Abstract
DNA methylation is an important epigenetic modification that alters gene expression; DNA hypomethylation contributes to tumorigenesis through multiple processes. In the present study, the methylation of long interspersed element-1 (LINE-1) in 95 gastric cancer (GC) tissues and matched adjacent normal tissues was investigated by pyrosequencing. LINE-1 methylation was compared with the expression of ubiquitin-like with PHD and ring-finger protein 1 (UHRF1), an essential regulator of DNA methylation, using reverse transcription-quantitative polymerase chain reaction. Significant hypomethylation of LINE-1 and overexpression of UHRF1 were observed in GC tissues compared with the matched controls (P<0.001 and P=0.001, respectively). LINE-1 hypomethylation was inversely correlated with UHRF1 overexpression in GC tissues (r=−0.026, P=0.028). In addition, LINE-1 hypomethylation in GC was significantly associated with Lauren's histological classification, tumor differentiation and background intestinal metaplasia (P=0.014, P=0.042 and P=0.034, respectively). These results suggest that LINE-1 hypomethylation may be a potential biomarker for GC and it is indirectly regulated by UHRF1 overexpression.
Keywords: gastric cancer, long interspersed nucleotide elements, DNA methylation, ubiquitin-like with PHD and ring-finger protein 1
Introduction
Epigenetic alterations are important in maintaining genomic stability. DNA methylation is the most common epigenetic modification, and is involved in various biological processes, including tumorigenesis, when aberrant DNA methylation occurring in promoter CpG islands can lead to gain of function in oncogenes and loss of function in tumor suppressor genes (1–4). The mechanism by which DNA hypomethylation contributes to tumorigenesis has been proposed to involve chromosomal instability (5,6), derepression of imprinted genes (7), and retrotransposon activation (8,9). Long interspersed element-1 (LINE-1) is a repetitive retrotransposon and a major constituent of interspersed DNA repeats. As it constitutes approximately 17% of the human genome, LINE-1 methylation is used as a surrogate marker of global DNA methylation (10). LINE-1 hypomethylation has been observed in several types of cancer, including gastric cancer (GC) (11–14). However, the mechanism by which LINE-1 methylation is regulated remains undefined.
Ubiquitin-like with PHD and ring-finger protein 1 (UHRF1) is a modular protein containing several functional domains. The SET and RING finger-associated domain plays an essential role in DNA methylation, transferring methylation patterns to daughter cells through the recruitment of DNA methyltransferase 1 (DNMT1) to newly synthesized strands during DNA replication (9,15–17). UHRF1 is overexpressed in several cancers (9,18–20). However, the relationship between LINE-1 hypomethylation and UHRF1 expression has not been reported in GC. Here, we have investigated LINE-1 methylation status and UHRF1 mRNA expression in GC tissues compared to matched adjacent normal tissues. In addition, we explored whether LINE-1 methylation is related with UHRF1 expression as well as clinicopathological features including age, gender, tumor location, Lauren's histologic classification, tumor differentiation, tumor stage, and accompanying atrophy and intestinal metaplasia.
Materials and methods
Ethics statement
The present study protocol was reviewed and approved by the Institutional Review Board of Chungnam National University Hospital approved the study protocol (IRB no. 2014-10-031). All patients enrolled in this study provided their written informed consent for tissue collection and use.
Patients and tissue samples
GC and matched adjacent normal specimens were obtained from 95 patients who underwent a gastrectomy at Chungnam National University Hospital between February 2010 and July 2015. The mean age of the patients (65 males and 30 females) was 65.4±11.0 (Table I). Adjacent normal mucosa was obtained at least 5.0 cm from the tumor margin. The biospecimens and data used for this study were provide by the Biobank of the Chungnam National University Hospital, a member of the Korea Biobank Network.
Table I.
