF350 was upregulated in lung adenocarcinoma and significantly downregulated in lung squamous cell carcinoma (P<0.0001), whereas HMGB1 was significantly downregulated in lung adenocarcinoma and significantly upregulated in lung squamous cell carcinoma (P<0.05, Figs. 6 and 7). Subsequently, the relationship between these genes and overall survival or disease-free survival of NSCLC was investigated. It was revealed that in lung squamous cell carcinoma, ZNF350 and PDE4D were significantly associated with disease-free survival (P<0.05). In lung adenocarcinoma, it was discovered that HNRNPU was associated with overall survival and disease-free survival (P<0.05), and PDE4D and HMGB1 were related to disease-free survival (P<0.05), and BHLHB9 was only associated with overall survival (P<0.05, Fig. 8). Additionally, the correlation between HOXA11-AS expression and the 6 target genes was analysed, and a weak negative correlation was revealed between HOXA11-AS and PDE4D or ZNF350 in lung adenocarcinoma and squamous cell carcinoma, whereas HNRNPU and SYNCRIP were positively correlated with HOXA11-AS. BHLHB9 and HMGB1 were revealed to have a weak positive correlation with HOX11-AS in lung adenocarcinoma, and a weak negative correlation with HOX11-AS in squamous cell carcinoma (Table II).
Figure 6.
Differential expression of ZNF350, HNRNPU, HMGB1, PDE4D, SYNCRIP, and BHLHB9 between lung adenocarcinoma and normal lung tissues based on the Cancer Genome Atlas database. Differential expression of (A) ZNF350, (B) HNRNPU, (C) HMGB1, (D) PDE4D, (E) SYNCRIP and (F) BHLHB9. ZNF350, zinc finger protein 350; HNRNPU, heterogeneous nuclear ribonucleoprotein U; HMGB1, high mobility group box 1; PDE4D, phosphodiesterase 4D; SYNCRIP, synaptotagmin binding cytoplasmic RNA interacting protein; BHLHB9, basic helix-loop-helix family member B9.
Figure 7.
Differential expression of ZNF350, HNRNPU, HMGB1, PDE4D, SYNCRIP and BHLHB9 between lung squamous cell carcinoma and normal lung tissues based on the Cancer Genome Atlas database. Differential expression of (A) ZNF350, (B) HNRNPU, (C) HMGB1, (D) PDE4D, (E) SYNCRIP and (F) BHLHB9. ZNF350, zinc finger protein 350; HNRNPU, heterogeneous nuclear ribonucleoprotein U; HMGB1, high mobility group box 1; PDE4D, phosphodiesterase 4D; SYNCRIP, synaptotagmin binding cytoplasmic RNA interacting protein; BHLHB9, basic helix-loop-helix family member B9.
Figure 8.
Kaplan-Meier curves for ZNF350, HNRNPU, HMGB1, PDE4D and BHLHB9 expression in lung adenocarcinoma and squamous cell carcinoma based on The Cancer Genome Atlas database. (A) Disease-free survival of patients with different ZNF350 expression levels in lung squamous cell carcinoma. Patients with high ZNF350 expression had a significantly poorer prognosis (85.14±7.98) compared with those with low expression (105.02±5.76) (P=0.039). (B) Disease-free survival of patients with differing PDE4D expression levels in lung squamous cell carcinoma. Patients with high PDE4D expression had a significantly poorer prognosis (83.15±8.29) than those with low expression (105.61±6.14) (P=0.012). (C) Disease-free survival of patients with different PDE4D expression levels in lung adenocarcinoma. Patients with low PDE4D expression had a significantly poorer prognosis (153.38±12.61) than those with high expression (112.77±14.03) (P=0.046). (D) Overall survival of patients with different HNRNPU expression levels in lung adenocarcinoma. Patients with high HNRNPU expression had a significantly poorer prognosis (70.68±9.93) than those with low expression (102.72±11.13) (P=0.029). (E) Disease-free survival of patients with different HNRNPU expression levels in lung adenocarcinoma. Patients with high HNRNPU expression had a significantly poorer prognosis (101.69±17.09) than those with low expression (142.28±14.76) (P=0.002). (F) Disease-free survival of patients with different HMGB1 expression levels in lung adenocarcinoma. Patients with high HMGB1 expression had a significantly poorer prognosis (92.53±14.88) than those with low expression (150.23±15.44) (P=0.009). (G) Overall survival of patients with different BHLHB9 expression levels in lung adenocarcinoma. Patients with low BHLHB9 expression had a significantly poorer prognosis (109.70±14.43) than those with high expression (71.41±7.96) (P=0.047). ZNF350, zinc finger protein 350; HNRNPU, heterogeneous nuclear ribonucleoprotein U; HMGB1, high mobility group box 1; PDE4D, phosphodiesterase 4D; SYNCRIP, synaptotagmin binding cytoplasmic RNA interacting protein; BHLHB9, basic helix-loop-helix family member B9.
