Abstract
A genome sequence is worthless if it cannot be deciphered, therefore efforts to describe — or ‘annotate’ — genes began as soon as DNA sequences became available. Whereas early work focused on individual protein-coding genes, the modern genomic ocean is a complex maelstrom of alternative splicing, non-coding transcription and pseudogenes. Scientists — from clinicians to evolutionary biologists — need to navigate these waters, and this has led to the design of high-throughput, computationally driven annotation projects. The catalogues produced are key resources for genome exploration, especially as they become integrated with expression, epigenomic and variation datasets. Their creation, however, remains challenging.
Introduction
The core output of a gene annotation project could be described as an in silico transcriptome: a collection of ‘models’ referred to here as a genebuild. However, genebuilds are found in a data server, not in the cell; they are only representations of the transcriptome that exists in nature. This fact has profound implications for the study of biology: gene annotation is a key mechanism through which information is leveraged from genome sequences, and deficiencies in genebuilds will be propagated into downstream analyses. How close then are our genebuilds to actual transcriptomes? Every publication seems to describe an entity that is larger, more dynamic and functionally diverse than previously thought — as illustrated in Figure 1 — and this picture becomes even more complicated when considering the genomic sequences that regulate genes. In fact, the sheer complexity of the transcriptome may cause one to ask whether it is even possible that it could ever be completely described in silico. However, we are begging the question of whether we need to fully capture this complexity. A key annotation question concerns the portion of the transcriptome that contributes to cellular function, and it could be argued that the goal of annotation projects should be to describe only this ‘functional transcriptome’; to extract the signal from the noise.
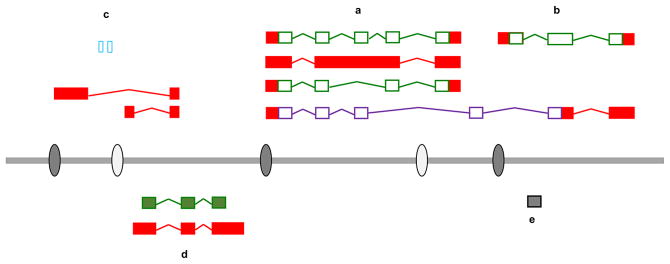
Figure 1. A modern view of the genomic landscape.
This hypothetical diagram illustrates the major types of genes and transcripts found in eukaryotic genomes. Two protein-coding genes are illustrated, (a) and (b). Coding sequences (CDS) are shown as open green boxes, untranslated regions as filled red boxes. Whereas locus (b) appears to generate a single CDS transcript, locus (a) generates two distinct protein isoforms through the differential incorporation of a central exon. Locus (a) also has a retained intron associated, while an additional ‘read-through’ transcript incorporates exons from (a) and (b). This transcript is subjected to NMD (unfilled lilac boxes). Gene (c) is a long non-coding RNA (lncRNA) with two transcripts (red boxes), although three small RNAs are also transcribed from within one of its introns (open blue boxes). Loci (d) and (e) are unprocessed (filled green boxes) and processed (grey box) pseudogenes respectively. Locus (d) is transcribed. A series of promoter regions (filled grey ovals) and enhancer regions (open ovals) are indicated. Promoters are associated with transcription start sites (TSSs) for the various loci, whereas enhancers are found some distance from the gene or genes they regulate.
Here, we review the current state of play in higher eukaryotic gene annotation, and attempt to take genebuilds out of the ‘black box’ for the benefit of annotation users. Firstly, we explain the key principles by which these resources are made, and why annotation projects are proceeding along alternative lines for different genomes. Inevitably, more work has been spent on human than any other genome, and many aspects of genome annotation are most effectively explained in this context. However, while human workflows are frequently reused in the description of other genomes, such projects are not truly analogous. This is because their scientific goals are often substantially different — typically more limited in scope — but also because the resources available to support annotation have changed dramatically in recent years. Secondly, even human genebuilds have ‘blind-spots’, and we wish to help users appreciate the biological information that is missing in such resources and how this can affect their work. These largely reflect biological questions that remain unanswered, in particular regarding the issue of transcript functionality. Nonetheless, our biological understanding of the transcriptome is developing rapidly, and leaps in understanding are also being made in neighbouring fields of molecular biology including proteomics, gene regulation and epigenomics. We shall explain how gene annotation projects are coordinating efforts to combine such datasets into fully integrated views of genomic organization. Even so, it is clear that increasing genbeuild complexity presents a considerable practical challenge to scientists, and we end by discussing the problems faced by annotation projects in improving their usability.
What is gene annotation?
Annotation targets
Figure 2 summarizes the core principles of gene annotation workflows. Although numerous strategies have been used to describe different genomes and gene features, each ultimately represents the unification of two processes. Firstly, annotation defines the structure of a transcript — e.g. its exon–intron architecture — and secondly it provides inferences into its potential function, for example whether it is protein-coding. We refer to this second aspect as ‘functional annotation’. However, it is vital to appreciate that ‘gene’ and ‘transcript’ are not equivalent terms in annotation. This is illustrated by the fact that most genes generate multiple, distinct RNAs, for example through alternative splicing (AS)1. Transcripts are the major target of annotation projects; we regard ‘gene annotation’ as a process that creates ‘transcript models’. As we shall see, our modern understanding of transcriptional complexity within genes is driving the evolution of annotation strategies, as is the knowledge that eukaryotic genomes contain not only protein-coding genes, but also pseudogenes and long non-coding RNAs (lncRNAs), as well as small RNA families including transfer RNAs, PIWI-interacting RNAs (piRNAs) and small nucleolar RNAs (snoRNAs)2. They may even contain RNA categories that remain to be discovered. In short, this complexity presents a substantial challenge to annotation projects (Fig. 1).
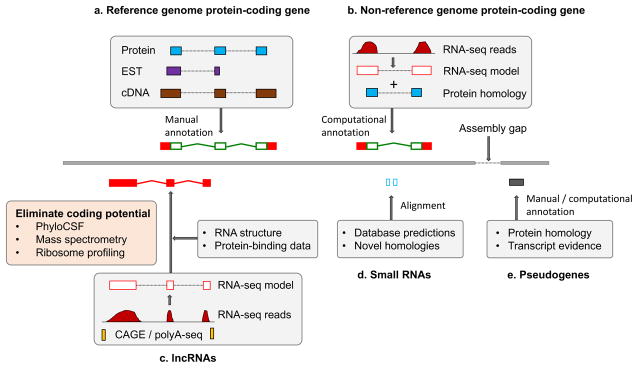
Figure 2. The core annotation workflows for different gene types.
These workflows illustrate general annotation principles, not the specific pipelines of any particular genebuild. a) Protein-coding genes within reference genomes were largely annotated based on the computational genomic alignment of Sanger-sequenced transcripts and protein-coding sequences, followed by manual annotation via interface tools such as Zmap1, WebApollo26, Artemis126 or the Integrative Genomics Viewer127. Transcripts were typically taken from GenBank128, proteins from Swiss-Prot12. b) Protein-coding genes within non-reference genomes are usually annotated based on fewer resources; here, RNA sequencing (RNA-seq) data are used in combination with protein homology information extrapolated from a closely-related genome. RNA-seq pipelines for read alignment include STAR129 and TopHat130, whereas model creation is commonly performed by Cufflinks22. c) Long non-coding RNA (lncRNA) structures can be annotated in a similar manner to protein-coding transcripts as for (a) and (b), although coding potential must be ruled out. This is typically done by examining sequence conservation with phyloCSF131 or using experimental datasets such as mass spectrometry or ribosome profiling. Here, 5’ Cap Analysis of Gene Expression (CAGE)45 and polyA-seq data46 are also incorporated to obtain true transcript endpoints. Designated lncRNA pipelines include PLAR48. d) Small RNAs are typically added to genebuilds by mining repositories such as RFAM132 or miRBase133. However, these entries can be used to search for additional loci based on homology. e) Pseudogene annotation is based on identification of loci with protein-homology to either paralogous or orthologous protein-coding genes. Computational annotation pipelines include PseudoPipe52, although manual annotation is more accurate53. Finally, all annotation methods can be thwarted by the existence of sequence gaps in the genome assembly (right-angled arrow).
