Abstract
Gastric cancer has the fourth highest morbidity rate of all cancers worldwide. Genetic factors including alterations in oncogenes and tumor suppressor genes serve an important role in gastric cancer development and progression. The P53 gene acts as a tumor suppressor gene by regulating the cell cycle, DNA transcription and repair, apoptosis, senescence and genome stability. In addition to somatic P53 mutations in cancer development, germline polymorphisms are also involved in different malignancies. The polymorphism of P53 at codon 72 (Arg72Pro) is established as a common variant that increases susceptibility to various cancers. The present case-control study was conducted to evaluate the possible association between this P53 polymorphism and gastric cancer in the Iranian population. A total of 59 patients with gastric cancer and 59 healthy controls were enrolled in the present study. Genomic DNA was extracted from peripheral blood mononuclear cells and genotype analysis was performed using a polymerase chain reaction-based restriction fragment length polymorphism assay. Genotype frequencies did not differ significantly between the patients and controls (P=0.4); the frequencies of the three genotypes Arg/Arg, Arg/Pro and Pro/Pro in gastric cancer patients were 28.8, 49.2 and 22.0%, and in controls were 37.3, 49.2 and 13.6%. Additionally, there were no differences in genotype frequencies based on tumor location, histological differentiation or tumor stage. Based on these findings, it may be concluded that the P53 codon 72 polymorphism does not contribute to gastric cancer susceptibility in Northern Iran.
Keywords: gastric cancer, P53, polymorphism, Northern Iran
Introduction
Gastric cancer has the third highest mortality and fourth highest morbidity rates of all cancers worldwide (1). In 2012, GloboCan statistics reported almost 1 million new cases of gastric cancer, and more than 700,000 mortalities caused by gastric cancer (1). Gastric cancer is a multifactorial disorder, in which genetic and environmental interactions serve an important role in development and progression (2). Increasing age, gender, lifestyle, dietary regime, environmental factors and Helicobacter pylori infections are among the known risk factors for stomach cancer (3,4). While dietary regime and lifestyle are the most recognized factors, more effective identification of the genetic risk factors is expected to improve understanding of the basic molecular events involved in tumorigenesis (5). Various genetic and epigenetic changes that have the potential to convert normal epithelial cells in the stomach into malignant neoplasms may be responsible for the development of both familial and sporadic gastric cancer (6,7). Studies performed recently have demonstrated that a high number of genes and various environmental factors are the causal agents of gastric cancer, and the presence of different forms of alleles in genes (polymorphisms) may promote the development of cancers; in this regard, the P53 gene has been a research focus due to its role as a major tumor suppressor gene (8,9). The P53 gene is located on the short arm of chromosome 17 and includes 11 exons (10). The P53 codon 72 (Arg72Pro) polymorphism, located in exon 4, has been investigated in numerous types of cancer (11). Substitution of a guanine base for cytosine in this codon leads to the replacement of an arginine amino acid to proline, which influences the activity of the resulting protein (12). The proline variant is effective in the repair of DNA damage, while the arginine variant leads to a strong induction of apoptosis (13). To date, studies have been performed on the P53 codon 72 polymorphism in breast, colorectal, skin and stomach cancers; however, no comprehensive result has been obtained (13–16). Considering the controversial results regarding the role of P53 gene polymorphism in gastric cancer, dependence of variants on geographical conditions, racial differences and genetic differences probably exists in different communities. This is indicated when considering the increase in incidence of stomach cancer and associated mortalities in Iran, particularly in northern parts of the country (17–20). Therefore, the aim of the present study was to determine the association of P53 gene polymorphism with gastric cancer in Northern Iran as a high-risk region.
Materials and methods
Study population
This was a case-controlled study intending to determine the association between P53 gene polymorphism and gastric cancer in patients referred to the Tuba Clinic (Academic Referral Center for the Mazandaran Province) from October 2016 to April 2017 in Sari, Iran, compared with non-cancer patients. The patients with gastric cancer (n=59) were diagnosed by oncologists and confirmed by pathological examination, while the non-cancer or control subjects (n=59) were healthy patients referred to the Tuba Clinic for routine laboratory tests. The exclusion criteria were the following: Presence of a tumor in a site other than the stomach and a non-adenocarcinoma type of gastric cancer in the case group, and a history of cancer and/or pregnancy in the control group. Study and control subjects were compared regarding age and gender, and study subjects were compared regarding age (≤55 vs. >55 years), gender and tumor differentiation (well vs. moderate vs. poor), stage (I/II vs. III/IV) and location (proximal vs. distal) (21). All data for the study population, including age, gender, clinical and laboratory diagnosis, were collected based on related checklists of the above variables. The study received approval from the Ethics Committee of Mazandaran University of Medical Sciences (Sari, Iran) and written informed consent was obtained from all subjects following full disclosure of the study objectives and procedures.