Table II.
Correlations between HOXA11 antisense RNA expression and the six target genes in lung adenocarcinoma and squamous cell carcinoma based on The Cancer Genome Atlas via Spearman's test.
| Gene name | |||||||
|---|---|---|---|---|---|---|---|
| Carcinoma type | Value | Zinc finger protein 350 | Heterogeneous nuclear ribonucleoprotein U | High mobility group box 1 | Phosphodiesterase 4D | Synaptotagmin-binding cytoplasmic RNA interacting protein | Basic helix-loop-helix family member B9 |
| Lung adenocarcinoma | R-value | −0.004 | 0.307 | 0.286 | −0.124 | 0.303 | 0.128 |
| P-value | 0.949 | <0.0001 | <0.0001 | 0.049 | <0.0001 | 0.041 | |
| Squamous cell carcinoma | R-value | −0.067 | 0.200 | −0.055 | −0.068 | 0.187 | −0.093 |
| P-value | 0.639 | 0.160 | 0.704 | 0.637 | 0.188 | 0.517 | |
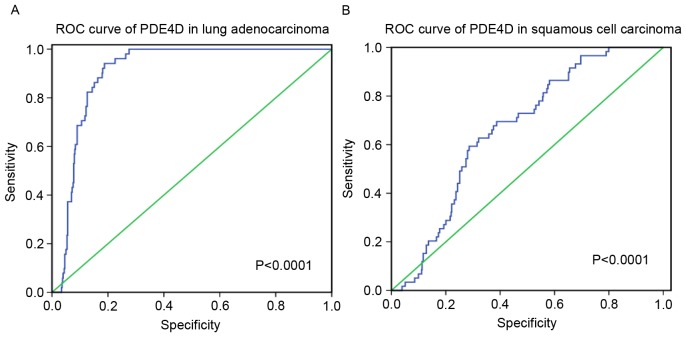
Amongst these results, PDE4D was downregulated in lung adenocarcinoma and squamous cell carcinoma, and a weak negative correlation was revealed between HOXA11-AS and PDE4D. Furthermore, the diagnostic value of PDE4D level in lung cancer was assessed and it was revealed that the AUC of PDE4D was 0.905 (95% CI, 0.879–0.931) for patients with lung adenocarcinoma and 0.665 (95% CI, 0.606–0.725) for patients with squamous cell carcinoma (P<0.0001; Fig. 9).
Figure 9.
ROC curve for PDE4D in lung adenocarcinoma and squamous cell carcinoma based on The Cancer Genome Atlas database. (A) ROC curve for PDE4D in lung adenocarcinoma. (B) ROC curve for PDE4D in squamous cell carcinoma. ROC, receiving operator characteristic; PDE4D, phosphodiesterase 4D.
Discussion
LncRNAs are non-protein-coding RNA molecules and the length of lncRNAs varies from 200 nucleotides to 100 kb (20). Numerous lncRNAs have been confirmed to have important functions in transcriptional regulation, epigenetic gene regulation or disease development (13–15). Additionally, lncRNAs have been associated with the tumorigenesis and progression of NSCLCs. To date, different lncRNAs have been reported to perform different functions in NSCLC, including lncRNA AK126698, GAS5-AS1 and TUSC7, which may be associated with cell proliferation, metastasis and a poor prognosis in NSCLC (56–58). Multiple previous reports have demonstrated that lncRNAs function in lung cancer by regulating the expression of miRNAs (23–25,59). HOXA11-AS is a member of the homeobox family of genes. To the best of our knowledge, only two previous studies have reported the relationship between HOXA11-AS and cancer. Richards et al (9) conducted various functional experiments and analysed genome-wide data, and revealed that HOXA11-AS may inhibit the oncogenic phenotype of epithelial ovarian cancer, which may be enhanced by the T allele. Wang et al (10) used a high-throughput microarray and gene set enrichment analysis to demonstrate that HOXA11-AS may have a growth-promoting function in glioma via the regulation of cell cycle progression. However, the specific pathogenesis of HOXA11-AS in NSCLC remains unclear. Thus, the present study was designed using A549 cells to investigate the expression profile changes of miR-642b-3p following HOXA11-AS knockdown and the potential molecular mechanisms of HOXA11-AS in NSCLC.
