Abstract
HuT-102 cells are considered one of the most representable human T-lymphotropic virus 1 (HTLV-1)-infected cell lines for studying adult T-cell lymphoma (ATL). In our previous studies, genome-wide screening was performed using the GeneChip system with Human Genome Array U133 Plus 2.0 for transforming growth factor-β-activated kinase 1 (TAK1)-, interferon regulatory factor 3 (IRF3)- and IRF4-regulated genes to demonstrate the effects of interferon-inducible genes in HuT-102 cells. Our previous findings demonstrated that TAK1 induced interferon inducible genes via an IRF3-dependent pathway and that IRF4 has a counteracting effect. As our previous data was performed by manual selection of common interferon-related genes mentioned in the literature, there has been some obscure genes that have not been considered. In an attempt to maximize the outcome of those microarrays, the present study reanalyzed the data collected in previous studies through a set of computational rules implemented using ‘R’ software, to identify important candidate genes that have been missed in the previous two studies. The final list obtained consisted of ten genes that are highly recommend as potential candidate for therapies targeting the HTLV-1 infected cancer cells. Those genes are ATM, CFTR, MUC4, PARP14, QK1, UBR2, CLEC7A (Dectin-1), L3MBTL, SEC24D and TMEM140. Notably, PARP14 has gained increased attention as a promising target in cancer cells.
Keywords: adult T-cell lymphoma, human T-lymphotropic virus 1, microarray, interferon-inducible genes, computational biology
Introduction
In 1977, a group of scientists from Japan identified adult T-cell lymphoma (ATL) as a helper T-cell malignancy (1). A year later, HuT-102 cells were developed from a patient with cutaneous T-cell Lymphoma (2). HuT-102 cells were identified as a subset of helper T-cells with high expression of IL-2 receptor and constitutive IL-2 production, aiding in an autonomous proliferation (2). In 1980, the human T-lymphotropic virus 1 (HTLV-1) was identified in HuT-102 cells, followed by its discovery in cells from ATL patient. Analysis of serum samples from around the world provided information of endemic areas for HTLV-1, and its role as an etiologic agent for ATL (1).