Association between LINE-1 methylation and clinicopathological features.
| LINE-1 methylation (%) | ||||
|---|---|---|---|---|
| Feature | N (%) | Mean | SEM | P-value |
| Age, years (mean ± SD) | 65.4±11.0 | |||
| <65 | 40 (52.6) | 69.5 | 1.6 | 0.695 |
| ≥65 | 45 (47.4) | 69.1 | 1.2 | |
| Sex | ||||
| Male | 65 (68.4) | 68.8 | 1.2 | 0.366 |
| Female | 30 (31.6) | 70.4 | 1.6 | |
| Tumor location | ||||
| Upper | 27 (28.4) | 71.6 | 1.6 | 0.056 |
| Lower | 68 (71.6) | 68.4 | 1.2 | |
| Background atrophy | ||||
| No | 68 (71.6) | 69.5 | 1.2 | 0.514 |
| Yes | 27 (28.4) | 68.7 | 1.7 | |
| Background intestinal metaplasia | ||||
| No | 12 (12.6) | 74.0 | 1.6 | 0.042 |
| Yes | 83 (87.4) | 68.6 | 1.1 | |
| Tumor differentiation | ||||
| Differentiated | 47 (49.5) | 66.7 | 1.6 | 0.034 |
| Undifferentiated | 48 (50.5) | 71.8 | 0.9 | |
| Lauren's histologic classification | ||||
| Intestinal | 67 (70.6) | 67.6 | 1.3 | 0.014 |
| Diffuse | 14 (14.7) | 72.9 | 1.6 | |
| Mixed | 14 (14.7) | 73.7 | 1.6 | |
| T classification | ||||
| T1/T2 | 29 (30.5) | 71.1 | 1.6 | 0.290 |
| T3/T4 | 66 (69.5) | 68.5 | 1.2 | |
| N classification | ||||
| N0 | 29 (30.5) | 69.9 | 1.5 | 0.796 |
| N1 | 24 (25.3) | 68.2 | 2.3 | |
| N2 | 17 (17.9) | 68.1 | 2.4 | |
| N3 | 25 (26.3) | 70.3 | 1.9 | |
| Tumor stage | ||||
| I (A+B) | 20 (21.1) | 69.0 | 2.0 | 0.111 |
| II (A+B) | 20 (21.1) | 73.0 | 1.5 | |
| III (A+B+C) | 46 (48.4) | 68.0 | 1.6 | |
| IV | 9 (9.5) | 73.0 | 2.4 | |
LINE-1, long interspersed element-1; SEM, standard error of the mean; SD, standard deviation.
DNA and bisulfite treatment
Genomic DNA (gDNA) was extracted using a DNeasy Blood & Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. DNA quality (A260/280 and A260/230) was assessed using a Nanodrop 2000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). The isolated DNA was treated with sodium bisulfite using an EZ DNA Methylation kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer's instructions.
Pyrosequencing
The methylation status of four CpG islands in the LINE-1 promoter (position 305, 318, 321, and 328 in GenBank accession no. X58075) was measured by pyrosequencing. A 50 µl volume PCR was performed using 50 ng of bisulfite-treated gDNA, Taq DNA polymerase, 50 pmol of the forward (5′-TTTTGAGTTAGGTGTGGGATATA-3′) and reverse (5′-biotin-AAAATCAAAAAATTCCCTTTC-3′) primers, and 0.3 µM of pyrosequencing primer (5′-AGTTAGGTGTGGGATATAGT-3′) with a Veriti Thermal Cycler (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) using the following PCR conditions: 95°C for 10 min; 45 cycles of 95°C for 30 sec, 50°C for 30 sec, 72°C for 30 sec; and 72°C for 5 min. Pyrosequencing was performed using a PyroMark Gold Q24 reagent kit and the Pyromark ID system (both from Qiagen, Germantown, MD, USA). Data were analyzed using PyroMark Q24 2.0.6 software (Qiagen). The methylation level was calculated as the percent of 5-methylated cytosine divided by the total cytosine, and represents the mean of the four sites.