Annotation strategies
Numerous factors come into play when choosing an annotation strategy for a genome. (Figs. 2, 3). Obviously, financial considerations can place major constraints on the availability of human resources and computational power, as well as in the generation of experimental data to provide ‘evidence’ for model construction (Box 1). However, the strategy also depends on what it is hoped to achieve. For human, genebuilds support scientific enquiries across a broad range of disciplines, and annotation resources are required to be as comprehensive as possible. The same is true for the projects of ’classic’ laboratory species such as Mus musculus, Arabidopsis thaliana, Caenorhabditis elegans and Drosophila melanogaster. Other genomes may be sequenced to ask more specific scientific questions. For example, a common goal of sequencing projects within evolutionary biology is to find genes subjected to positive selection. In this scenario, a high premium is placed on the identification of protein-coding sequences; features such as pseudogenes and small RNAs may even be completely ignored. Meanwhile, the Functional Annotation of Animal Genomes (FAANG) consortium plans to sequence and annotate livestock genomes in order to further our understanding of quantitative phenotypes3.
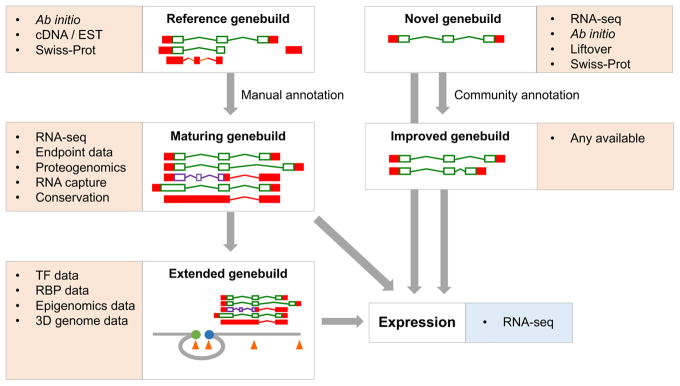
Figure 3. High-level strategies for gene annotation projects.
This schematic details the annotation pathways for reference and novel genomes. Coding sequences (CDS) are outlined in green, nonsense-mediated decay (NMD) is shown in purple and untranslated regions (UTRs) are filled in red. The core evidence sets used at each stage are listed, although their availability and incorporation vary across different projects. The types of evidence used for reference genebuilds have evolved over time: RNA sequencing (RNA-seq) has replaced Sanger sequencing, conservation-based methodologies have become more powerful and proteogenomic datasets are now available. By contrast, novel genebuilds are constructed based on RNA-seq and/or ab initio modelling, in combination with the projection of annotation from other species (known as liftover) and the usage of other species evidence sets. In fact, certain novel genebuilds such as pig and rat now incorporate a modest amount of manual annotation, and could perhaps be described as ‘intermediate’ in status between ‘novel’ and ‘reference’. Furthermore, such genebuilds have also been improved by community annotation; this process typically follows the manual annotation workflows for reference genomes, although at a smaller scale. While all reference genebuilds are ‘mature’ in our view, progress into the ‘extended genebuild’ phase is most advanced for human. A promoter is indicated by the blue circle, an enhancer is indicated by the orange circle, and binding sites for transcription factors (TFs) or RNA-binding proteins (RBPs) are shown as orange triangles. Gene expression can be analyzed on any genebuild regardless of quality, although it is more effective when applied to accurate transcript catalogues. Clearly, the results of expression analyses have the potential to reciprocally improve the efficacy of genebuilds, although it remains to be seen how this will be achieved in practice (indicated by ‘?’).
Box 1. A description of gene annotation experimental datasets.
Transcript sequencing
Sanger sequenced transcripts
RNAs obtained by traditional chain-termination methodologies. These are either cDNAs of approximately 1,000–2,000bp in size (depending in part on the mRNA size) or expressed sequence tags (ESTs), which are single cDNA sequencing reads of approximately 500bp.
Short-read RNA-seq
Whole-transcriptome shotgun-sequenced RNAs obtained as enormous libraries, typically on the Illumina platform11. The read-length depends on the protocol used, although the most common datasets available are under 200bp. Reads are commonly generated as ‘paired ends’, where sequence is obtained from both ends of an RNA.
Long-read RNA-seq
The next wave of RNA-seq methods, generating longer sequences although at lower throughput. The Roche 454 platform provides reads of up to 1000bp, while the Iso-Seq methodology from Pacific Biosciences can capture whole RNAs.
Cap Analysis of Gene Expression (CAGE)
Produces enormous ~27bp fragment libraries extracted from the 5’ capped end of whole transcriptome RNA molecules when coupled to next-generation sequencing platforms121.
RNA Annotation and Mapping of Promoters for Analysis of Gene Expression (RAMPAGE)
122. Similar to CAGE, although provides longer paired-end reads as opposed to short sequence tags, with the size dependent on the short-read RNA-seq platform used.
PolyA-seq
Captures RNA sequence immediate upstream of the polyA tail. The protocol reported by Derti et al. generates amplicons of 200–500bp, although the size of the tags obtained will depend on the sequencing strategy used46.
CaptureSeq
Uses strategically designed oligonucleotide probes to pull-down target RNA from a sample. The captured RNAs can be sequenced using any common platform43.
Functionality
Mass spectrometry (MS)
Most commonly applied through the combination of liquid chromatography and tandem MS/MS, which produces large numbers of peptide spectral graphs based on their mass-to-charge ratio. Spectra are typically interpreted by comparison against a set of theoretical peptides extrapolated from an in silico CDS database74.
Ribosome profiling (RP)
Identifies regions of transcripts that are undergoing translation. Cellular RNA is chemically degraded, allowing for RNA fragments that are ‘protected’ by ribosome binding to be recovered for high-throughput sequencing 80,81. Also known as ‘Ribo-seq’.
UV cross-linking immunoprecipitation followed by sequencing (CLIP-seq)
Ribosome-binding proteins are bound to their target RNAs, which are recovered and subjected to high-throughput sequencing97. It has the resolution to reveal binding sites within the RNA.
The extended gene
Hi-C
A massively high-throughput version of chromosome conformation capture methodologies. DNA is crosslinked across the sites of chromosome loops using chemical treatment, and these linkage sites are recovered and sequenced on next-generation sequencing platforms103.
[b2] Chromatin Interaction Analysis Paired-end Tag Sequencing (ChIA-PET)
An adapted form of Hi-C that enriches for specific DNA–protein complexes using ChIP-seq. It can thus be used to investigate the role of specific proteins in chromosome looping123.
[b2] Chromatin immunoprecipitation followed by sequencing (ChIP-seq)
A method for analyzing DNA–protein interactions in a cell124. It produces libraries of target DNA sites that are bound to a protein of interest, which are then mapped back to the genome to identify protein-binding regions.
Of course, it is broadly true that the more valuable a genome is to the scientific community, the more resources have been committed to its annotation. Thus, human1, 4, mouse5, 6, A. thaliana7, C. elegans8 and D. melanogaster9 have each been subjected to large-scale annotation projects over many years, involving numerous scientific institutes and sequencing centres (Table 1). In fact, the human and mouse genomes even have overlapping annotation resources that are independently produced, such as the genebuilds created by the RefSeq4, 5 and GENCODE1, 6 projects. Finally, we note that genome quality is an important factor when strategizing. One cannot create high-quality genebuilds on poor-quality genomes, and even modest genome assembly improvements can be massively beneficial to annotation projects, as demonstrated for honey bee10. Indeed, annotation and sequencing have been carried out for human and model organisms genomes in a reciprocal manner, and we refer to them as ‘reference’ genomes and genebuilds (Fig. 2, 3).
Table 1.