Similar to freshly obtained cells from ATL patients, HuT-102 cells is a helper T-cell cell that express Tax protein, which promote these cells particularly as an excellent model for studying ATL (3). Our previous findings have identified the interferon regulatory factor (IRF) family members as efficient mediators that modulate cytokines profile in those cells, hence providing therapeutic solutions (3). Although the microarray data upon which we based our previous studies were used to elaborately cover the roles of IRFs, particularly IRF3 and IRF4, on Interferon-inducible genes, it was performed by manual selection of related genes mentioned in the literature (3,4). Therefore, we try in the current study to reanalyze the previous data through more efficient computational tools. The aim of browsing our previous data for further analysis is to find important genes that we might have passed away, in order to discover promising opportunities against ATL.
Materials and methods
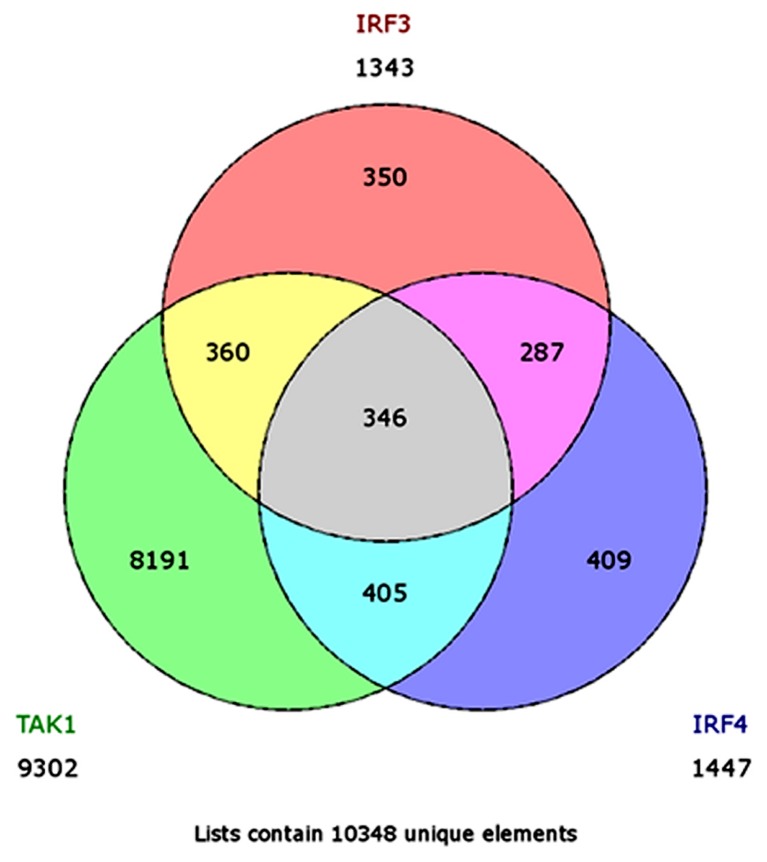
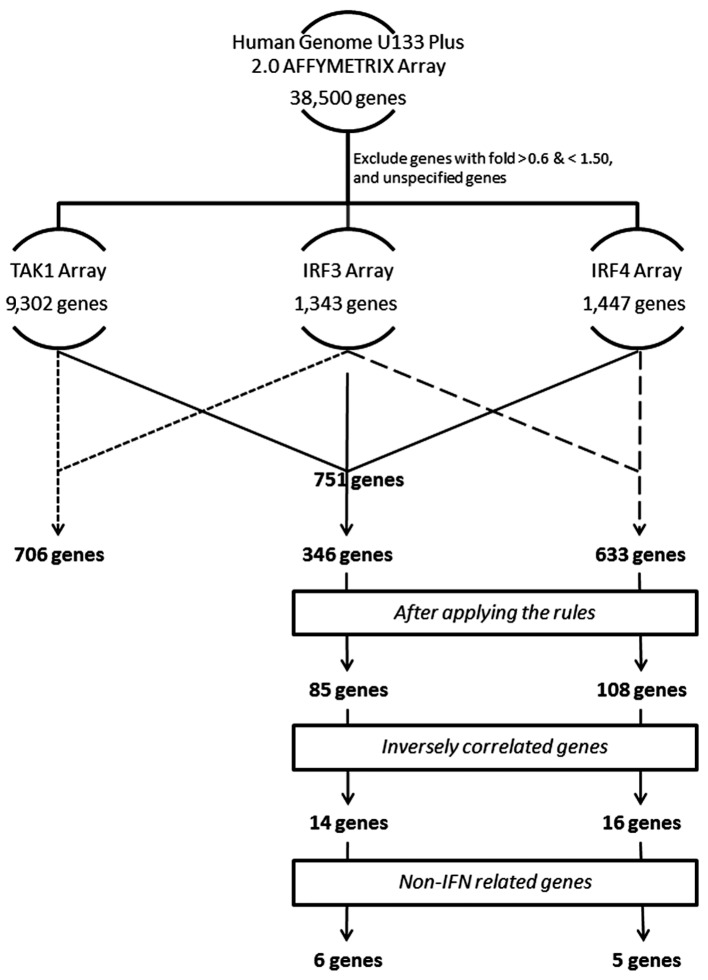
Gene expression was analyzed using a GeneChip system with Human Genome Array U133 Plus2.0 (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA) as described previously (3,4). The arrays were performed independently in two studies as follow, a total of four arrays for the first study: Two for HuT102-shLuc cells and two for HuT102-shTAK1 cells (deposited in the GEO database under accession no. GSE16219). In the second study, six arrays were used: Two for HuT102-siLuc cells, two for HuT102-siIRF4 cells, and two for HuT102-siIRF3 cells (deposited in the GEO Database under accession no. 22036). The findings of both studies are summarized in Fig. 1. In order to better analyze our study, we restricted the number of the analyzed genes. Our criteria was to focus on the genes which have markedly changed. We primarily excluded genes with values between 0.60 and 1.5 fold changes. Genes equal to or less than 0.60 are considered downregulated, and genes equal to or more than 1.50 are considered upregulated. The number of genes remaining after this step was 9,302, 1,343, and 1,447 genes for TAK1, IRF3, and IRF4 arrays, respectively. Next, we used GeneVenn (genevenn.sourceforge.net) to generate a Venn chart demonstrating the set of common genes between the different sets of arrays. The Venn chart indicated a list of 706 common genes between TAK1 and IRF3, 751 common genes between TAK1 and IRF4, 633 common genes between IRF3 and IRF4, and finally 346 common genes between the three datasets. Finally, R software for statistical computing of data sets was used, as explained below, to restrict our selection to the most promising genes, as indicated.