DNA extraction
Samples of 5–10 ml fasting venous blood were obtained from the subjects and transferred to two tubes: A serum-separating tube and a tube containing the anticoagulant EDTA. Subsequently, the samples were transferred to 15 ml Falcon tubes and brought to a total volume of 15 ml with lysis buffer I (DynaBio™ Blood/Tissue DNA Extraction Mini kit, cat. no. KI0015; Unilabs, Geneva, Switzerland). The tubes were agitated, incubated for 5 min at room temperature and centrifuged (4,000 × g for 10 min at 4°C). The upper layer of the solution was removed, the tubes were re-filled with 15 ml of buffer I and the steps listed above were repeated three times. A total of 2 ml buffer II was added to the tubes, which were then incubated for 30 min in a 45°C water bath. During incubation, the tubes were agitated periodically to ensure that the sediment dissolved. A total of 0.5 ml 5 M sodium perchlorate was added to the tubes and allowed to combine for 2–3 min. Subsequently, 2 ml cold chloroform was added to each tube under a hood and the tubes were centrifuged at 4,000 × g for 5–7 min at 4°C. A total of 3 ml cold ethanol (99% v/v) were added to the upper layer of the solution containing the DNA, and precipitated DNA was observed and allowed to settle. Finally, DNA was removed from the solution using a Pastor pipet and dried in the open air to allow the ethanol to evaporate. DNA was eluted in 100–200 ml sterilized distilled water.
Amplification of the P53 region
The region of the P53 gene containing the codon 72 (Arg72Pro) polymorphism on exon 4 was amplified by polymerase chain reaction (PCR) using the following specific primers: Forward, 5′-TTGCCGTCCCAAGCAATGGATGA-3′ and reverse, 5′-TCTGGGAAGGGACAGAAGATGAC-3′. The PCR reaction mixture contained 10 pmol of each primer, 200 ng genomic DNA, 1 U Taq DNA polymerase (Denazist Asia Co., Mashhad, Iran), 1.5 µmol MgCl2, 200 mM of each dNTP and nuclease-free water to a final volume of 25 ml. The reaction mixtures were preincubated for 10 min at 94°C. The PCR conditions were 94°C for 30 sec and 55°C for 1 min, followed by 72°C for 1 min for 40 rounds. After confirmation of an amplified fragment of the expected size (199 bp) (20) on a 1.5% agarose gel with ethidium bromide staining, the PCR products were digested with 0.1 µl (10 U/µl) BstUI restriction enzyme (Fermentas, Vilnius, Lithuania) at 60°C for 16 h. The DNA fragments were electrophoresed through a 2% agarose gel containing gel stain (DNA Green Viewer™; Parstous Biotechnology, Mashhad, Iran). The Pro allele is not cleaved by BstUI at codon 72 and has a single band with length of 199 bp. The Arg allele is cleaved by BstUI and digested products are separated as two fragments of 113 and 86 bp in length. The heterozygote genotype has 3 bands of 199, 113 and 86 bp in length (22).
Statistical analysis
Based on the results of a recent study (23) which estimated the frequencies of the proportions of a polymorphism in case and control groups (P1 and P2, respectively) with 95% confidence (α=0.05) and 80% test power (β=0.20), the sample size of the present study for the gastric cancer and non-cancer groups was at least 100 subjects (gastric cancer, n=59 and non-cancer, n=59). Continuous variables were expressed as the mean ± standard deviation, and categorical values were expressed as frequencies. Statistical analyses by unpaired Student's t-test and one-way analysis of variance were performed using SPSS software version 19 (IBM Corp., Armonk, NY, USA). The correlation of gastric cancer with P53 gene codon 72 polymorphism was assessed by logistic regression analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
In the present study, 59 patients with gastric cancer and 59 healthy controls from Northern Iran were assessed for P53 codon 72 genotype. The demographic characteristics of all subjects are listed in Table I. The sex distribution was not significantly different between the two groups (P=0.056); however, the mean age of patients with cancer was higher than that of the controls (P<0.001).