In the present study, miR-642b-3p and its target genes (ZNF350, HNRNPU, HMGB1, PDE4D, SYNCRIP, BHLHB9) were analysed. To the best of our knowledge, only one previous study has reported on the relationship between miR-642b-3p and cancer. Hamam et al (60) revealed that miR-642b-3p was upregulated in breast cancer, and miR-642b-3p in combination with eight other upregulated miRNAs may be used for the early detection of breast cancer. Similarly, to this previous study, miR-642b-3p was upregulated in lung adenocarcinoma and squamous cell carcinoma, but the specific association between miR-642b-3p and NSCLC remains to be verified by functional experiments. Then, six target genes of miR-642b-3p were investigated and it was revealed that all six target genes were differentially expressed in lung adenocarcinoma and squamous cell carcinoma. Additionally, miR-642b-3p, ZNF350, HNRNPU, HMGB1, PDE4D and BHLHB9 were associated with the overall survival or disease-free survival of patients with lung adenocarcinoma or patients with squamous cell carcinoma based on TCGA database. Numerous previous studies have confirmed the association between these target genes and NSCLC. Deng et al (61) used a western blot analysis to determine that miR-193a-3p may inhibit the metastasis of lung cancer cells by de-regulating the expression of HNRNPU and other tumour-associated proteins. Numerous studies have assessed HMGB1. HMGB1 has been associated with cell migration, invasion, apoptosis, sensitivity to chemotherapy drugs and the prognosis of NSCLC (62–64). Karachaliou et al (65) revealed that PDE4D was associated with resistant epidermal growth factor receptor (EGFR)-mutant cancer cell lines, and the combination of EGFR tyrosine kinase inhibitors with PDE4D inhibitors may be an effective therapy for patients with EGFR-mutant NSCLC.
In the findings of the present study, PDE4D was downregulated in lung adenocarcinoma and squamous cell carcinoma, and a weak negative association was established between HOXA11-AS and PDE4D. Additionally, PDE4D was associated with disease-free survival of lung adenocarcinoma and squamous cell carcinoma, which indicated that PDE4D may influence the prognosis of adenocarcinoma. Therefore, the present study hypothesized that miR-642b-3p, which is regulated by lncRNA HOXA11-AS and targets PDE4D, may have a function in NSCLC tumorigenesis and deterioration.
Numerous studies have demonstrated that PDE4D may function in cancer via different pathways, including the EGFR/phosphoinositide 3-kinase/protein kinase B signalling pathway, MAPK pathway and Hedgehog pathway (66–68). However, further exploration is required to clarify the potential molecular mechanisms of PDE4D in NSCLC.
Additionally, there are limitations in the present study. The present study was conducted based on several online tools, including DAVID, STRING, Cytoscape and TCGA, which may only be used for reference. Furthermore, the specific molecular mechanisms of miR-642b-3p and genes it is associated with, require further elucidation through functional experiments. To assess this hypothesis, a series of experiments may be designed using various molecule, cell, tissue and animal models (including RT-qPCR, western blot analysis, proliferation, invasion and metastasis assays, dual luciferase reporter assay, RNA pull-down, chromatin immunoprecipitation, chicken embryo chorioallantoic membrane and nude mouse models). The present study, which focused on the function of the HOXA11-AS/miR-642b-3p/PDE4D axis, suggests a novel target for a clinical therapeutic strategy for NSCLC. In conclusion, HOXA11-AS may influence the expression of miR-642b-3p in the different biological processes of NSCLC by targeting the expression of PDE4D or other target genes. Raw data from TCGA confirmed the oncogenic function of miR-642b in lung adenocarcinoma and squamous cell carcinoma. The findings of the present study lay the foundation for future studies on the relationship between HOXA11-AS and the potential molecular mechanisms in NSCLC tumorigenesis and progression.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. NSFC81560469 and NSFC81360327), the Scientific Research Project of Basic Ability Promoting for Middle Age and Youth Teachers of Guangxi Universities (grant no. KY2016YB077), the Natural Science Foundation of Guangxi, China (grant no. 2015GXNSFCA139009) and the Guangxi Provincial Health Bureau Scientific Research Project (grant nos. Z2013201 and Z2014055).