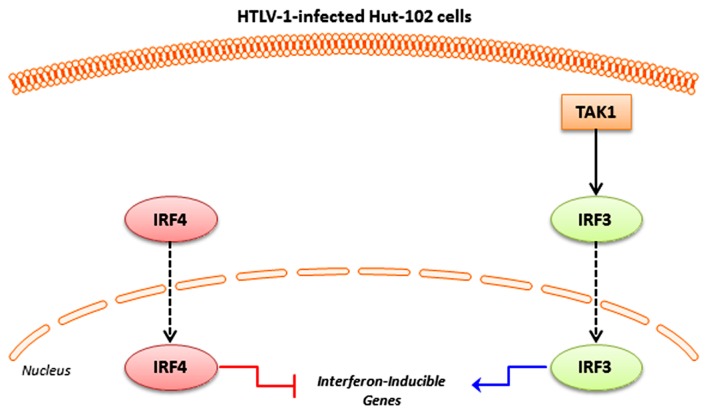
Figure 1.
Schematic diagram showing the reversed action of TAK1-IRF3 against IRF4 on the transcription of Interferon-inducible genes. TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor; HTLV-1, human T-lymphotropic virus 1.
Results
Rules applied on the identified datasets
In this study, we focused on IRF4 compared to both TAK1 and IRF3, or IRF3 only. Therefore, we continued our analysis using either the 346 common genes between the three datasets (Set A), or the 633 common genes between IRF3 and IRF4 (Set B) (Fig. 2). To filter down the list of genes generated from the Venn chart, we applied simple rules for the inclusion of genes. We firstly denoted the basic level of a given gene as the average of luciferase transfected sample in the given arrays. We then identified the basic level with different class categories, and applied rules of inclusion for genes according to Tables I and II. Through the applied criteria, we obtained 85 genes out of Set A, and 108 genes out of Set B (Figs. 3 and 4). The next stage was to identify genes inversely correlated between IRF4 against both TAK1 and IRF3, and IRF4 against IRF3 (Fig. 4). From this step, we obtained 14 genes from Set A, and 16 genes from Set B. As our goal from this analysis is to identify new genes inversely controlled by the IRF4 axis vs. the TAK1-IRF3 axis, we excluded unspecified genes, and genes related to interferon for being already covered in previous studies (3,4) (Tables III and IV, and Fig. 4).
Figure 2.
Venn diagram identifying different subsets of intersection within the microarrays data. TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor.
Table I.
Rules implemented for set A (IRF4#IRF3/TAK1) and the perspective number of genes.
| Basic level | Class | No. of genes | Rule | No. of genes after applying the rule |
|---|---|---|---|---|
| >100 | 5 | 13 | Include all | 13 |
| 50–100 | 4 | 18 | Include if 2 arrays are below 0.5 or more than 2 | 15 |
| 25–50 | 3 | 94 | Include if 2 arrays are below 0.4 or more than 4 | 32 |
| 10–25 | 2 | 132 | Include if 2 arrays are below 0.3 or more than 6 | 21 |
| 0–10 | 1 | 89 | Include if 2 arrays are below 0.2 or more than 8 | 4 |
| Total | 346 | 85 |
TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor.
Table II.
Rules implemented for set B (IRF4#IRF3) and the perspective number of genes.
| Basic level | Class | No. of genes | Rule | No. of genes after applying the rule |
|---|---|---|---|---|
| >100 | 5 | 13 | Include all | 13 |
| 50–100 | 4 | 72 | Include if 2 arrays are below 0.5 or more than 2 | 49 |
| 25–50 | 3 | 186 | Include if 2 arrays are below 0.4 or more than 4 | 38 |
| 10–25 | 2 | 201 | Include if 2 arrays are below 0.3 or more than 6 | 8 |
| 0–10 | 1 | 161 | Include if 2 arrays are below 0.2 or more than 8 | 0 |
| Total | 633 | 108 |
IRF, interferon regulatory factor
Figure 3.