Table I.
Demographic characteristics of subjects in the patient and control groups.
| Group | |||
|---|---|---|---|
| Variable | Gastric cancer | Control | P-value |
| Sex, n (%) | |||
| Male | 36 (61.0) | 45 (76.3) | 0.056 |
| Female | 23 (39.0) | 14 (23.7) | |
| Age, mean ± SD | 62.9±11.8 | 29.8±13.2 | <0.001 |
SD, standard deviation.
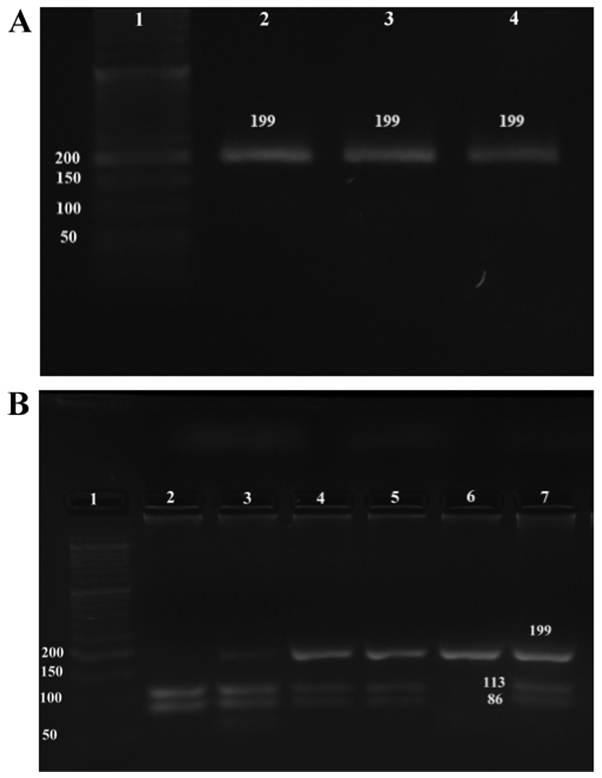
PCR-RFLP products of P53 codon 72 variants
Band visualization confirmed restriction digestion of the P53 gene. The electrophoretic pattern of the P53 gene segments were obtained following BstUI digestion and 2% agarose separation (Fig. 1). Electrophoresis of the undigested amplified P53 product detected the expected 199 bp fragment (Fig. 1A). In the presence of proline, the P53 allele remains unchanged on cleavage of the PCR product with 10 U/µl BstUI, and the allele is detected as undigested product. Thus, the homozygote genotype (Pro/Pro) yielded a single 199 bp band (Fig. 1B, lane 6). By contrast, cleavage of the arginine homozygote genotype (Arg/Arg) generated two bands at 113 and 86 bp (Fig. 1B, lane 2). The Arg/Pro heterozygote genotype yielded all three fragments (Fig. 1B, lanes 3–5 and 7).
Figure 1.
Representative results of polymerase chain reaction-restriction fragment length polymorphism analysis of P53 codon 72 polymorphism. (A) Lanes 2–4: Undigested amplified P53 product was detected as a single band at 199 bp (lane 1, 50-bp ladder). (B) Undigested proline homozygote genotype (Pro/Pro) also yielded a single 199 bp band (lane 6); homozygote arginine genotype (Arg/Arg) was cleaved into two fragments of 113 and 86 bp (lane 2); Arg/Pro heterozygote genotype yielded all three fragments (lanes 3–5 and 7). Lane 1, 50-bp ladder.