A selection of publicly available gene annotation resources for reference genomes
| Resource and url | Description | Primary institutions |
|---|---|---|
| RefSeq www.ncbi.nlm.nih.gov/refseq |
Enormous integrated database of genome sequences, transcripts and proteins, covering all domains of life. Gene annotation is primarily based on the in-house computational Gnomon pipeline, while models for key species such as human have been subjected to extensive manual curation. | National Center for Biotechnology Information (NCBI) |
| GENCODE www.gencodegenes.org |
A multi-institute project providing gene annotation for human and mouse, initially as part of the larger ENCODE project. The genebuilds are a merge of manually-annotated models produced by the HAVANA group with computational models generated by Ensembl. Further experimental and in silico validation for models is provided by other groups. | Wellcome Trust Sanger Institute; European Bioinformatics Institute; University of Lausanne; Centre de Regulacio Genomica; University of California, Santa Cruz; Massachusetts Institute of Technology; Yale University; Spanish National Cancer Research Centre. |
| Ensembl www.ensembl.org |
Multifaceted genome annotation resource, providing genebuilds alongside other annotations, such as regulatory and disease data. It also provides the Ensembl genome browser for integrated visualization. Gene annotation is based on the in-house Ensembl analysis pipeline. | European Bioinformatics Institute (EBI) |
| UCSC Genome Browser https://genome.ucsc.edu/ | Online tool supporting the visualization of genome annotations for numerous vertebrate and invertebrate species. Includes genebuilds from RefSeq, GENCODE and Ensembl alongside other gene annotations such as AUGUSTUS, CCDS and LRG. Certain groups have provided access to their own RNA-seq model collections as ‘Track Data hubs’. | University of California, Santa Cruz (UCSC) |
| WormBase www.wormbase.org |
Database providing biogical information – including genes and genome sequence - for the nematode Caenorhabditis elegans alongside other nematode species. While all C. elegans gene models were initially created computationally, each has now been subject to manual curation. Gene annotations for most other nematodes are generated computationally by the MAKER2 pipeline. | European Bioinformatics Institute; Wellcome Trust Sanger Institute; Ontario Institute for Cancer Research; Washington Univerity, St. Louis; California Institute of Technology. |
| FlyBase www.flybase.org |
Central repository for genetics information relating to the insect family Drosophilidae, including a browser for gene annotations. Effectively all gene annotations have now been manually curated. | Harvard University; Indiana Universty, University of Cambridge. |
| The Arabidopsis Information Resource (TAIR) www.arabidopsis.org |
Database of genetic and molecular data for the model plant Arabidopsis thaliana, including gene annotation. Models were initially produced by the Arabidopsis Genome Initiative, improved by The Insitute for Genomic Research before being further improved and maintained by TAIR. The models have been subject to extensive manual curation, and community-annotation is now facilitated via Web Apollo. | Phoenix Bioinformatics |
| UniProtKB www.uniprot.org |
A unified protein repository incorporating the Swiss-Prot and TrEMBL databases of protein sequences. Swiss-Prot is manually annotated by expert curators (based on literature and manual gene curation), whereas TrEMBL contains computationally analyzed entries largely extracted from computataionally-derived transcript models. | European Bioinformatics Institute; Swiss Institute of Bioinformatics; the Protein Information Resource. |
| Roadmap Epigenomics Project www.roadmapepigenomics.org |
Multi-institute collaboration developing a resource for the presentation and processing of human experimentally-derived epigemomics data. It aims to generate reference epigenomes across a large variety of cell types. It includes data on gene expression, histone modification, DNA methylation and chromatin accessibility. | The National Institute of Health Epigenomics Mapping Consortium |
| The ENCODE encyclopedia https://encodeproject.org/data/annotations/ |
Computational analysis pipeline being developed by the multi-institute ENCODE project to summarize the findings of experimental datasets across the genome sequence, including RNA-seq, Hi-C, ChIP-seq and histone marks (and incorporating data from the Roadmap Epigenomics Project). For example, it can help users extrapolate whether a given region looks like an enhancer. | The ENCODE consortium |
| Functional ANnoTation Of The Mamalian genome (FANTOM) http://fantom.gsc.riken.jp/ |
International research consortium seeking to obtain further knowledge of the human and mouse genomes and transcriptomes. Since 2000, the project has shifted its focus from cDNA annotation, to transcription start and promoter anlaysis, and onto the description of lncRNAs. | Coordinated by RIKEN Yokohama. |
This table is an entry point for exploring eukaryotic annotation resources in more detail. It focuses on resources discussed in the main text, and is not intended to be comprehensive; the complete list of projects and groups that have contributed to gene annotation in the genome-sequencing era would be exceptionally large. Furthermore, it has not been possible to list individual groups contributing to the FANTOM and ENCODE projects due to space limitations.
Abbreviations CCDS, consensus coding sequence; ChIP-seq, chromatin immunoprecipitation followed by sequencing; ENCODE, Encyclopedia of DNA Elements; HAVANA, Human and Vertebrate Analysis and Annotation; LRG, Locus Reference Genomic; lncRNAs, long non-coding RNAs; RNA-seq, RNA sequencing.
Annotation evidence
Regardless of the scientific context of an annotation project, the most important factors influencing the genebuild produced are the evidence and methodologies used for model construction (Box 2; Fig. 2, 3). It is informative to consider how these elements have changed as reference annotation projects have become outnumbered by non-reference projects. In terms of evidence, the obvious difference is that Sanger-based transcript sequencing has been superseded by short-read RNA-sequencing (RNA-seq) 11. Thus, while the bulk of models in reference genebuilds were constructed on cDNA or EST evidence, such libraries are typically absent for other genomes. This fact is important when it comes to annotation. Most obviously, although RNA-seq is cheaper and more high-throughput than earlier protocols, the RNA sequences obtained are shorter and more prone to error. This creates notable problems for annotation, as we shall discuss, and it still remains easier to build accurate models on longer RNA sequences.
Box 2. Defining functionality within the genome and transcriptome.
There has been much debate about the portion of the eukaryotic genome that is truly functional, largely through disagreements on how ‘functionality’ should actually be defined. Evolutionary biologists have traditionally placed high value on the maxim that ‘conservation equals function’, and may thus doubt the functionality of non-conserved bases86. More recently, human experimental biology projects such as ENCODE have used a biochemical definition based on the use of high-throughput assays including RNA-seq and immunoprecipitation techniques102. However, the proportion of the genome that participates in transcription and epigenomics is far larger than that which displays conservation, hence these definitions appear irreconcilable. The process of genome annotation can provide useful insights into this debate, as it approaches things from a different direction. Here, the initial focus is not on individual base-pairs, rather on whole sequence elements such as transcripts, and it is of course transcripts that are the primary effectors of genomic information.
The question can therefore be restated as: which transcripts are functional and how do they function? In this context, the word ‘functional’ primarily concerns the role of the transcript in the cell; whether it is translated, for example, or actually makes no contribution to physiology. It would therefore seem reasonable to describe an mRNA as a ‘functional transcript’, and a transcript that is simply stochastic noise as ‘non-functional’. The more challenging ground is found between these two poles. For example, it is debatable whether the AIRN lncRNA transcript is a functional molecule given that it is ultimately a by-product of a regulatory pathway91, and the same question could be asked of regulatory non-productive transcripts (NPTs) found within protein-coding genes. Certainly, the generation of these transcripts directly mediates functional processes, and for this reason we prefer to regard them as functional molecules. The question ‘how do transcripts function?’ in fact has a further layer of complexity. A typical mRNA contains a variety of sequence features: most obviously the coding sequence (CDS), but potentially also regulatory sequences such as trans-factor binding sites, secondary structures and upstream open reading frames (uORFs). These are all potential targets for annotation, which suggests that we should regard an mRNA more properly as a ‘functional transcript that contains a number of distinct functional features’. It is also interesting to note that mRNAs may contain sequences that are not functional according to the strict evolutionary definition; this is in fact more dramatically the case for well-studied lncRNAs such as HOTAIR125. While this discussion may seem esoteric, it is actually of great practical importance. Gene annotation is of particular value in the clinic, where it is often used to aid the interpretation of disease-associated variants. A clinician would like to know not only that a given mutation is associated with a functional transcript, but also which sequence features it affects within that transcript. While sequence conservation can be a useful aid to the prioritization of variants, annotation processes are ultimately required to convert such information into actual biological features.
The second key source of evidence is protein sequences. Here, the situation is more complicated, as the field of experimental protein sequencing lags far behind that for RNA or DNA. Thus, the earliest annotation projects described coding sequences (CDS) based on curated protein sequences from Swiss-Prot12 (Table 1) and through the use of ab initio ‘open reading frame (ORF)-finders’13,14. The ORF-finding strategy sought to identify CDS through a combination of codon frequency usage and ORF size, although many translations were subsequently judged as spurious by manual curation. Today, most non-reference genomes still lack substantial numbers of high-quality protein sequences, although ORF-finder efficacy has increased markedly as more genome sequences have become available. This is because a powerful way to find CDS is through the ratio of synonymous to non-synonymous substitutions within a prospective ORF15; i.e. to identify regions of DNA evolving as protein-coding sequence.