Reverse transcription-quantitative polymerase chain reacvtion (RT-qPCR)
Total RNA was isolated with TRIzol (Thermo Fisher Scientific, Inc.) and cDNA was synthesized using a SuperScript III First Strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.), both according to the manufacturer's instructions. PCR amplification was performed using cDNA, TaqMan Universal Master Mix II with UNG (Applied Biosystems; Thermo Fisher Scientific, Inc.), and a TaqMan Gene Expression Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.); the genes assayed were UHRF1 (Hs00273589_m1) and GAPDH (Hs99999905_m1). Amplification reactions were performed in triplicate with a StepOne Plus system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C. The relative mRNA expression was calculated as the difference in quantification cycle (ΔCq) between the triplicate mean Cq for UHRF1 and the triplicate mean Cq for GAPDH from the same sample.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM). Two group comparisons were performed using a Paired Samples t-test. The Wilcoxon test was used to compare LINE-1 methylation or UHRF1 mRNA expression between tumors and adjacent normal tissues. Mann-Whitney U and Kruskal-Wallis tests were used to evaluate associations between LINE-1 methylation and clinicopathological parameters. The Spearman correlation test was used to evaluate correlation between LINE-1 methylation and UHRF1 mRNA expression in tumor tissues. A Two-sided P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS (SPSS, Inc., Chicago, IL, USA), version 22.0 for Windows.
Results
Correlation between LINE-1 methylation and UHRF1 expression in GC
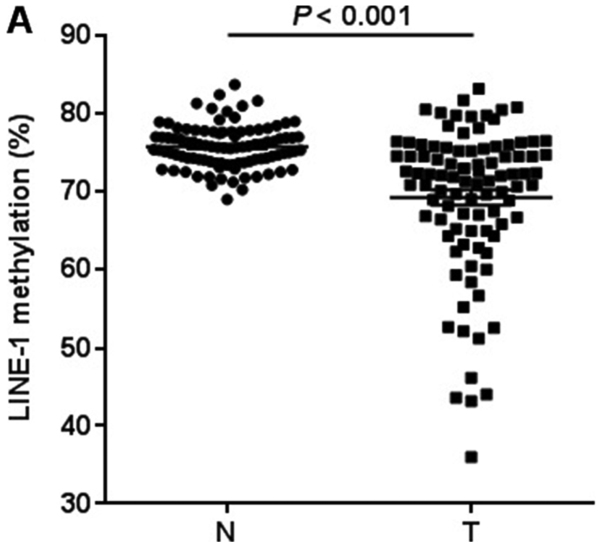
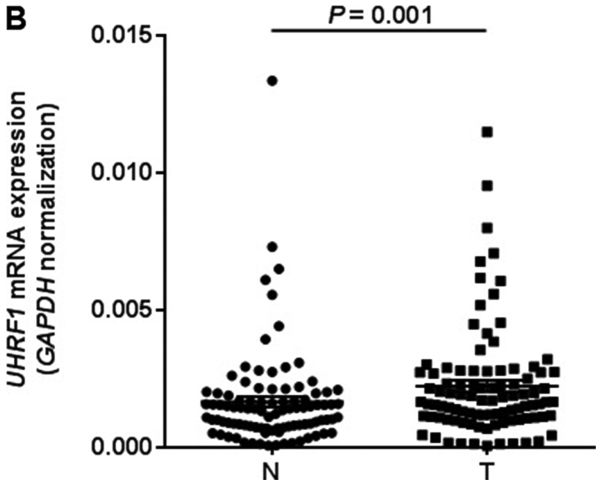
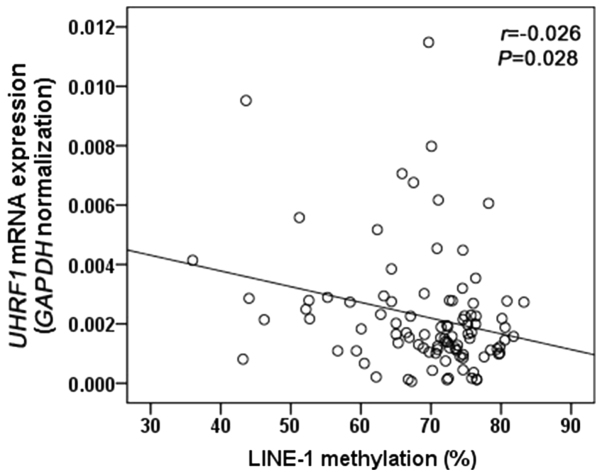
We first quantified LINE-1 methylation in 95 GC tissues and matched adjacent normal tissues by pyrosequencing analysis (Fig. 1). LINE-1 was significantly hypomethylated in GC tissues compared with matched controls (75.8±0.3 vs. 69.3±1.0%, P<0.001; Fig. 2A). We next measured UHRF1 mRNA expression in 95 GC tissues and matched adjacent normal tissues by RT-qPCR analysis. UHRF1 was highly expressed in GC tissues compared to adjacent normal tissues (P=0.001; Fig. 2B). To evaluate whether UHRF1 overexpression is related to LINE-1 hypomethylation, we analyzed the correlation between UHRF1 mRNA expression and LINE-1 methylation in GC tissues. LINE-1 methylation was inversely correlated with UHRF1 mRNA expression (Spearman r=−0.026, P=0.028; Fig. 3).