Distribution of P53 codon 72 polymorphism variants
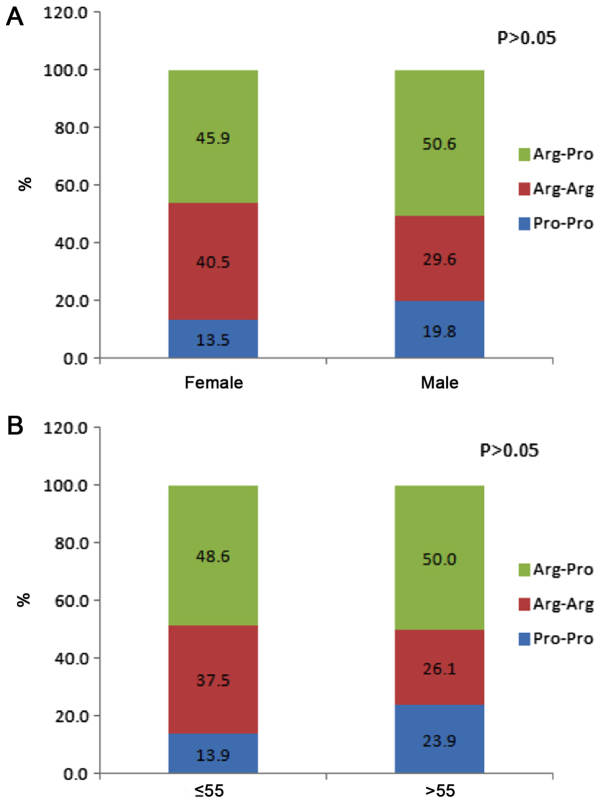
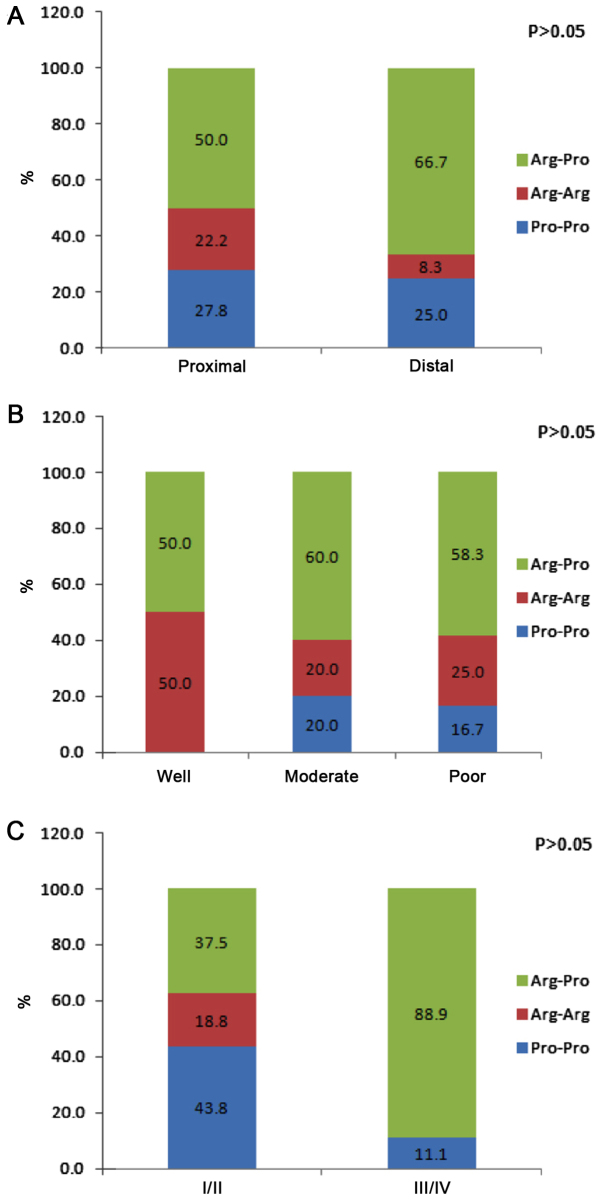
The genotype frequencies of the P53 codon 72 polymorphism in Iranian gastric cancer cases and controls are summarized in Table II. Genotype frequencies did not differ significantly between the patients and controls (P=0.4); the frequencies of the three genotypes Arg/Arg, Arg/Pro and Pro/Pro in gastric cancer patients were 28.8, 49.2 and 22.0%, and in controls were 37.3, 49.2 and 13.6%. When the patients with gastric cancer were classified by sex and age group (≤55 or >55 years), there were no significant differences in the genotype distributions between males and females or patient age groups (Fig. 2). Fig. 3 presents genotype frequency based on tumor location, histological differentiation and tumor stage. The location of the tumor in 60% of patients was proximal and in 40% was distal. Histological differentiation of the tumor samples was defined as well, moderate and poor in 8.3, 41.7 and 50.0% of the gastric cancer cases, respectively. There were no significant differences in genotype frequencies based on tumor location, histological differentiation or tumor stage.