Annotation workflows
The annotation ‘workflow’ chosen illustrates a second key difference between reference and non-reference genebuilds. Whereas all whole-genome annotation is highly dependent on computational processing, the projects for reference genomes have supplemented these processes with manual analysis. Generally, this involves teams of curators who either create transcript models from scratch, or else ‘curate’ sets of computationally generated models1, 4, 7–9, and can involve interactions with external groups such as UniProt12 or gene nomenclature committees16. ‘Manual annotation’ is regarded as ‘gold standard’ 17, and is one of the core workflows that allows genebuilds to be classified as ‘mature’ when performed to a significant degree (Fig. 3). Nonetheless, such labour-intensive work cannot cope with the number of species genomes becoming available, and most new genebuilds are generated entirely in silico.
Computational annotation has three core processes, depending on the resources available (Fig. 2). The first is based on the alignment of transcript evidence. The second is comparative annotation, whereby the evolutionary closeness of two species allows for annotation — commonly the CDS — to be ‘projected’ from one genome to another, or for evidence from one species to be used to build models on the other. The third is ab initio annotation, whereby algorithm-based ‘gene finders’ such as GENSCAN14 or AUGUSTUS13 construct models based on a priori knowledge of their likely sequence. Pure ab initio annotation is actually now uncommon in higher eukaryotic genomes, and these strategies are most often used in combination., The RefSeq Gnomon pipeline is a modified form of GENSCAN that can perform purely ab initio annotation, although it can also integrate RNA and protein homology data when available [www.ncbi.nlm.nih.gov/genome/annotation_euk/process/]. Ensembl have adapted their pipeline in a similar manner, and the less species-specific evidence is available for a given genome, the more annotation will be based on a combination of projection and ab initio modelling. Similarly, WormBase are combining projection from C. elegans with ab initio modelling in the annotation of other nematode genomes8.
Even the largest annotation projects cannot yet describe genomes by the thousand, and researchers must often produce their own genebuilds. The Avian Genome Consortium, which aims to describe hundreds of bird genomes, are achieving this by working closely with Ensembl18. Annotation is being generated by the Beijing Genome Institute via the projection of existing bird and human Ensembl models, and is displayed in ‘Avianbase’: a modified form of the Ensembl schema19. RefSeq have also worked with external collaborators on specific genebuilds4. For researchers with fewer resources, numerous software tools can be used to perform truly independent gene annotation. AUGUSTUS remains a popular choice; although it was developed as an ab initio tool for the Human Genome Project, its modern incarnation can incorporate transcript libraries and comparative evidence, albeit with a cost in terms of speed and ease of use13, 20, 21. For such practical reasons researchers often annotate their genome using a simpler RNA-seq assembly pipeline such as Cufflinks22. Besides suffering from the RNA-seq assembly problems discussed below, such methods are severely limited by the fact that they do not produce true functional annotation (see below), and, in common with ab initio builders, will typically generate a single model per gene. We do not regard these catalogues as true genebuilds.
Community annotation
For genomes such as rat it is has become clear that computational genebuilds cannot meet the needs of the community, and yet adequate resources are not available to follow the RefSeq or GENCODE reference annotation model. One solution is to manually improve the annotation in a systematic, collaborative manner based on ‘crowdsourcing’23 (Fig. 3). Either the interested parties meet in person and perform a large amount of annotation over a short period of time (a ‘jamboree’)24, or else they work remotely over a longer of period, following the same annotation criteria25 and using software such as WebApollo, which allows for ‘live’ annotation to be shared remotely26. This latter workflow has been central to the annotation efforts of VectorBase, which is a community effort seeking to describe the genomes of invertebrates that transmit disease to humans27. Nonetheless, the output of most projects cannot match reference curation teams in scale, and the focus is often limited to a particular biological theme, e.g. the annotation of porcine immunology-related genes28.
Annotation in population genomics
It is now commonplace to generate multiple genome sequences from the same species, especially to aid the study of variation. Human studies have inevitably led the way, with projects such as the UK10K generating genomes by the thousands29, although ‘population genomics’ has now been performed for species as diverse as rice30 and killer whale31. Do these genomes require annotation? If DNA variation is of primary interest, single nucleotide polymorphisms (SNPs) can simply be extracted and displayed against the main assembly for that species. Furthermore, if users wish to ‘browse’ additional genomes then transcript models can be ‘projected’ from the main assembly. Projection is part of the annotation strategy of the Mouse Genomes Project (MGP) — who have released 36 genome sequences of laboratory mice and wild-derived strains — in combination with ab initio modelling32.
Nonetheless, the MGP also illustrates scenarios for which manual intervention is desirable. For example, when genes do not project successfully then manual curation can resolve whether this is due to variation or genome sequence error6, and it can also be used to judge the quality of ab initio models. Manual annotation can also be essential when investigating structural variants (SVs), which are of great interest to biologists due their association with disease and evolution33. The Genome Reference Consortium (GRC) [www.ncbi.nlm.nih.gov/projects/genome/assembly/grc] continue to improve the human and mouse genome assemblies, and have created a series of ‘alternative (alt) loci’ for both species that target allelic variation as well as SV regions containing genes that are subject to copy number variation (CNV)34. For example, the GRCh38 human genome assembly contains 8 haplotypes for the major histocompatibility complex (MHC)35 and 35 for the leukocyte receptor complex (LRC)36. The interpretation of CNV gene families can be difficult: gene copies are often highly similar or even identical, and a protein-coding gene in one genome may be pseudogenized in another. It is impossible to simply extract this information for display against a reference genome, and such regions can be difficult to resolve without manual intervention.
When is a genebuild complete?
Identifying missing transcripts
Having discussed progress in gene annotation, we now turn our attention to the limitations of existing genebuilds. Users should understand that even human genebuilds are works in progress, and we now consider how far into the distance the finishing line for such endeavours might be found. Logically, a complete genebuild would contain all the transcripts that a genome produces, with accurate functional information attached to each model. Certainly, an attempt to identify all transcripts may be considered a key goal in the generation of a mature genebuild (Fig. 3). However, multicellular organisms have almost as many transcriptomes as they have cells, and an emerging goal for annotation projects is to provide information on where and when transcripts are expressed. In practice, this depends on the prior creation of unified transcript catalogues, i.e. where transcripts from all sources are combined. Meanwhile transcripts may be absent from genebuilds for three reasons: existing models may be incomplete, i.e. truncated at one or both ends; whole transcripts could be missing within existing genes; or entire genes could be absent. Obviously, the relevant RNAs may not be present in transcript libraries, which is most likely for transcripts with restricted expression. Nonetheless, additional transcripts clearly exist in libraries that are not yet incorporated into genebuilds; human projects routinely describe thousands of novel models37, in common with targeted efforts on other reference genebuilds such as A. thaliana38.
Unfortunately, RNA-seq continues to confound annotators39. The most common protocols generate ‘short’ reads under 200bp in size (Box 1), which is far shorter than the average mRNA. Reads are aggregated to predict full-length transcripts, although this process is challenging11. RNA-seq models are emphatically predictions, and have not been incorporated wholesale into most reference genebuilds due to quality concerns40. As noted above, they have instead proved a frequent necessity for annotating genomes lacking Sanger-sequenced transcript libraries. Meanwhile, ‘long-read’ RNA-seq libraries are becoming available to improve annotation (Box 1). It is easier to align longer reads with accuracy, although the sequencing quality is still not comparable to Sanger protocols41. An interesting development is SLR-seq, which circumnavigates the problem of short-read transcript assembly by generating ‘synthetic’ long-reads via the reconstruction of fractionated and barcoded short RNA fragments 42. Efforts are also being made to complete transcript catalogues based on targeted methodologies. ‘CaptureSeq’ involves the usage of genomic hybridization arrays to ‘pull down’ portions of the transcriptome for sequencing43. It is effective at isolating transcripts expressed at low levels, which may be ‘drowned out’ in whole RNA assays44. CaptureSeq is typically used to identify novel genes (see Figure 4), and to target partial models for completion. The experimental set-up is laborious, however, and its usage is thus far currently limited to human and mouse.