References
- 1.Xu YJ, Du Y, Fan Y. Long noncoding RNAs in lung cancer: What we know in 2015. Clin Transl Oncol. 2016;18:660–665. doi: 10.1007/s12094-015-1448-y. [DOI] [PubMed] [Google Scholar]
- 2.Kang CG, Lee HJ, Kim SH, Lee EO. Zerumbone suppresses osteopontin-induced cell invasion through inhibiting the FAK/AKT/ROCK pathway in human non-small cell lung cancer A549 cells. J Nat Prod. 2016;79:156–160. doi: 10.1021/acs.jnatprod.5b00796. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 4.Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Chen J, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe C, Shin YS, Kim C, Choi SJ, Lee J, Kim SY, Cho YB, Kim J. Crosstalk with cancer-associated fibroblasts induces resistance of non-small cell lung cancer cells to epidermal growth factor receptor tyrosine kinase inhibition. Onco Targets Ther. 2015;8:3665–3678. doi: 10.2147/OTT.S89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 7.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards EJ, Permuth-Wey J, Li Y, Chen YA, Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–34757. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R, Pu P, Kang C, Han L. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373:251–259. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 11.Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther. 2017 Oct 11; doi: 10.1080/15384047.2017.1385679. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu CH, Sun QY, Zhang FM, Jia YM. Long non-coding RNA ROR is a novel prognosis factor associated with non-small-cell lung cancer progression. Eur Rev Med Pharmacol Sci. 2017;21:4087–4091. [PubMed] [Google Scholar]
- 13.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan X, Wang J, Tang X, Li Y, Xia P, Gao X. Berberine ameliorates nonalcoholic fatty liver disease by a global modulation of hepatic mRNA and lncRNA expression profiles. J Transl Med. 2015;13:24. doi: 10.1186/s12967-015-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Wei Y, Niu B. Role of MALAT1 as a prognostic factor for survival in various cancers: A systematic review of the literature with meta-analysis. Dis Markers. 2015;2015:164635. doi: 10.1155/2015/164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: Roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285–2295. [PubMed] [Google Scholar]
- 20.Xie X, Pan J, Wei L, Wu S, Hou H, Li X, Chen W. Gene expression profiling of microRNAs associated with UCA1 in bladder cancer cells. Int J Oncol. 2016;48:1617–1627. doi: 10.3892/ijo.2016.3357. [DOI] [PubMed] [Google Scholar]
- 21.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L, Xu H, Luo F, Liu X, Lu X, Yang Q, Xue J, Chen C, Shi L, Liu Q. Epigenetic silencing of miR-218 by the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle transition in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2016;304:30–41. doi: 10.1016/j.taap.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 26.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Tang Y, Guo W, Du Y, Wang Y, Li P, Zang W, Yin X, Wang H, Chu H, et al. Up-regulation of microRNA-138 induce radiosensitization in lung cancer cells. Tumour Biol. 2014;35:6557–6565. doi: 10.1007/s13277-014-1879-z. [DOI] [PubMed] [Google Scholar]
- 31.Tang R, Liang L, Luo D, Feng Z, Huang Q, He R, Gan T, Yang L, Chen G. Downregulation of MiR-30a is associated with poor prognosis in lung cancer. Med Sci Monit. 2015;21:2514–2520. doi: 10.12659/MSM.894812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S, Shen W, Pan Y, Zhu M, Xie K, Geng L, Wang Y, Liang Y, Xu J, Cao S, et al. Genetic variations in key MicroRNAs are associated with the survival of nonsmall cell lung cancer. Medicine (Baltimore) 2015;94:e2084. doi: 10.1097/MD.0000000000002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang T, She K, Peng G, Wang W, Huang J, Li J, Wang Z, He J. MicroRNA-186 suppresses cell proliferation and metastasis through targeting MAP3K2 in non-small cell lung cancer. Int J Oncol. 2016;49:1437–1444. doi: 10.3892/ijo.2016.3637. [DOI] [PubMed] [Google Scholar]