Table II.
Frequency of P53 codon 72 polymorphism genotypes in the patient and control groups.
| Group frequency, n (%) | |||
|---|---|---|---|
| Genotype | Gastric cancer | Control | P-value |
| Proline-Proline | 13 (22.0) | 8 (13.6) | |
| Arginine-Arginine | 17 (28.8) | 22 (37.3) | 0.4 |
| Arginine-Proline | 29 (49.2) | 29 (49.2) | |
| Total | 59 (100.0) | 59 (100.0) | |
Figure 2.
Distribution of P53 codon 72 polymorphism genotypes according to (A) sex and (B) age group.
Figure 3.
Distribution of P53 codon 72 polymorphism genotypes according to the (A) location, (B) differentiation and (C) stage of tumors.
Discussion
In the present study, the genotype frequencies of a P53 polymorphism in Iranian patients with gastric cancer did not differ significantly to those in healthy controls; the frequencies of the three genotypes, Pro/Pro, Arg/Arg and Arg/Pro were 22, 28.8 and 49.2% in the case group and 13.6, 37.3 and 49.2% in the control group. Similar to the present study, Chung et al (24) reported that there was no specific genotype of P53 polymorphism in a gastric cancer cohort compared with other groups with or without H. pylori-associated chronic gastritis in a Korean population, though another study by Liu et al identified a significant difference between P53 expression in primary tumor and non-tumor tissue in gastric cancer patients (25). In the gastric cancer group in the current study, the male to female ratio was 1.56, and there was no significant difference in sex distribution between the case and control groups. However, the mean age of patients in the case group was significantly (P<0.0001) higher compared with that of the control group. Furthermore, the location of the tumor in 60% of patients was proximal (cardia, body, fundus and curve of the stomach) and in 40% was distal (antrum of the stomach). Histological differentiation of the tumor samples was well, moderate and poor in 8.3, 41.7 and 50% of gastric cancer patients. These results are in agreement with the study by Chung et al (24). Additionally, 64% percent of the gastric cancer cases were stage I/II and 36% were stage III/IV. However, the study failed to identify significant differences in P53 polymorphism variants between different tumor locations, histological differentiations or tumor stages in Iranian gastric cancer cases.
Zhou et al (26) analyzed the association between P53 codon 72 polymorphism and gastric cancer among a Chinese population. High frequencies of Pro/Pro in cardia gastric cancer patients and Arg/Arg in advanced gastric cancer patients suggested that this polymorphism was associated with the location and stage of gastric cancer. Shen et al (27) identified that the P53 Arg allele in homozygote and heterozygote genotypes was associated with increased risk of gastric cancer. Furthermore, a meta-analysis including 1,665 gastric cancer patients and 2,358 controls revealed high frequencies of the Arg/Arg allele in advanced gastric cancer patients, while the Pro/Pro allele was significantly higher in patients with cardia gastric cancer compared with healthy controls. They came to the conclusion that P53 codon 72 polymorphism is likely associated with gastric cancer among Asian populations (28). The same results were reported in a meta-analysis by Liu et al (29); they also demonstrated an association between polymorphism in P53 at codon 72 and gastric cancer among Asian populations.
The association of polymorphism at codon 72 of P53 is not restricted to gastric cancer, and has been discussed in various types of carcinoma. Buyru et al (9) reported that the Arg/Arg genotype was notably correlated with breast cancer. In contrast, a Chinese population contradicted any possible association of P53 codon 72 Pro/Arg polymorphism with ovarian cancer (30). Tang et al (31) conducted a meta-analysis on the role of P53 codon 72 polymorphism in colorectal cancer. They determined no association between the Pro/Arg variant and the risk of colorectal cancer in their study population. Meanwhile, a different systematic analysis identified high frequencies of the Pro/Pro allele in colorectal cancer patients (32). The risk of oral cancer with P53 codon 72 polymorphism was investigated in a study by Jing et al (33). They reported that the Arg genotype was associated with a reduced risk of oral cancer, and a high frequency of the Pro/Pro allele in oral cancer patients. Furthermore, the Arg/Arg genotype and reduced cancer risk has also been reported (34). A study performed in a Japanese population revealed that the Pro/Pro genotype at codon 72 was associated with increased risk of prostate cancer and its progression (35). Two separate meta-analyses by Jia et al (36) and Lao et al (37) suggested that the homozygote and heterozygote genotypes of Pro participated in the development of endometriosis in Asian and Caucasian populations. Analysis of this variant among an Iranian population revealed that the Arg genotype increased the risk of breast cancer while Pro served as a protective factor (38).