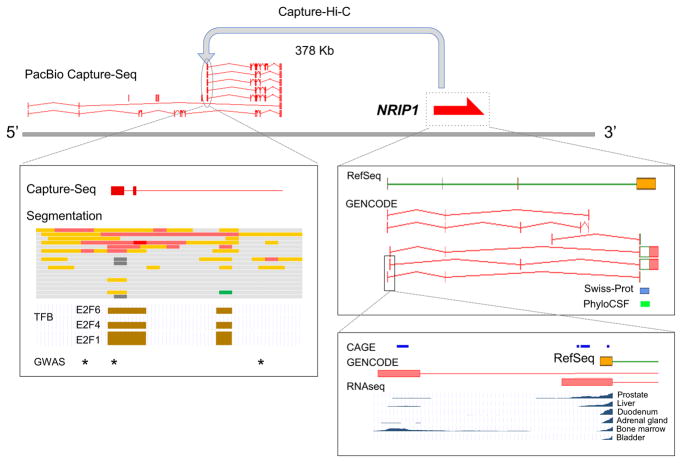
Figure 4. Transcriptional complexity in the NRIP1 locus.
a) Capture Hi-C105 indicates that the nuclear receptor interacting protein 1 (NRIP1) locus on human chromosome 21 forms a loop with a previously unannotated region nearby. Pacific Biosciences (PacBio) CaptureSeq data could be aligned here (R. Johnson, personal communication), leading to the annotation of lncRNA OTTHUMG00000488671 in GENCODE. b)| A long non-coding RNA (lncRNA) transcription start site (TSS) falls within an ENCODE-defined enhancer 102 (red and orange blocks; processed by Ensembl134). Three transcription factor binding (TFB) regions — E2F1, E2F4 and E2F6 — co-localize based on ENCODE chromatin immunoprecipitation followed by sequencing (ChIP-seq) data102. In combination, these data suggest an ‘extended gene model’ for NRIP1, which may aid the interpretation of three genome-wide association study (GWAS) signals linked to Crohn’s disease (rs2823286, rs1297265 and rs1736020; shown as asterisks) as previously noted by Mifsud et al.105 c) NRIP1 contains one transcript in RefSeq and 6 in GENCODE. The coding sequence (CDS; shown as an open green box) has Swiss-Prot support, and a PhyloCSF conservation signal131. (The untranslated regions (UTRs) are shown as filled red boxes.) d) Two distinct first exons of NRIP1 are annotated, both supported by 5’ Cap Analysis of Gene Expression (CAGE) data45. RNA-seq from Uhlen et al.115 indicates differential expression, with usage of the upstream exon apparently limited to bone marrow (and adipose; not shown). This TSS is dominant in white blood cells, which are bone-marrow-derived. RNA-seq and CAGE support a more general expression profile for the downstream first exon, with evidence of TSS variability.
Annotating transcript endpoints
How can you tell if a model is precisely full length, i.e. contains the transcription start site (TSS) and transcript end site (TES) of the RNA? TESs can be identified from the 3’ polyadenylation tail, although there is no consistent diagnostic sequence for TSS so it is difficult to know if a transcript is 5’ truncated. Such ambiguity is problematic, because confident functional annotation depends on accurate structures. Whereas a CDS may be obvious on a full-length transcript, it could be missed on a truncated version, especially if sequencing has not encompassed the translation initiation site (TIS) or STOP codon. The implications of this are particularly concerning in disease genetics, where CDS annotation is the key dataset through which identified genetic variants are interpreted. This problem appears to be solvable, however, given the advent of modified RNA-seq assays to sequence endpoints (Box 1). Notably, FANTOM5 have generated of millions of 5’ Cap Analysis of Gene Expression (CAGE) sequences from over 400 hundred tissues or cell lines for human and mouse45. While the major goal of this project is to study transcript expression, these data are also proving highly useful for manual curation efforts (Figs. 2, 3)6, 39.
However, genes display considerable variability in their endpoints45, 46 — even within the same exon — which challenges our assumptions about the relationship between transcript models and cellular RNA. Annotation projects utilizing these datasets can try to represent this diversity or else attempt to summarize it. The key issue is whether this complexity is biologically meaningful. This may not be the case for endpoint ‘wobble’, which could reflect stochastic variability in the binding of the RNA polymerase II or polyadenylation complexes. If a project favours simplicity, ‘gene-boundary’ data can be converted into single base-pair sites and incorporated into computational workflows. For example, Boley et al. used CAGE and polyA-seq data in the generation of D. melanogaster RNA-seq-based models47, while the PLAR pipeline incorporated polyA-seq in the annotation of 17 vertebrate genomes48. However, differential endpoint usage can have important functional consequences, especially linked to gene regulation as discussed below.
Functional annotation
When RNA-seq protocols can produce accurate, full-length transcripts the need to curate these structures will diminish. Instead, the legacy of genebuilds is likely to be their functional annotation. Traditionally, functional annotation centered on the question ‘which models encode protein?’ Today, we know that non-coding genes and untranslated transcripts can function in many different ways. Indeed, the definition of ‘functional’ remains controversial in genomics, as we discuss in Box 2. Nonetheless, a survey for protein-coding loci remains a common starting point for the annotation of novel genomes, while efforts to annotate the complete set of translated regions are ongoing even in reference genebuilds.
Distinguishing protein-coding genes and pseudogenes
As discussed, CDS annotation is typically based on the incorporation of curated protein sequences, as well the computational processing of protein homologies and conservation signals. However, a genuine signal does not confirm that a region is coding, rather that it has been coding at some point in its history. This distinction is crucial, as eukaryotic genomes contain large numbers of pseudogenes49 (Fig. 1, 2). Pseudogenes are a major confounding factor for computational CDS annotation: they may contain large ORFs and are frequently transcribed, while duplicated or retrotransposed pseudogenes with high sequence similarity to the parent locus can complicate both CDS projection and RNA-seq mapping50. Even their manual interpretation is complicated, which is a major reason why GENCODE, RefSeq and UniProt do not agree on the number of human protein-coding genes. For example, retrotransposition can generate intact copies of the parental CDS51, and whereas GENCODE have annotated over 300 ‘retrogenes’ as protein-coding, the functionality of those that do not exhibit conservation remains speculative. Alternatively, while duplicated copies of a parent gene may have disrupted CDS, it can be unclear whether this causes loss of function (LoF)50. These ambiguities are exacerbated in lower-quality genome sequences: CDS disablements in prospective pseudogenes — and LoF mutations in resequenced genomes — could instead be sequencing errors. While there are a limited number of dedicated tools for the computational analysis of pseudogenes, including PseudoPipe52, manual annotation remains preferable53.
The coding potential of alternative splicing
A second complication in CDS annotation is that protein-coding genes can make distinct proteins (‘isoforms’) through AS54, 55. However, although AS is ubiquitous among multi-exon genes, the extent to which it generates proteomic diversity is debatable56, 57. Indeed, it should be emphasized that the bulk of CDS annotation in eukaryotes is based on extrapolation as opposed to experimental evidence, and this fact is likely to have profound implications across the field of biology. Certainly, AS does not always generate isoforms, and we refer to transcripts from protein-coding genes that do not generate mature proteins as non-productive transcripts (NPTs). Distinguishing coding transcripts and NPTs is a major goal of maturing annotation projects39, although RefSeq and GENCODE approach the problem from different directions. RefSeq have traditionally focused on models considered likely to be protein-coding based on additional evidence, e.g. Swiss-Prot. While GENCODE annotate such models along similar lines, they ultimately aim to provide functional annotation for all identified transcripts. The coding potential of these additional transcripts are judged in comparison to a model within the gene known to be protein-coding. Thus, an ‘exon-skipping’ transcript is likely to be annotated as coding if it does not contain a frameshift. Such ‘first principles’-based annotations are speculative. However, GENCODE reappraise their human CDS based on scoring provided by the annotation of principal and alternative splice isoforms (APPRIS) pipeline58, which combines CDS conservation alongside predictions into the effects of AS on known protein domains. APPRIS has generated annotation for six mammals, as well as C. elegans and D. melanogaster. Finally, we note that GENCODE and RefSeq in fact collaborate on the ongoing Consensus CDS (CCDS) project, whose core goal is to produce CDS sets that are unified between different annotation projects (Table 1)59.