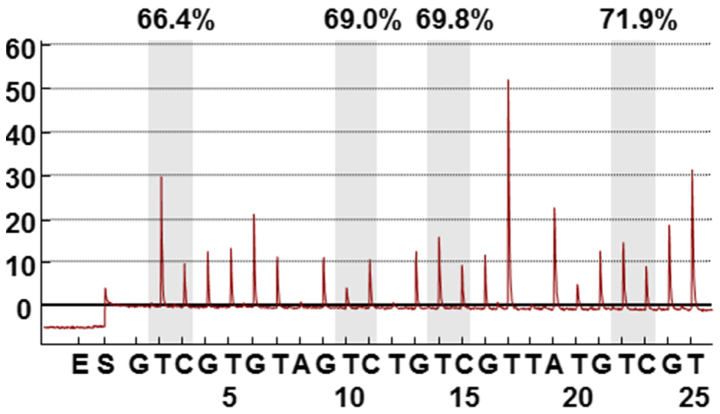
Figure 1.
Pyrosequencing assay to measure LINE-1 methylation levels. Percentage is the proportion of cytosine at each CpG site after bisulfite conversion. Overall LINE-1 methylation level is the average of proportion of cytosine at the four CpG sites. LINE-1, long interspersed element-1.
Figure 2.
LINE-1 methylation and UHRF1 expression comparision between 95 gastric cancer tissues and matched adjacent normal tissues. (A) LINE-1 methylation levels. The cancer tissues showed significantly lower levels of methylation than matched normal tissues (P<0.001 by Wilcoxon signed-rank test). (B) UHRF1 expression levels. UHRF1 mRNA expression levels were significantly higher in cancer tissues than matched normal tissues (P=0.001 by Wilcoxon signed-rank test). The mRNA levels were normalized to those of GAPDH. Horizontal lines represent the mean ± standard error of the mean as determined by triplicate assays. N, adjacent normal tissues; T, tumor tissues; LINE-1, long interspersed element-1; UHRF1, ubiquitin-like with PHD and ring-finger protein 1.
Figure 3.
Correlation between LINE-1 methylation and UHRF1 expression in 95 gastric cancer tissues. r, Spearman's correlation coefficient; UHRF1, ubiquitin-like with PHD and ring-finger protein 1; LINE-1, long interspersed element-1.
Association between LINE-1 methylation and clinicopathological features
We also evaluated possible correlations between LINE-1 methylation and clinicopathological features of the GCs. The level of LINE-1 methylation was significantly related to the Lauren's histologic classification, tumor differentiation, and the presence of background intestinal metaplasia (P=0.014, P=0.042, and P=0.034, respectively; Table I). However, there were no significant associations between LINE-1 methylation and other clinicopathological features of GC.
Discussion
Pyrosequencing is a reliable assay to measure LINE-1 methylation (21,22). In this study, we investigated LINE-1 methylation and UHRF1 mRNA expression in GC tissues and matched controls using pyrosequencing and RT-qPCR, respectively. LINE-1 hypomethylation and UHRF1 overexpression were observed in GC tissues compared to adjacent normal tissues (Fig. 2A and B).
Several studies have reported LINE-1 hypomethylation and UHRF1 overexpression in various cancers, including GC (9,11–14,18–20,23,24). However, there have been no reports demonstrating that LINE-1 hypomethylation is correlated with UHRF1 overexpression in GC. Here, we show that LINE-1 hypomethylation is inversely correlated with UHRF1 overexpression, indicating that LINE-1 methylation may be regulated by UHRF1, which need to be confirmed molecular mechanism by further studies.