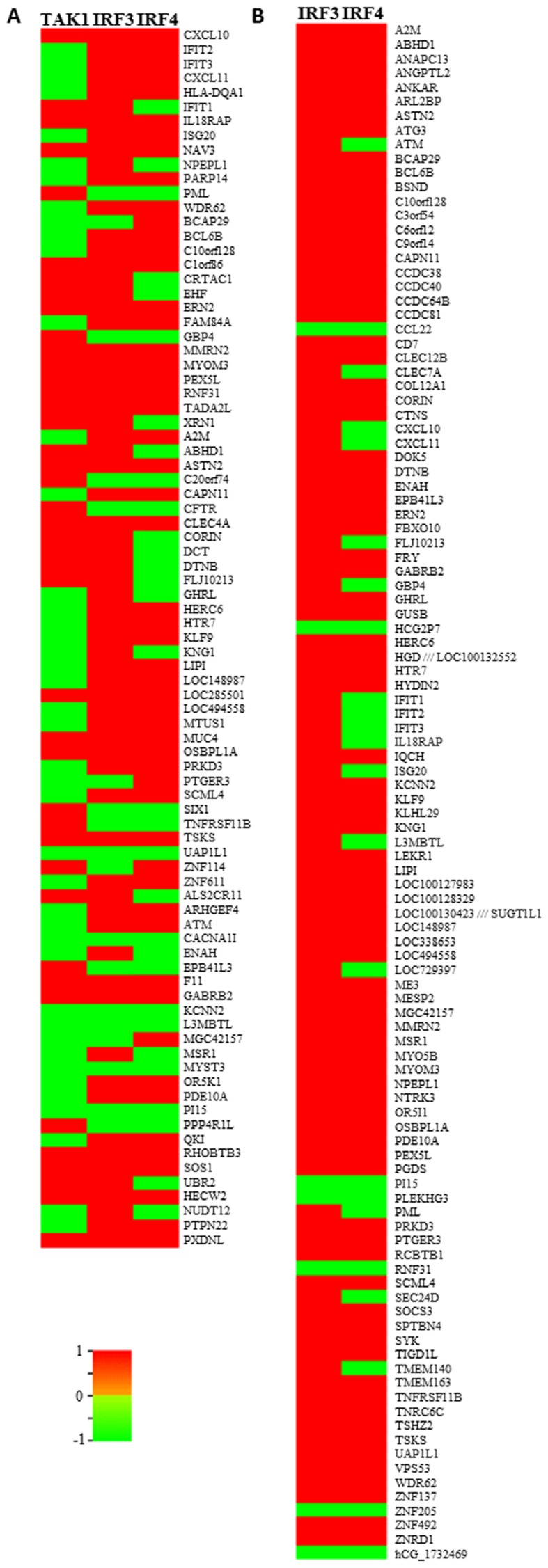
(A) Heat map showing the expression of genes intersecting in using either the 346 common genes between the three datasets (Set A), or (B) the 633 common genes between IRF3 and IRF4 (Set B). TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor.
Figure 4.
Flow chart identifying the process of gene selection in the current study. TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor.
Table III.
Final list of inversely correlated genes created from set A (IRF4#IRF3/TAK1).
| Gene symbol | Gene name | Array 1 (TAK1/luc) | Array 2 (IRF3/luc) | Array 3 (IRF4/luc) | Class |
|---|---|---|---|---|---|
| ATM | Ataxia telangiectasia mutated | 0.52 | 0.26 | 12.87 | 2 |
| CFTR | Cystic fibrosis transmembrane conductance regulator | 4.31 | 2.11 | 0.34 | 3 |
| CXCL10 | Chemokine (C-X-C Motif) ligand 10 | 0.35 | 0.19 | 6.55 | 5 |
| CXCL11 | Chemokine (C-X-C Motif) ligand 11 | 0.21 | 0.19 | 8.98 | 5 |
| FLJ10213 | Hypothetical protein LOC55096 | 0.58 | 0.16 | 4.23 | 3 |
| GBP4 | Guanylate binding protein 4 | 0.51 | 0.45 | 7.51 | 4 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 0.36 | 0.33 | 5.27 | 5 |
| IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | 0.5 | 0.29 | 2.49 | 5 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | 0.48 | 0.43 | 2.23 | 5 |
| IL18RAP | Interleukin 18 receptor accessory protein | 0.22 | 0.49 | 2.21 | 5 |
| MUC4 | Mucin 4, cell surface associated | 1.66 | 3.29 | 0.31 | 3 |
| PARP14 | Poly (ADP-ribose) polymerase family, member 14 | 0.1 | 0.58 | 2.94 | 5 |
| QKI | QKI, KH domain containing, RNA binding | 4.38 | 5.4 | 0.59 | 2 |
| UBR2 | Ubiquitin protein ligase E3 component N-recognin 2 | 0.29 | 0.23 | 1.94 | 2 |
TAK1, transforming growth factor-β-activated kinase 1; IRF, interferon regulatory factor.