Non-productive transcription and untranslated regions in protein-coding genes
If transcripts within protein-coding genes do not make proteins, what do they do? All cellular machines are error-prone, and intron retention (IR), for example, could simply be due to spliceosome failure39, 56. Furthermore, the sequence motifs that govern transcription, splicing and translation are typically basic, and ‘cryptic’ sites throughout the genome can act as competitors to canonical sites. Such knowledge recontextualizes the question of when a transcript catalogue is complete, and it is generally accepted that a proportion of the transcriptome is aberrant ‘noise’, although the size of this portion is debated56, 60–63. However, NPT can impart gene regulation. Many protein-coding genes reduce their protein output not by ‘switching off’, rather by directing their transcription into non-productive pathways. The best characterized mechanism by which this occurs is AS-linked nonsense-mediated decay (NMD)64. Although NMD was originally understood as a mechanism for the degradation of aberrant transcripts, many genes use this pathway to dampen their output in a regulated manner 65, typically through the splicing of a ‘poison’ exon that contains a termination codon.
Regulation can also be imparted through IR, which is emerging as a key control mechanism in haematopoiesis66. In fact, up to three quarters of mammalian genes exhibit systematic IR, especially in cell types where expression is not anticipated; IR may be ‘functionally tuning’ these cells67. The contribution of IR to gene regulation is particularly well established in A. thaliana38. Regulatory NPT can also be invoked through differential TSS usage, although in this scenario the transcripts are probably ‘less productive’ rather than non-productive. For example, human GRN gene produces two transcripts with highly different rates of translation, even though they have the same CDS68. The weakly translated form has a longer 5’ untranslated region (UTR), which incorporates a short ‘upstream’ ORF (uORF) that competes for ribosome binding with the regular CDS. TSS switching to the short 5’ UTR form thus increases protein production. Most human and mouse first exons contain multiple TSS regions according to FANTOM45, and most 5’ UTRs contain uORFs (personal observation). Could transcripts that differ in their 5’ UTRs have precisely the same translational efficiency? Certainly, the regulatory importance of uORFs is recognized69, although they remain a blindspot for even reference genebuilds.
The situation for differential TESs usage is at least superficially similar, and there is evidence that this process can modulate RNA stability and localization by creating transcripts that differ in their secondary structure or response to trans factors70, 71. Just as for TSSs, annotation projects generally extend models to the maximum 3’ distance supported by transcriptional evidence, and do not annotate additional models based solely on alternative TES.
NPTs are not simply a late-stage target for mature genebuilds; such transcripts will also be sucked into the annotation pipelines for novel genebuilds, and are at risk of mis-annotation. Nonetheless, if such knowledge could be captured it may radically change the way users perceive their genes of interest. An obvious question is how to distinguish models that invoke NPT as part of regulatory programs from those that arise as stochastic noise. Currently, this is being achieved by low-throughput laboratory studies, and it is notable that the differential TSS usage in GRN is currently not represented in GENCODE or RefSeq. However, global insights can sometimes be gained from comparative analyses; poison exons, for example, are often highly conserved in vertebrates65, 72. It may be that the blueprints for such phenomena can ultimately be read in the genome, e.g. in the form of binding sites for trans-acting factors73.
Annotating proteins with experimental data
CDS annotation is interpretive because the chemistry of the protein molecule makes it far less amenable to sequencing than RNA. However, recent advances in mass spectrometry (MS) have given birth to ‘proteogenomics’: the identification of CDS through the integration of peptide data and genomic or transcriptomic sequences74. The experimental parameters for this emerging technique are still being established75–78. Above all, it is a completely different paradigm to RNA sequencing (Box 1): peptide identification depends not on mapping, rather on the correlation between spectra observed in the experiment and those predicted to be produced within a CDS search space defined in silico. The design of this ‘search space’ has a substantial bearing on the results, and the false-discovery rates for proteogenomics assays are notoriously difficult to gauge74. Also, peptides are frequently too short distinguish isoforms. Furthermore, not all proteins are amenable to MS due to their chemistry or cellular location, and it is harder to capture proteins with low expression74. Nonetheless, the utility of this technique for CDS identification and validation is clear79.
Ribosome profiling (RP) identifies RNA regions that are undergoing translation (Box 1)80,81. At present, there is no community consensus on how RP datasets should be used in annotation, and there are outstanding technical questions regarding their production and interpretation82. It seems that genuine RP regions do not necessarily highlight actual CDS, i.e. that RNA–ribosome interactions do not always lead to the production of mature proteins83. This could be because certain interactions are transient as opposed to truly functional84. Alternatively, there is evidence that lncRNAs and protein-coding genes can utilize ribosome binding to impart regulation, for example via NMD64. Nonetheless, others suspect that RP datasets truly identify significant numbers of typically small proteins that do not display conservation or homology to known proteins85. The concept of ‘lineage specific’ biology provokes strong opinions86, and this debate is important from an annotation perspective, where conservation is a key proxy for functionality. While RP has been performed on at least six other eukaryotic genomes so far — as collated by the RPFdb resource87 — these data have not yet been incorporated in the computational annotation pipelines of reference genomes.
Long non-coding RNA annotation
LncRNAs present similar challenges to annotation projects as NPTs; they can have functional roles in mammalian cells88, although it has been argued that many are transcriptional noise89. Pertinently, lncRNAs are typically weakly conserved in comparison to CDS, and show high evolutionary turnover90. Nonetheless, it may be misguided to judge lncRNA functionality solely by analogy to protein-coding transcription, since the base-pair content of these transcripts is not always coupled to their functionality in an obvious way. For example, the AIRN lncRNA regulates the activity of the IGF2R locus on the opposite strand not through the activity of its transcript — which is apparently a byproduct — rather through the act of its transcription91. This emerging perspective on functionality represents a paradigm shift for annotation projects (Box 2).
It is difficult to infer lncRNA functionality through annotation alone; true understanding comes from the laboratory. Nonetheless, annotation does play an important role in judging translation, and most lncRNA models within genebuilds (or generated by pipelines such as PLAR48) are simply transcripts that are not protein-coding, pseudogenes or small RNAs. It may also be useful to sub-classify models based on their genomic location92. This could aid scientists investigating particular lncRNA categories; enhancer-associated ‘e-lncRNAs’, for example, are of interest in the field of regulatory genomics93, as is the bidirectional transcription commonly observed from protein-coding gene promoters94. However, lncRNA functional annotation may become more proactive: sequences such as microRNA binding sites95 and RNA structures96 are beginning to be described, while UV cross-linking immunoprecipitation followed by sequencing (CLIP-seq) can identify RNAs interacting with RNA-binding proteins97. In the meantime, genebuilds can incorporate laboratory-gained knowledge of functionality. LncRNAdb is a database attempting to catalogue functional lncRNAs based on literature curation98. Presently, it contains entries for 287 lncRNAs from a variety of eukaryotic species. Other repositories seek to build larger consolidated lncRNA catalogues, including LNCipedia99 which focuses on human, and NONCODE100 which contains information from 16 species. Meanwhile, the RNAcentral database101 contains 8.1 million RNA sequences, representing all major functional classes of non-coding RNAs from a selection of species; a key goal is to resolve the redundancy between the lncRNA datasets produced from different annotation groups.
Annotating the extended gene
Human genetics is faced with a substantial problem: trait-associated variants are commonly found outside gene sequences and thus defy interpretation. Annotation projects are therefore turning their attention to the genomic elements that control gene activity, the best studied of which are promoters and enhancers. Both are controlled by transcription factor (TF) binding, and each has its own characteristic (albeit imprecisely understood) epigenomic profile. ENCODE especially have provided enormous datasets on these sequences, largely through the use of immunoprecipitation techniques (Box 1)102. Furthermore, it has been known for decades that chromosomes exhibit ‘loops’, which can be indicative of transient enhancer–promoter interactions, as well as more stable chromosome 3D structures known as ‘topologically associated domains’ (TADs). Modern assays such as Hi-C103 and chromatin interaction analysis paired-end tag sequencing (ChIA-PET)104 capture the DNA fragments flanking these loops, allowing them to be mapped onto the genome.