- 34.Lu YJ, Liu RY, Hu K, Wang Y. MiR-541-3p reverses cancer progression by directly targeting TGIF2 in non-small cell lung cancer. Tumour Biol. 2016;37:12685–12695. doi: 10.1007/s13277-016-5241-5. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Wang X, Fu B, Meng L, Lang B. Differentially expressed genes and microRNAs in bladder carcinoma cell line 5637 and T24 detected by RNA sequencing. Int J Clin Exp Pathol. 2015;8:12678–12687. [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Ge X, Xu X, Zhong Y, Qie Z. Comparison of the gene expression profiles between gallstones and gallbladder polyps. Int J Clin Exp Pathol. 2014;7:8016–8023. [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang CM, Wang XH, Shu J, Yang WX, Fu P, Zhuang LL, Zhou GP. Analysis of differentially expressed genes based on microarray data of glioma. Int J Clin Exp Med. 2015;8:17321–17332. [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Zhuo D, Chen J, Yuan H. Screening feature genes of lung carcinoma with DNA microarray analysis. Int J Clin Exp Med. 2015;8:12161–12171. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database Issue):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server Issue):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database Issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bornstein S, Schmidt M, Choonoo G, Levin T, Gray J, Thomas CR, Jr, Wong M, McWeeney S. IL-10 and integrin signaling pathways are associated with head and neck cancer progression. BMC Genomics. 2016;17:38. doi: 10.1186/s12864-015-2359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093. doi: 10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu X, Li H, Liu C, Hu B, Li T, Wang Y. Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther. 2016;9:3815–3827. doi: 10.2147/OTT.S100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Jin Y, Ren H, Ma X, Wang B, Wang Y. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res. 2016;8:680–687. [PMC free article] [PubMed] [Google Scholar]
- 59.You J, Zhang Y, Liu B, Li Y, Fang N, Zu L, Li X, Zhou Q. MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer. 2014;51(Suppl 3):e77–e81. doi: 10.4103/0019-509X.154055. [DOI] [PubMed] [Google Scholar]
- 60.Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, Alajez NM. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng W, Yan M, Yu T, Ge H, Lin H, Li J, Liu Y, Geng Q, Zhu M, Liu L, et al. Quantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancer. Cell Physiol Biochem. 2015;35:1677–1688. doi: 10.1159/000373981. [DOI] [PubMed] [Google Scholar]
- 62.Zhang C, Ge S, Hu C, Yang N, Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 2013;45:1055–1061. doi: 10.1093/abbs/gmt109. [DOI] [PubMed] [Google Scholar]
- 63.Zhang R, Li Y, Wang Z, Chen L, Dong X, Nie X. Interference with HMGB1 increases the sensitivity to chemotherapy drugs by inhibiting HMGB1-mediated cell autophagy and inducing cell apoptosis. Tumour Biol. 2015;36:8585–8592. doi: 10.1007/s13277-015-3617-6. [DOI] [PubMed] [Google Scholar]
- 64.Feng A, Tu Z, Yin B. The effect of HMGB1 on the clinicopathological and prognostic features of non-small cell lung cancer. Oncotarget. 2016;7:20507–20519. doi: 10.18632/oncotarget.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karachaliou N, Codony-Servat J, Teixidó C, Pilotto S, Drozdowskyj A, Codony-Servat C, Giménez-Capitán A, Molina-Vila MA, Bertrán-Alamillo J, Gervais R, et al. BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer. Sci Rep. 2015;5:17499. doi: 10.1038/srep17499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu T, Wu S, Yuan Y, Yan G, Xiao D. Knockdown of phosphodiesterase 4D inhibits nasopharyngeal carcinoma proliferation via the epidermal growth factor receptor signaling pathway. Oncol Lett. 2014;8:2110–2116. doi: 10.3892/ol.2014.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Powers GL, Hammer KD, Domenech M, Frantskevich K, Malinowski RL, Bushman W, Beebe DJ, Marker PC. Phosphodiesterase 4D inhibitors limit prostate cancer growth potential. Mol Cancer Res. 2015;13:149–160. doi: 10.1158/1541-7786.MCR-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge X, Milenkovic L, Suyama K, Hartl T, Purzner T, Winans A, Meyer T, Scott MP. Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. Elife. 2015;4 doi: 10.7554/eLife.07068. [DOI] [PMC free article] [PubMed] [Google Scholar]