Table IV.
Final list of inversely correlated genes created from set B (IRF4#IRF3).
| Gene symbol | Gene name | Fold 1 (IRF3/luc) | Fold 2 (IRF4/luc) | Class |
|---|---|---|---|---|
| ATM | Ataxia telangiectasia mutated | 0.26 | 12.87 | 3 |
| CLEC7A | C-type lectin domain family 7, member A | 0.14 | 4.62 | 2 |
| CXCL10 | Chemokine (C-X-C Motif) ligand 10 | 0.19 | 6.55 | 5 |
| CXCL11 | Chemokine (C-X-C Motif) ligand 11 | 0.19 | 8.98 | 5 |
| FLJ10213 | (unidentified) | 0.16 | 4.23 | 3 |
| GBP4 | Guanylate binding protein 4 | 0.45 | 7.51 | 4 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 0.33 | 5.27 | 5 |
| IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | 0.29 | 2.49 | 5 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | 0.43 | 2.23 | 5 |
| IL18RAP | Interleukin 18 receptor accessory protein | 0.49 | 2.21 | 5 |
| ISG20 | Interferon stimulated exonuclease gene 20 kDa | 0.45 | 2.24 | 5 |
| L3MBTL | Lethal(3)malignant brain tumor-like gene | 0.13 | 4.84 | 3 |
| LOC729397 | Hypothetical LOC729397 | 0.31 | 2.23 | 4 |
| PML | Promyelocytic leukemia | 0.53 | 2.06 | 5 |
| SEC24D | SEC24 family member D | 0.31 | 3.24 | 3 |
| TMEM140 | Transmembrane protein 140 | 0.35 | 2.61 | 4 |
IRF, interferon regulatory factor.
List of genes identified as possible future therapeutic targets
The final list we obtained consists of 10 genes that we highly recommend as potential candidate for therapies targeting the HTLV-1 infected cancer cells, and might be potential in other cancers as well.
i) Ataxia telangiectasia mutated (ATM) is a Serine/threonine protein kinase with a distinct role in double-strand breaks. Its activation causes the phosphorylation and the activation of several downstream DNA damage and cell cycle arrest checkpoints including the histone variant H2ax, the effector protein kinases, and the tumor suppressor p53 (5–7). Germ-line mutations of ATM lead to ataxia-telangiectasia and shows a high risk of breast cancer (8).
ii) Cystic Fibrosis Transmembrane conductance Regulator (CFTR) is Chloride channel membrane protein (9). It is a member of the ABC transporter superfamily and is activated by phosphorylation by PKA. CFTR conducts anions to flow down their electrochemical gradient and facilitate the passive movement of the positively charged ions. The CFTR gene has been reported to be mutated in patients with cystic fibrosis (CF) and has been a potential therapeutic target. In 2012, the FDA approved Ivacaftor as the first targeted therapy for patient with CF (10). Recently CFTR was identified as s a tumor suppressor including intestinal, prostate and ovarian cancer (11–13). Moreover, CFTR was shown as a potent suppressor of epithelial-to-mesenchymal transition (EMT) breast cancer cells and was associated with poor prognosis in patients (14).
iii) Mucin 4 (MUC4) is a high molecular weight glycosylated protein, which plays various roles in promoting cancer progression (15). Overexpression of mucins has been shown to reduce of cell adhesion and promote cancer cell migration and metastasis. MUC4 also phosphorylate the ErbB2 leading to increasing the tumor cell proliferation independent on the activation of neither the MAPK nor AKT pathways. Importantly, MUC4 has been shown as a promising biomarker for diagnoses of pancreatic cancer with undetectable levels in normal pancreas (16).