Such datasets offer the potential to create ‘extended gene’ models, as illustrated for NRIP1 in Figure 4. From a human perspective, an obvious benefit of linking genes to regulatory elements is that it increases the space within which disease-associated variants can interpreted, although it should be emphasized that such efforts are in their infancy. A problem is that Hi-C and ChIA-PET highlight enormous numbers of loops, raising questions about the signal to noise ratio103, 104. This noise could be biological as well as artefactual106, and when ‘capture’ methods are used to target known promoters105 it is unclear what proportion of genuine loops actually demarcate enhancers. Presently, the ENCODE enhancer sets — extrapolated from biochemical data102 — are far larger than those which have been functionally validated in the laboratory107. The fact that gene regulation is spatiotemporal, complicates the situation, and it is known that genes can be controlled by multiple enhancers, while enhancers can control multiple genes108. Extended genes would be more useful if they could also integrate the TF-binding sites found within enhancers and promoters. TF annotation has traditionally proved difficult: binding motifs are typically short (~6bp) and imprecise, thwarting genome-mining efforts109. However, Chromatin immunoprecipitation followed by sequencing (ChIP-seq) datasets are now available for dozens of TFs, highlighting in vivo regions of DNA occupancy while allowing for more accurate consensus motifs to be deduced110. If such information can be combined with chromosome conformation and chromatin immunoprecipitation datasets, more precise extended genes may be obtained. This is well demonstrated for CTCF, a factor known to play a key role in loop formation111, 112. The challenge for genebuilds is how to integrate and display such data alongside their transcript models. A description of extended genes is a core goal of the developing ENCODE ‘encyclopedia’ resource [www.encodeproject.org], while tools to visualize 3D datasets on the genome are becoming available110 (http://www.3dgenome.org).
Improving the usability of genebuilds
The incorporation of transcript expression data
As genebuilds provide more precise representations of the transcriptome, they inevitable become more complex. This point has important repercussions for users. For example, many scientists are focused on human BRCA1 due to its association with breast cancer, and may wonder what to make of the fact that GENCODE has 30 transcript models whereas RefSeq has 6. In practice, annotation resources are utilized in many different ways, especially for human. Whereas some scientists would like to use all transcripts associated with a given gene — for example when designing hypothesis-driven experiments within a single locus — a common desire is for simplification. In fact, users often wish to work with a single transcript model per gene, for example to streamline the experimental design of whole-transcriptome studies. One way to perform ‘transcript prioritization’ is by measuring RNA expression, i.e. to identify the ‘dominant’ transcript in a gene. While it remains challenging to resolve individual transcripts based on RNA-seq, the fact that most human protein-coding genes have a dominant transcript indicates that there is value to expression-based filtering113. However, dominance can ‘switch’ between cell types, and expression changes are typically analogue rather than simply ‘on’ or ‘off’113–115. An additional point of profound importance is that RNA is ultimately a proxy for the measurement of protein output within protein-coding genes. In reality, the relationship between RNA and protein output remains imprecisely understood116, and correlations between the two are frequently not strong117. Although this may have striking consequences for RNA-based expression studies, the maturing field of quantitative proteomics does not yet provide precise guidance for annotation projects.
The prioritization of functional transcripts
Reference genebuilds do not explicitly highlight principal transcripts based on quantitative evidence at present, and the description of spatiotemporal expression comes instead from ‘downstream’ endeavours such as the Genotype–Tissue Expression (GTEx) project114. One could anticipate that such information will soon be leveraged in reference genebuilds, influencing perhaps how models are displayed in genome browsers. Nonetheless, it is debatable how much expression data can tell us about transcript functionality (Box 2): transcripts with lower expression are not necessarily nonfunctional (or even less functional), and in fact the expression of numerous genes appears to be dominated by NPT113. When it comes to genebuild usability, it is this question of functionality that is of paramount importance, most obviously when it comes to CDS annotation. For example, it is important to predict the molecular and clinical consequences of variation within BRCA1, and the processing of variant datasets typically begins with a comparison against gene annotation118. This allows variants to be stratified according to their potential mechanistic consequences, for example whether they disrupt a CDS or fall within an intron.
Clearly, there is a close relationship between the quality of gene annotation and the accuracy of variant interpretation, and yet many aspects of annotation — especially functional annotation — remain putative. This is particularly true for genebuilds such as GENCODE, which attempt to annotate all transcripts. Putative functional annotation can introduce false positives into variant-calling workflows, e.g. where LoF mutations are called in CDS exons that are not in reality coding. GENCODE attempt to reduce this problem by providing ‘Basic’: a ~50% reduced build in comparison to the ‘Comprehensive’ set, resulting especially from the removal of models with truncated CDS. As discussed, GENCODE also uses APPRIS to highlight coding models of probable functionality based on conservation58. By contrast, the use of smaller genebuilds could introduce false negatives into variant analyses, i.e. where consequential variants are missed or misinterpreted because they fall outside the gene annotation. However, RefSeq allow users of their core genebuilds to work with sets of more prospective transcripts, in the form of their uncurated (XM) ‘in silico’ models. Finally, Ensembl and the NCBI are collaborating in the Locus Reference Genomic (LRG) project119. The remit of this work is to standardize the gene annotation used in the clinic, with a key aim being to select a set of transcript models for core disease genes. These models are manually selected, in order to include what appear to be the key functional elements for a given gene.
Conclusions
The complexity of gene annotation projects reflects the complexity that exists in eukaryotic cells, and, since we do not fully understand the transcriptome at the present time, all of our genebuilds are incomplete. Present ambiguities are most keenly felt in our own species, where nothing less than a total understanding of biology is demanded. For other projects, the ‘finish line’ may not be so far into the distance, and the length of journey taken will in many ways reflect the value of that genome to science. However, all genebuilds face challenges in how they present their resource to the public; most obviously, they must find ways to make sure that increasing complexity does not correlate with decreasing usability.
Acknowledgments
Research reported in this publication was supported by the National Human Genome Research Institute of the National Intitutes of Health under award number U41HG007234-004. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank A. Frankish for informative discussions.
Glossary
Definitions of core annotation targets and concepts
- Gene
Redefined for the modern era by Gerstein et al as ‘a union of genomic sequences encoding a coherent set of potentially overlapping functional products [i.e. RNAs or proteins]’120.
- Transcript
any form of RNA molecule that is transcribed from the genome sequence,.
- Coding sequence (CDS)
the region of a transcript that is translated, i.e. contains the information that encodes a protein sequence.
- Translation initiation site (TIS)
The codon that is translated to give the first amino acid of peptide; almost always [ATG]; also known as a START codon.
- STOP codon
The final codon of a protein translation; almost always [TAG], [TAA] or [TGA]; also known as a translation termination site or codon.
- Polyadenylation tail
A sequence of adenosine monophosphates attached to the 3’ end of an RNA as transcription terminates, beginning at the polyA site.
- Transcription start site (TSS)
The base-pair on the genome where transcription begins.
- Untranslated regions (UTRs)
Non-coding sequences on CDS transcripts found between the transcription start site and the translation initiation site (5’ UTR), and the STOP codon and polyA site (3’ UTR).
- Intron retention (IR)
Occurs when a transcript does not splice out one or more introns, i.e. this sequence is left incorporated into the mature RNA.
- Nonsense-mediated decay (NMD)
Cellular ‘surveillance’ mechanism that targets transcripts for destruction. Imprecisely understood, though transcripts featuring termination codons more than 50bp upstream of splice junctions are thought likely to be substrates.
- Poison exon
An exon that prevents correct CDS translation when incorporated into the transcript of a protein-coding gene, either by causing a frame-shift or through the introduction of a premature termination codon.
- Alternative splicing (AS)
Process by which a gene makes distinct transcripts through the usage of different splice sites or exon combinations; these are referred to as alternative transcripts or transcript variants.
- Isoforms
Protein molecules that differ in their amino acid composition from other translations made from the same gene, for example due to alternative splicing.
- Long non-coding RNAs (lncRNAs)
Genes that do not contain protein-coding transcripts and are not pseudogenes or small RNAs; a 200bp size cut-off is typically applied to distinguish them from small RNAs.
- Pseudogenes
‘Broken’ genes derived from protein-coding loci. Can be formed by retrotransposition (‘processed’), duplication (‘unprocessed’) or inactivation (‘unitary’, which may be polymorphic). All forms may be transcribed.
- Small RNA
Member of one of several known families of small RNA molecules. Includes the classical tRNA and rRNA families alongside more recent discoveries such as PIWI-interacting RNAs (piRNAs), microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs).
- Promoter
The region immediately upstream of the transcription start site where the RNA polymerase complex attaches in order to initiate transcription.