As study limitations, the two population groups were not matched based on age or gender, and there was no evaluation of H. pylori infections, lifestyle or dietary regime. Other limitations included the small sample size and sampling of individuals of the same geographical region and race. Therefore, to confirm the results, further studies considering different geographical locations and races and a larger number of participants are necessary. Nonetheless, the present findings failed to indicate an association between P53 codon 72 polymorphism and gastric cancer risk.
In conclusion, the present study identified no significant association between Arg72Pro at codon 72 of P53 and gastric cancer risk in North Iranian patients. Additionally, there were no differences in genotype frequencies based on tumor location, histological differentiation or tumor stage.
Acknowledgements
Not applicable.
Funding
The current study was supported by Mazandaran University of Medical Sciences (grant no. MAZUMS.2649).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AH and RA were responsible for study conception and design. GJ, VO, YH, OA and MT were responsible for acquisition of data. RA performed analysis of the data. AH and RA were responsible for drafting of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Mazandaran University of Medical Sciences, Sari, Iran (approval no. IR.MAZUMS.IMAMHOSPITAL.REC.1396.2649). Written informed consent was obtained from all subjects following full disclosure of the study objectives and procedures.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang C, Zhang J, Cai M, Zhu Z, Gu W, Yu Y, Zhang X. DBGC: A Database of Human Gastric Cancer. PLoS One. 2015;10:e0142591. doi: 10.1371/journal.pone.0142591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 3.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–628. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eybpoosh S, Talebkhan Y, Saberi S, Esmaeili M, Oghalaie A, Ebrahimzadeh F, Karimi T, Abdirad A, Nahvijou A, Mohagheghi MA, et al. Mohammadi M. Age-specific gastric cancer risk indicated by the combination of Helicobacter pylori sero-status and serum pepsinogen levels. Iran Biomed J. 2015;19:133–142. doi: 10.7508/ibj.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zali H, Rezaei-Tavirani M, Azodi M. Gastric cancer: Prevention, risk factors and treatment. Gastroenterol Hepatol Bed Bench. 2011;4:175–185. [PMC free article] [PubMed] [Google Scholar]
- 6.Yasui W, Sentani K, Sakamoto N, Anami K, Naito Y, Oue N. Molecular pathology of gastric cancer: Research and practice. Pathol Res Pract. 2011;207:608–612. doi: 10.1016/j.prp.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Arjmand Kolukhi Z, Salehi Z, Mashayekhi F, Najafi B, Mirpoor S. Analysis of Glu298Asp eNOS Gene polymorphism in patients with Gastric Cancer in the Guilan population. Analysis. 2014;17:53–62. [Google Scholar]
- 8.Hossein Pour Feizi MA, Ravanbakhsh Gavgani R, Pourahmad R, Pouladi N, Azarfam P, Montazeri V. Association of p53 Arg/Pro polymorphism at codon 72 with risk of breast cancer in east azerbaijani women. J Babol Univ Med Sci. 2012;14:31–38. [Google Scholar]
- 9.Buyru N, Tigli H, Dalay N. P53 codon 72 polymorphism in breast cancer. Oncol Rep. 2003;10:711–714. [PubMed] [Google Scholar]