iv) Poly (ADP-ribose) polymerase 14 (PARP14) is a member of the PARP family of proteins, which involves in DNA repair and programmed cell death. PARP14 was shown to regulate the STAT6-dependent transcription (17). Moreover, Iansante et al has reported the vital role of PARP14 to mechanistically link apoptosis to metabolism. Through inhibiting the JNK1/PKMa2 regulatory axis, PARP14 potentially promoted Warburg effect in hepatocellular carcinoma (18).
v) Quaking homolog, KH domain RNA binding (QK1) is a RNA-binding protein. It is a member of the Signal Transduction and Activation of RNA (STAR) proteins family. QK1 was reported to play a distinct role in schizophrenia via regulating the myelin-related genes activity (19). QKI has been recently shown to be downregulated in lung cancer resulted in poorer prognosis (20). Additionally, it suppresses glioblastoma by stabilizing microRNA-20a leading to regulating TGFβ pathway (21).
vi) UBR2 is an E3 ubiquitin-protein ligase, which acts by recognizing and binding to the N-terminal residues-carrying proteins leading to their ubiquitination and degradation. Notably, UBR2 together with UBR1 were shown as potential negative regulators of mammalian target of rapamycin (mTOR) pathway via degrading the Leucine protein, and therefore its expression can be associated with cancer elimination (22).
vii) C-type lectin domain family 7 member A (CLEC7A, also known as Dectin-1) is a glycoprotein with a distinct role in regulating innate immunity. It is predominantly expressed in macrophages, neutrophils and dendritic cells and binds both CD4+ and CD8+ T cells (23). Dectin-1 also recognizes several fungal species, which triggers the induction of numerous cytokines and chemokines including TNF-α, IL-2, IL-6, and IL-23 (24). Mechanistically, Dectin-1 ligand has been shown to act via recruiting and activating the NFkB inflammatory pathway (25). Importantly, activation of dectin-1 on macrophages induces an adaptive immune suppression and promotes pancreatic cancer progression (26).
viii) Lethal (3) malignant brain tumor-like protein (L3MBTL) is a polycomb group protein (PcG) that recognizes and binds methyllysine residues on the target proteins leading to post-translational modifications. L3MBTL was shown to be associated with myeloid malignancies (27).
ix) SEC24D is a member of the SEC23/SEC24 family, which is involved in vesicle trafficking at the endoplasmic reticulum (28).
x) TMEM140 is the transmembrane protein 140, shown as a promising prognostic marker for patients with glioma where its overexpression strongly correlates with tumor size and overall patients' survival rates. Importantly, silencing TMEM140 suppressed the viability, migration, and invasion of glioma cells suggesting its importance as an attractive therapeutic target (29).
Discussion
In this study, we have identified a set of 10 genes relevant to interferon signaling. To date, interferon based therapy is widely used to treat ATL. Some of the identified genes might have wider scope of functions irrespective to the interferon-related signaling, per se. With the exception of SEC24D, which has very limited data, the remaining genes in the list has several roles in cancer progression through their overexpression or mutation. As a matter of interest, PARP-14 is of an exceptionally rising importance in cancer research and metabolism. PARP-14 and other molecular targets identified in this study can act as potential therapeutic targets for Cancer (18,30).