Accumulating evidence suggests that UHRF1 regulates gene expression through epigenetic mechanisms including DNA methylation, histone methylation (25), histone deacetylation (26), and possibly histone ubiquitination (27). A previous study demonstrated that plasmid-mediated UHRF1 overexpression delocalized and destabilized zebrafish Dnmt1, causing DNA methylation (18). In addition, they demonstrated that oncogenic UHRF1 overexpression induces global DNA hypomethylation, causing hepatocellular carcinoma in zebrafish (18). The relationship between UHRF1 expression and LINE-1 methylation has been investigated in esophageal cancer (19). Consistent with our results, UHRF1 mRNA expression was negatively associated with LINE-1 methylation in esophageal cancer tissues (19); however, the mechanism by which UHRF1 overexpression causes global DNA hypomethylation remains to be fully elucidated. Mechanisms including DNMT1 delocalization and destabilization (27), ubiquitination and degradation (24), and redistribution and/or sequestration of DNMT1 away from DNA (28) have been proposed.
We also evaluated the relationships between LINE-1 hypomethylation and clinicopathological features of the GC samples. Our analysis revealed that LINE-1 hypomethylation of GC was associated with Lauren's histologic classification, tumor differentiation, and background intestinal metaplasia. This is consistent with a previous study that demonstrated that LINE-1 methylation was significantly associated with histologic differentiation and Lauren's histologic classification (22). Our results showed no correlation with tumor stage, similar to previous studies (22,29). Some studies have demonstrated that LINE-1 is a potential prognostic biomarker (22,29); that is, LINE-1 hypomethylation in GC is related to unfavorable prognosis. However, in our study, there was no significant association between LINE-1 hypomethylation and prognosis (data not shown). The mechanism by which aberrant global DNA methylation results in poorer prognosis remains unclear. Several studies have suggested that global DNA methylation is associated with chromosomal instability and mitotic catastrophe (5,6,9,18,30). Large-scale studies are required to confirm LINE-1 methylation status as a useful prognostic factor. Additionally, we analyzed the association between UHRF1 gene expression and clinicopathological features of the GC samples but did not find any relation between them (data not shown).
GC develops by multistep progression, from chronic gastritis, atrophic gastritis, intestinal metaplasia, dysplasia, to GC (22). Previous studies have evaluated the genomic hypomethylation status at various stages in gastric tumorigenesis. One study showed that LINE-1 methylation, measured using pyrosequencing, progressively declined from chronic gastritis to gastric dysplasia (22). However, there were no additional changes in LINE-1 methylation during the progression from dysplasia to cancer (22). Meanwhile, another study based on combined bisulphite restriction analysis demonstrated that LINE-1 methylation progressively decreased from chronic gastritis to GC (31). In our study, LINE-1 methylation in GCs with background intestinal metaplasia was significantly lower than in GCs without background metaplasia. Meanwhile, the presence or absence of background atrophic gastritis in GC was not associated with LINE-1 methylation. Helicobacter pylori infection is closely related to premalignant lesions, including atrophic gastritis and intestinal metaplasia, in multistep gastric tumorigenesis. Therefore, the prevalence of H. pylori infection can influence methylation in premalignant gastric lesions. It has been suggested that quantitative data regarding the methylation of specific genes may be useful for predicting the risk of developing GC (32,33). Taken together, it can be inferred that LINE-1 may be a useful biomarker for understanding gastric tumorigenesis and for classifying GC types according to pathogenesis.
There are limitations in this study. First, in vitro study are required to elucidate the regulatory mechanism of LINE-1 methylation in GC. Second, the sample size was relatively small. Finally, we could not investigate the relationship between LINE-1 methylation and H. pylori infection status, owing to the lack of data.