- 10.Gloria-Bottini F, Banci M, Saccucci P, Neri A, Bottini E, Magrini A. p53 codon 72 polymorphism and coronary artery disease: Evidence of interaction with ACP1. Med Sci Monit. 2012;18:CR712–CR715. doi: 10.12659/MSM.883597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin HY, Huang CH, Wu WJ, Chang LC, Lung FW. TP53 codon 72 gene polymorphism paradox in associated with various carcinoma incidences, invasiveness and chemotherapy responses. Int J Biomed Sci. 2008;4:248–254. [PMC free article] [PubMed] [Google Scholar]
- 12.Kalemi TG, Lambropoulos AF, Gueorguiev M, Chrisafi S, Papazisis KT, Kotsis A. The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett. 2005;222:57–65. doi: 10.1016/j.canlet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 13.Weng Y, Lu L, Yuan G, Guo J, Zhang Z, Xie X, Chen G, Zhang J. p53 codon 72 polymorphism and hematological cancer risk: An update meta-analysis. PLoS One. 2012;7:e45820. doi: 10.1371/journal.pone.0045820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu KD, Di GH, Yuan WT, Fan L, Wu J, Hu Z, Shen ZZ, Zheng Y, Huang W, Shao ZM. Functional polymorphisms, altered gene expression and genetic association link NRH:quinone oxidoreductase 2 to breast cancer with wild-type p53. Hum Mol Genet. 2009;18:2502–2517. doi: 10.1093/hmg/ddp171. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zhang X, Han C, Wan G, Huang X, Ivan C, Jiang D, Rodriguez-Aguayo C, Lopez-Berestein G, Rao PH, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, et al. Swedish Low-risk Colorectal Cancer Study Group A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khatami F, Noorinayer B, Ghiasi S, Mohebi R, Hashemi M, Zali MR. Single nucleotide polymorphisms of DNA methyltransferase 1 gene and gastric cancer in iranian patients: a case control study. Asian Pac J Cancer Prev. 2009;10:1177–1182. [PubMed] [Google Scholar]
- 18.Amoori N, Mahdavi S, Enayatrad M. Epidemiology and trend of stomach cancer mortality in Iran. Int J Epidemiol Res. 2016;3:268–275. [Google Scholar]
- 19.Hedayatizadeh-Omran A, Rafiei A, Alizadeh-Navaei R, Tehrani M, Valadan R, Moradzadeh K, Panbechi M, Taghavi SM. Role of HER2 in brain metastasis of breast cancer: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16:1431–1434. doi: 10.7314/APJCP.2015.16.4.1431. [DOI] [PubMed] [Google Scholar]
- 20.Hedayatizadeh-Omran A, Rafiei A, Khajavi R, Alizadeh-Navaei R, Mokhberi V, Moradzadeh K. Association between ghrelin gene (Leu72Met) polymorphism and ghrelin serum level with coronary artery diseases. DNA Cell Biol. 2014;33:95–101. doi: 10.1089/dna.2013.2218. [DOI] [PubMed] [Google Scholar]
- 21.Uchino S, Noguchi M, Ochiai A, Saito T, Kobayashi M, Hirohashi S. p53 mutation in gastric cancer: A genetic model for carcinogenesis is common to gastric and colorectal cancer. Int J Cancer. 1993;54:759–764. doi: 10.1002/ijc.2910540509. [DOI] [PubMed] [Google Scholar]
- 22.Matei MC, Negură L, Liliac L, Negură A, Azoicăi D. Validation of PCR-RFLP techniques for the evaluation of codon 72 of p53 and CYP1A1 gene's polymorphisms in relation with ovarian cancer in a Romanian population. Rom J Morphol Embryol. 2012;53:47–54. [PubMed] [Google Scholar]
- 23.Pandey R, Misra V, Misra SP, Dwivedi M, Misra A. Helicobacter pylori infection and a P53 codon 72 single nucleotide polymorphism: A reason for an unexplained Asian enigma. Asian Pac J Cancer Prev. 2014;15:9171–9176. doi: 10.7314/APJCP.2014.15.21.9171. [DOI] [PubMed] [Google Scholar]