- Enhancer
Sequence that regulates a promoter from a distal site on the chromosome, probably brought into close proximity via DNA looping.
- Genebuild
Term used by GENCODE and Ensembl for a collection of transcript models generated by computational or manual annotation across an entire genome sequence. Protein-coding genes, lncRNAs, small RNAs and pseudogenes may be included.
- Functional annotation
The process of defining or predicting functional roles for transcript models during gene annotation.
- Manual annotation
When a person constructs a transcript model de novo after appraising the available evidence (typically using software tools), or examines and potentially validates (‘curates’) a model that has been created computationally.
- Computational annotation
The process of generating genebuilds through entirely in silico processes, i.e. by the use of computational algorithms.
References
- 1.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Andersson L, et al. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015;16:57. doi: 10.1186/s13059-015-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGarvey KM, et al. Mouse genome annotation by the RefSeq project. Mamm Genome. 2015;26:379–90. doi: 10.1007/s00335-015-9585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome. 2015;26:366–78. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berardini TZ, et al. The Arabidopsis information resource: Making and mining the "gold standard" annotated reference plant genome. Genesis. 2015;53:474–85. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe KL, et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–80. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attrill H, et al. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–92. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsik CG, et al. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conesa A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutet E, et al. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 13.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–25. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 14.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 15.Yandell M, Ence D. A beginner's guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13:329–42. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 16.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–85. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guigo R, et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl 1):S21–31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–20. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eory L, et al. Avianbase: a community resource for bird genomics. Genome Biol. 2015;16:21. doi: 10.1186/s13059-015-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32:767–9. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–44. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 22.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loveland JE, Gilbert JG, Griffiths E, Harrow JL. Community gene annotation in practice. Database (Oxford) 2012;2012:bas009. doi: 10.1093/database/bas009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennisi E. Ideas fly at gene-finding jamboree. Science. 2000;287:2182–4. doi: 10.1126/science.287.5461.2182. [DOI] [PubMed] [Google Scholar]
- 25.Archibald AL, et al. Pig genome sequence--analysis and publication strategy. BMC Genomics. 2010;11:438. doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giraldo-Calderon GI, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–13. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson HD, et al. Structural and functional annotation of the porcine immunome. BMC Genomics. 2013;14:332. doi: 10.1186/1471-2164-14-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UK10K Consortium. et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo L, Gao Z, Qian Q. Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice (N Y) 2014;7:4. doi: 10.1186/s12284-014-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foote AD, et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun. 2016;7:11693. doi: 10.1038/ncomms11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams DJ, Doran AG, Lilue J, Keane TM. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm Genome. 2015;26:403–12. doi: 10.1007/s00335-015-9579-6. [DOI] [PubMed] [Google Scholar]
- 33.Baker M. Structural variation: the genome's hidden architecture. Nat Methods. 2012;9:133–7. doi: 10.1038/nmeth.1858. [DOI] [PubMed] [Google Scholar]
- 34.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–83. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 35.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–23. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirayasu K, Arase H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet. 2015;60:703–8. doi: 10.1038/jhg.2015.64. [DOI] [PubMed] [Google Scholar]
- 37.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mudge JM, Frankish A, Harrow J. Functional transcriptomics in the post-ENCODE era. Genome Res. 2013;23:1961–73. doi: 10.1101/gr.161315.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10:1177–84. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H, et al. High-resolution transcriptome analysis with long-read RNA sequencing. PLoS One. 2014;9:e108095. doi: 10.1371/journal.pone.0108095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilgner H, et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 2015;33:736–42. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercer TR, et al. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat Protoc. 2014;9:989–1009. doi: 10.1038/nprot.2014.058. [DOI] [PubMed] [Google Scholar]
- 44.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–51. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The FANTOM Consortium et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–83. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boley N, et al. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat Biotechnol. 2014;32:341–6. doi: 10.1038/nbt.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hezroni H, et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–22. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sisu C, et al. Comparative analysis of pseudogenes across three phyla. Proc Natl Acad Sci U S A. 2014;111:13361–6. doi: 10.1073/pnas.1407293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frankish A, Harrow J. GENCODE pseudogenes. Methods Mol Biol. 2014;1167:129–55. doi: 10.1007/978-1-4939-0835-6_10. [DOI] [PubMed] [Google Scholar]
- 51.Carelli FN, et al. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016;26:301–14. doi: 10.1101/gr.198473.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, et al. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 2006;22:1437–9. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]
- 53.Pei B, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelemen O, et al. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell. 2016;164:805–17. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao Y, et al. Semi-supervised Learning Predicts Approximately One Third of the Alternative Splicing Isoforms as Functional Proteins. Cell Rep. 2015;12:183–9. doi: 10.1016/j.celrep.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez JM, et al. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013;41:D110–7. doi: 10.1093/nar/gks1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrell CM, et al. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014;42:D865–72. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derrien T, Guigo R, Johnson R. The Long Non-Coding RNAs: A New (P)layer in the "Dark Matter". Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most "dark matter" transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2014;13:1126–35. doi: 10.1128/EC.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lareau LF, Brenner SE. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol Biol Evol. 2015;32:1072–9. doi: 10.1093/molbev/msv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–95. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 67.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–86. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capell A, Fellerer K, Haass C. Progranulin transcripts with short and long 5' untranslated regions (UTRs) are differentially expressed via posttranscriptional and translational repression. J Biol Chem. 2014;289:25879–89. doi: 10.1074/jbc.M114.560128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeh HS, Yong J. Alternative Polyadenylation of mRNAs: 3'-Untranslated Region Matters in Gene Expression. Mol Cells. 2016;39:281–5. doi: 10.14348/molcells.2016.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69:3613–34. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mudge JM, et al. The origins, evolution, and functional potential of alternative splicing in vertebrates. Mol Biol Evol. 2011;28:2949–59. doi: 10.1093/molbev/msr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barash Y, Garcia JV. Predicting alternative splicing. Methods Mol Biol. 2014;1126:411–23. doi: 10.1007/978-1-62703-980-2_28. [DOI] [PubMed] [Google Scholar]
- 74.Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Methods. 2014;11:1114–25. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilming LG, et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–60. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ezkurdia I, Vazquez J, Valencia A, Tress M. Analyzing the First Drafts of the Human Proteome. J Proteome Res. 2014 doi: 10.1021/pr500572z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright JC, et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun. 2016;7:11778. doi: 10.1038/ncomms11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15:205–13. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 81.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson R, Standart N. The awesome power of ribosome profiling. RNA. 2015;21:652–4. doi: 10.1261/rna.049908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ingolia NT. Ribosome Footprint Profiling of Translation throughout the Genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raj A, et al. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife. 2016;5 doi: 10.7554/eLife.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mumtaz MA, Couso JP. Ribosomal profiling adds new coding sequences to the proteome. Biochem Soc Trans. 2015;43:1271–6. doi: 10.1042/BST20150170. [DOI] [PubMed] [Google Scholar]
- 86.Graur D, et al. On the immortality of television sets: "function" in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–90. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie SQ, et al. RPFdb: a database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res. 2016;44:D254–8. doi: 10.1093/nar/gkv972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–65. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kutter C, et al. Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 92.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lai F, Shiekhattar R. Enhancer RNAs: the new molecules of transcription. Curr Opin Genet Dev. 2014;25:38–42. doi: 10.1016/j.gde.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scruggs BS, et al. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol Cell. 2015;58:1101–12. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Furio-Tari P, Tarazona S, Gabaldon T, Enright AJ, Conesa A. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci. 2013;14:23672–84. doi: 10.3390/ijms141223672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 98.Quek XC, et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–73. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Volders PJ, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–8. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Consortium RN. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–9. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–76. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–9. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 106.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dickel DE, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat Methods. 2014;11:566–71. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–54. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 110.Zerbino DR, et al. Ensembl regulation resources. Database (Oxford) 2016;2016 doi: 10.1093/database/bav119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–84. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 112.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gonzalez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.The GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 116.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Battle A, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–7. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dalgleish R, et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gerstein MB, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–81. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 121.Takahashi H, Kato S, Murata M, Carninci P. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol. 2012;786:181–200. doi: 10.1007/978-1-61779-292-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013;23:169–80. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 125.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–9. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nawrocki EP, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–7. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]