Abstract
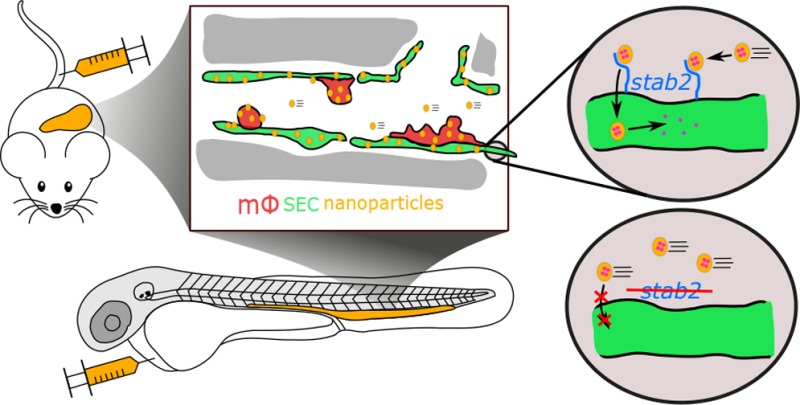
Up to 99% of systemically administered nanoparticles are cleared through the liver. Within the liver, most nanoparticles are thought to be sequestered by macrophages (Kupffer cells), although significant nanoparticle interactions with other hepatic cells have also been observed. To achieve effective cell-specific targeting of drugs through nanoparticle encapsulation, improved mechanistic understanding of nanoparticle–liver interactions is required. Here, we show the caudal vein of the embryonic zebrafish (Danio rerio) can be used as a model for assessing nanoparticle interactions with mammalian liver sinusoidal (or scavenger) endothelial cells (SECs) and macrophages. We observe that anionic nanoparticles are primarily taken up by SECs and identify an essential requirement for the scavenger receptor, stabilin-2 (stab2) in this process. Importantly, nanoparticle–SEC interactions can be blocked by dextran sulfate, a competitive inhibitor of stab2 and other scavenger receptors. Finally, we exploit nanoparticle–SEC interactions to demonstrate targeted intracellular drug delivery resulting in the selective deletion of a single blood vessel in the zebrafish embryo. Together, we propose stab2 inhibition or targeting as a general approach for modifying nanoparticle–liver interactions of a wide range of nanomedicines.
Keywords: endothelial cells, scavenger receptor, nanomedicine, liposomes, stabilin, zebrafish, targeted drug delivery
Cell-type specific targeting is a common goal in nanoparticle drug delivery. However, the inability to efficiently target subpopulations of cells, beyond the macrophages and monocytes of the mononuclear phagocyte system (MPS), has stymied progress of these technologies into clinical use.1−4 Up to 99% of systemically administered nanoparticles, of all shapes, sizes, and chemical compositions are cleared through the liver.5 While it is generally accepted that nanoparticles are taken up by liver-resident macrophages (Kupffer cells (KCs)),6 the principal cell type of the MPS in the liver, significant nanoparticle interactions with other hepatic cells, including liver sinusoidal endothelial cells (LSECs), hepatocytes, and hepatic B-cells, have also been observed.7−10 In these instances however, the cell-specific mechanisms underpinning these interactions have not been elucidated. A detailed understanding of exactly where and how nanoparticles are sequestered and cleared within the liver is crucial for the effective optimization of nanoparticle-mediated drug delivery.
The principle function of the liver is to maintain homeostasis. This includes the removal (“scavenging”) of macromolecular and colloidal waste and pathogens from the blood. Within the liver, scavenging function is primarily associated with the hepatic sinusoids,11 specialized blood vessels connecting the hepatic artery and portal vein (incoming blood flow) with the central vein (outgoing blood flow). In these vessels, scavenging function is facilitated by a >10-fold decrease in blood flow velocity.12 Hepatic sinusoids are primarily composed of LSECs (∼70%) and KCs (∼20%).13 Together these cells comprise the hepatic reticuloendothelial system (RES), a term originally proposed in the early 20th century by Aschoff14 to include specialized cells that accumulated vital stains. Since then, the term RES has been largely superseded by the MPS, which in the liver sinusoid includes KCs but not LSECs.
Cells with a scavenging function similar to mammalian LSECs have been identified in all vertebrates examined. However, in teleost fish, sharks, and lampreys these cells have not been found in the liver, but are identified in various other organs.15 Collectively, these cells are known as scavenger endothelial cells (SECs), a specialized endothelial cell type functionally defined as the major clearance site of endogenous macromolecules such as oxidized low-density lipoprotein (oxLDL) and hyaluronic acid (HA) from the blood.11 Mammalian LSECs have also been implicated in clearance of blood-borne viruses from circulation16−18 and are important cell-types of both the innate and adaptive immune system.19,20 In LSECs, clearance function is mediated through a relatively small number of pattern-recognition endocytosis receptors.11 Given the wide variety of macromolecules, colloids, and pathogens sequestered by LSECs, these receptors are clearly promiscuous with respect to potential binding partners. However, what general physicochemical properties direct materials to LSECs, to what extent are individual endocytosis receptors involved, and the significance of these interactions in the clearance of nanoparticles from circulation are not clearly defined.