References
- 1.Coffin JM. The discovery of HTLV-1, the first pathogenic human retrovirus. Proc Natl Acad Sci USA. 2015;112:15525–15529. doi: 10.1073/pnas.1521629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Refaat A, Zhou Y, Suzuki S, Takasaki I, Koizumi K, Yamaoka S, Tabuchi Y, Saiki I, Sakurai H. Distinct roles of transforming growth factor-beta-activated kinase 1 (TAK1)-c-Rel and interferon regulatory factor 4 (IRF4) pathways in human T cell lymphotropic virus 1-transformed T helper 17 cells producing interleukin-9. J Biol Chem. 2011;286:21092–21099. doi: 10.1074/jbc.M110.200907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki S, Zhou Y, Refaat A, Takasaki I, Koizumi K, Yamaoka S, Tabuchi Y, Saiki I, Sakurai H. Human T cell lymphotropic virus 1 manipulates interferon regulatory signals by controlling the TAK1-IRF3 and IRF4 pathways. J Biol Chem. 2010;285:4441–4446. doi: 10.1074/jbc.M109.031476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, Halicka HD, Darzynkiewicz Z. Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage (DNA double-strand breaks) Curr Protoc Cytom Chapter. 2004;7 doi: 10.1002/0471142956.cy0727s30. Unit 7.27. [DOI] [PubMed] [Google Scholar]
- 7.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene. 2006;25:5906–5911. doi: 10.1038/sj.onc.1209873. [DOI] [PubMed] [Google Scholar]
- 9.Gadsby DC, Vergani P, Csanády L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones AM, Helm JM. Emerging treatments in cystic fibrosis. Drugs. 2009;69:1903–1910. doi: 10.2165/11318500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 11.Than BLN, Linnekamp JF, Starr TK, Largaespada DA, Rod A, Zhang Y, Bruner V, Abrahante J, Schumann A, Luczak T, et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene. 2017;36:3504. doi: 10.1038/onc.2017.3. [DOI] [PubMed] [Google Scholar]
- 12.Xie C, Jiang XH, Zhang JT, Sun TT, Dong JD, Sanders AJ, Diao RY, Wang Y, Fok KL, Tsang LL, et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32:2282–2291, 2291.e1-7. doi: 10.1038/onc.2012.251. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Yong M, Li J, Dong X, Yu T, Fu X, Hu L. High level of CFTR expression is associated with tumor aggression and knockdown of CFTR suppresses proliferation of ovarian cancer in vitro and in vivo. Oncol Rep. 2015;33:2227–2234. doi: 10.3892/or.2015.3829. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JT, Jiang XH, Xie C, Cheng H, Da Dong J, Wang Y, Fok KL, Zhang XH, Sun TT, Tsang LL, et al. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochim Biophys Acta. 2013;1833:2961–2969. doi: 10.1016/j.bbamcr.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Jhala N, Jhala D, Vickers SM, Eltoum I, Batra SK, Manne U, Eloubeidi M, Jones JJ, Grizzle WE. Biomarkers in Diagnosis of pancreatic carcinoma in fine-needle aspirates. Am J Clin Pathol. 2006;126:572–579. doi: 10.1309/CEV30BE088CBDQD9. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehrotra P, Riley JP, Patel R, Li F, Voss L, Goenka S. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J Biol Chem. 2011;286:1767–1776. doi: 10.1074/jbc.M110.157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iansante V, Choy PM, Fung SW, Liu Y, Chai JG, Dyson J, Del Rio A, D'Santos C, Williams R, Chokshi S, et al. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6:7882. doi: 10.1038/ncomms8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci USA. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, Hui J. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kume K, Iizumi Y, Shimada M, Ito Y, Kishi T, Yamaguchi Y, Handa H. Role of N-end rule ubiquitin ligases UBR1 and UBR2 in regulating the leucine-mTOR signaling pathway. Genes Cells. 2010;15:339–349. doi: 10.1111/j.1365-2443.2010.01385.x. [DOI] [PubMed] [Google Scholar]
- 23.Taylor PR, Brown GD, Reid DM, Willment JA, Martinez-Pomares L, Gordon S, Wong SY. The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. 2002;169:3876–3882. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- 24.Huysamen C, Brown GD. The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol Lett. 2009;290:121–128. doi: 10.1111/j.1574-6968.2008.01418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijo S, Iwakura Y. Dectin-1 and Dectin-2 in innate immunity against fungi. Int Immunol. 2011;23:467–472. doi: 10.1093/intimm/dxr046. [DOI] [PubMed] [Google Scholar]
- 26.Activation of Dectin-1 on Macrophages Promotes Pancreatic Cancer. Cancer Discov. 2017;7:549. [Google Scholar]
- 27.Li J, Bench AJ, Vassiliou GS, Fourouclas N, Ferguson-Smith AC, Green AR. Imprinting of the human L3MBTL gene, a polycomb family member located in a region of chromosome 20 deleted in human myeloid malignancies. Proc Natl Acad Sci USA. 2004;101:7341–7346. doi: 10.1073/pnas.0308195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagano A, Letourneur F, Garcia-Estefania D, Carpentier JL, Orci L, Paccaud JP. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7840. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Huang MZ, Wang XQ, Tao BB, Zhong J, Wang XH, Zhang WC, Li ST. TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion. J Hematol Oncol. 2015;8:89. doi: 10.1186/s13045-015-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camicia R, Winkler HC, Hassa PO. Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: A comprehensive review. Mol Cancer. 2015;14:207. doi: 10.1186/s12943-015-0474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]