In conclusion, our results suggest that LINE-1 hypomethylation, which may be regulated by UHRF1 overexpression, may be a useful biomarker for GC. Further studies are needed to better clarify how UHRF1 regulates LINE-1 methylation and whether the mutation in UHRF1 gene and other methylation regulators affect LINE-1 methylation levels in GC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- LINE-1
long interspersed element-1
- GC
gastric cancer
- UHRF1
ubiquitin-like with PHD and ring-finger protein 1
- DNMT1
DNA methyltransferase 1
- gDNA
Genomic DNA
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SD
standard deviation
- SEM
standard error of the mean
Funding
This study was supported by Chungnam National University Hospital Research fund in 2014, by Chungnam National University research fund, by National Research Foundation of Korea (NRF) grant funded by the Korean goverment (MSIP) (2015R1C1A2A01052150), by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) grant funded by the Ministry of Health & Welfare (HI14C1063), and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B04033515).
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Author's contributions
JKS, JHH and EHJ designed the research; SK performed the experiments; KSS contributed the selection of subjects and clinical data acquisition; EHJ and JHH performed the data and statistical analysis; JKS and EHJ contributed to writing and revision of the manuscript; All authors read and approved final manuscript.
Ethics approval and consent to participate
All patients enrolled in this study provided their written informed consent for tissue collection and use. The Institutional Review Board of Chungnam National University Hospital approved the study protocol (IRB No. 2014-10-031).
Consent for publication
All patients provided their written informed consent for the publication of any associated data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 2.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Z, Jiang W, Wang L. Biomarkers for gastric cancer: Progression in early diagnosis and prognosis (Review) Oncol Lett. 2015;9:1502–1508. doi: 10.3892/ol.2015.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 6.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–8639. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 7.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 9.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 10.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese TL, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin Cancer Res. 2010;16:2418–2426. doi: 10.1158/1078-0432.CCR-09-2819. [DOI] [PubMed] [Google Scholar]
- 13.Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, Schernhammer ES, Hunter DJ, Giovannucci EL, Fuchs CS, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Kim HS, Cho MY. Different methylation profiles between intestinal and diffuse sporadic gastric carcinogenesis. Clin Res Hepatol Gastroenterol. 2014;38:613–620. doi: 10.1016/j.clinre.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 16.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura K, Baba Y, Kosumi K, Harada K, Shigaki H, Miyake K, Kiyozumi Y, Ohuchi M, Kurashige J, Ishimoto T, et al. UHRF1 regulates global DNA hypomethylation and is associated with poor prognosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7:57821–57831. doi: 10.18632/oncotarget.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daskalos A, Oleksiewicz U, Filia A, Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK, Liloglou T. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer. 2011;117:1027–1037. doi: 10.1002/cncr.25531. [DOI] [PubMed] [Google Scholar]
- 21.Irahara N, Nosho K, Baba Y, Shima K, Lindeman NI, Hazra A, Schernhammer ES, Hunter DJ, Fuchs CS, Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae JM, Shin SH, Kwon HJ, Park SY, Kook MC, Kim YW, Cho NY, Kim N, Kim TY, Kim D, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131:1323–1331. doi: 10.1002/ijc.27369. [DOI] [PubMed] [Google Scholar]
- 23.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 24.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 25.Kim KB, Son HJ, Choi S, Hahm JY, Jung H, Baek HJ, Kook H, Hahn Y, Kook H, Seo SB. H3K9 methyltransferase G9a negatively regulates UHRF1 transcription during leukemia cell differentiation. Nucleic Acids Res. 2015;43:3509–3523. doi: 10.1093/nar/gkv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 29.Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, Ishimoto T, Iwatsuki M, Baba H. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16:480–487. doi: 10.1007/s10120-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surg Today. 2013;43:1355–1364. doi: 10.1007/s00595-012-0451-y. [DOI] [PubMed] [Google Scholar]
- 31.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima T, Maekita T, Oda I, Gotoda T, Yamamoto S, Umemura S, Ichinose M, Sugimura T, Ushijima T, Saito D. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317–2321. doi: 10.1158/1055-9965.EPI-06-0436. [DOI] [PubMed] [Google Scholar]
- 33.Kaise M, Yamasaki T, Yonezawa J, Miwa J, Ohta Y, Tajiri H. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacter. 2008;13:35–41. doi: 10.1111/j.1523-5378.2008.00572.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding author on reasonable request.