- 24.Chung WC, Lee KM, Lee BI, Chun JS, Lee SY, Chang UI, Park SH, Yang JM, Choi KY, Chung IS. P53 genetic polymorphism of gastric cancer in Korea. Korean J Intern Med. 2006;21:28–32. doi: 10.3904/kjim.2006.21.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Wang S, Xia X, Chen Y, Zhou Y, Wu X, Zhang J, He S, Tan Y, Qiang F, et al. Synergistic role between p53 and JWA: Prognostic and predictive biomarkers in gastric cancer. PLoS One. 2012;7:e52348. doi: 10.1371/journal.pone.0052348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Li N, Zhuang W, Wu X. p53 Codon 72 polymorphism and gastric cancer risk in a Chinese Han population. Genet Test Mol Biomarkers. 2010;14:829–833. doi: 10.1089/gtmb.2010.0115. [DOI] [PubMed] [Google Scholar]
- 27.Shen H, Solari A, Wang X, Zhang Z, Xu Y, Wang L, Hu X, Guo J, Wei Q. P53 codon 72 polymorphism and risk of gastric cancer in a Chinese population. Oncol Rep. 2004;11:1115–1120. [PubMed] [Google Scholar]
- 28.Zhou Y, Li N, Zhuang W, Liu GJ, Wu TX, Yao X, Du L, Wei ML, Wu XT. P53 codon 72 polymorphism and gastric cancer: A meta-analysis of the literature. Int J Cancer. 2007;121:1481–1486. doi: 10.1002/ijc.22833. [DOI] [PubMed] [Google Scholar]
- 29.Liu KJ, Qi HZ, Yao HL, Lei SL, Lei ZD, Li TG, Zhao H. An updated meta-analysis of the p53 codon 72 polymorphism and gastric cancer risk. Mol Biol Rep. 2012;39:8265–8275. doi: 10.1007/s11033-012-1674-0. [DOI] [PubMed] [Google Scholar]
- 30.Kang S, Duan LH, Zhang JH, Guo W, Wang N, Li Y. Association of p53 gene polymorphism with susceptibility to ovarian cancer. Zhonghua Fu Chan Ke Za Zhi. 2004;39:754–758. (In Chinese) [PubMed] [Google Scholar]
- 31.Tang NP, Wu YM, Wang B, Ma J. Systematic review and meta-analysis of the association between P53 codon 72 polymorphism and colorectal cancer. Eur J Surg Oncol. 2010;36:431–438. doi: 10.1016/j.ejso.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Qin H, Zhang Y, Shi T, Liu B, Sun Y, Ma Y. P53 codon 72 polymorphism and colorectal cancer: A meta-analysis of epidemiological studies. Hepatogastroenterology. 2011;58:1926–1929. doi: 10.5754/hge10215. [DOI] [PubMed] [Google Scholar]
- 33.Jing G, Lv K, Jiao X. The p53 codon 72 polymorphism and the risk of oral cancer in a Chinese Han population. Genet Test Mol Biomarkers. 2012;16:1149–1152. doi: 10.1089/gtmb.2012.0138. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Chen W, Zhang W, Jiang Q, Liu C, Lin Y, Hu Z, Yu S, Xu G. Genetic polymorphisms of p53 codon 72 and bladder cancer susceptibility: A hospital-based case-control study. Genet Test Mol Biomarkers. 2011;15:337–341. doi: 10.1089/gtmb.2010.0230. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki K, Matsui H, Ohtake N, Nakata S, Takei T, Nakazato H, Okugi H, Koike H, Ono Y, Ito K, et al. A p53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci. 2003;10:430–435. doi: 10.1007/BF02256434. [DOI] [PubMed] [Google Scholar]
- 36.Jia S, Xu L, Chan Y, Wu X, Yang S, Yu H, Yang H, Luo Y, Tang W. p53 codon 72 polymorphism and endometriosis: A meta-analysis. Arch Gynecol Obstet. 2012;285:1657–1661. doi: 10.1007/s00404-012-2226-5. [DOI] [PubMed] [Google Scholar]
- 37.Lao X, Chen Z, Qin A. p53 Arg72Pro polymorphism confers the susceptibility to endometriosis among Asian and Caucasian populations. Arch Gynecol Obstet. 2016;293:1023–1031. doi: 10.1007/s00404-015-3923-7. [DOI] [PubMed] [Google Scholar]
- 38.Soleimani A, Rahmani Y, Farshchian N, Delpisheh A, Khassi K, Shahmohammadi A, Amirifard N. The evaluation of p53 polymorphism at codon 72 and association with breast cancer in Iran: A systematic review and meta-analysis. J Cancer Prev. 2016;21:1–293. doi: 10.15430/JCP.2016.21.4.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.