Here, we show a specific part of the zebrafish embryonic vasculature displays functional homology to the mammalian liver sinusoid and includes macrophages/monocytes and functional SECs. Using this model, we are able to study which general properties of nanoparticles result in their uptake by each of these cell types after intravenous injection. For SECs, we reveal an important molecular mechanism required for nanoparticle clearance, involving the transmembrane receptor stabilin-2, which can be both inhibited and exploited to guide cell-specific nanoparticle-mediated drug delivery.
Results and Discussion
A Zebrafish Model for Liposome Biodistribution
Of the myriad nanoparticles reported as potential drug delivery vectors, liposomes are the most widely investigated and the major class of nanoparticles approved for clinical use.21,22 So far, the ability to predict the fate of liposomes following intravenous injection based on lipid composition alone has been limited. Furthermore, the opacity of mammalian models precludes comprehensive assessment of the dynamic behavior of liposomes in vivo. Recent studies have shown that the small and transparent zebrafish embryo allows for the direct observation of circulating nanoparticles, including liposomes, and their interactions with cells.23−26 These studies show key aspects of nanoparticle behavior, including uptake by the MPS, are conserved between zebrafish and mammals. We therefore selected this model to identify the influence of lipid composition on liposome biodistribution and the mechanisms of liposome uptake by cells.
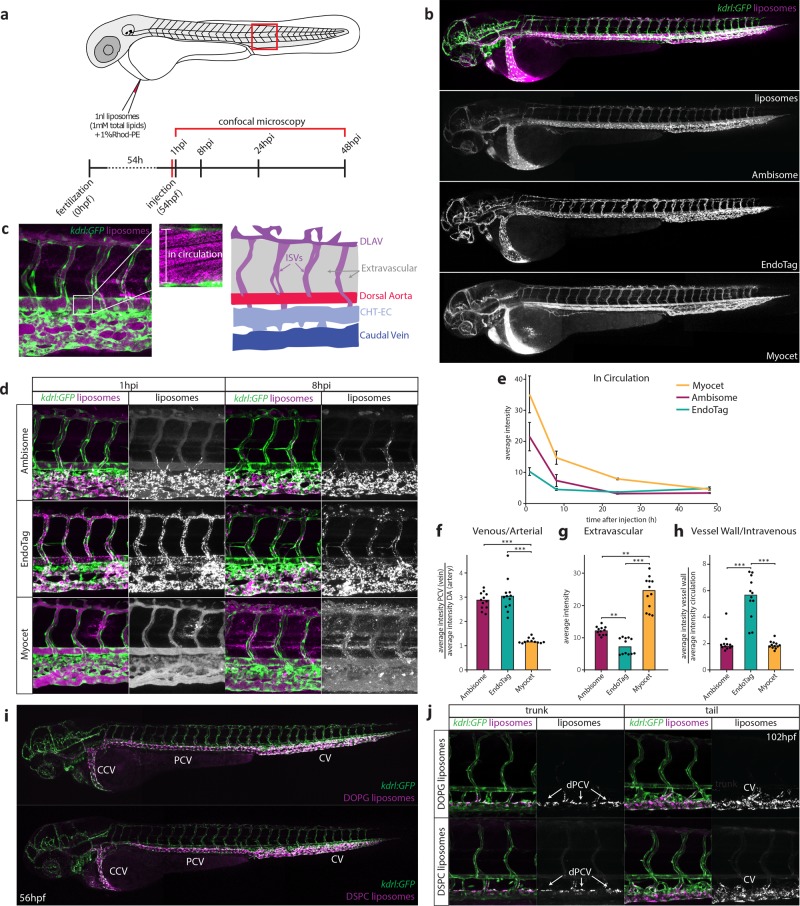
Three liposome formulations, either approved for clinical use or under development (Myocet, EndoTAG-1, and AmBisome),27−29 were initially selected for intravenous injection into zebrafish embryos. These formulations were specifically chosen to assess the influence of contrasting nanoparticle surface charge. Myocet is a neutral liposomal-doxorubicin formulation showing extravasation in tumors.27 EndoTAG-1 is a positively charged liposomal-paclitaxel formulation targeting actively growing tumor blood vessels.28 AmBisome is a negatively charged liposomal-amphotericin B formulation used to treat severe fungal infections.29 Fluorescently labeled liposomes (∼100 nm in diameter and without encapsulated drugs) based on the lipid composition of these formulations (Table S1) were injected intravenously into the duct of Cuvier of zebrafish embryos at 54 h post-fertilization (hpf), a stage at which most organ systems are established. Injected embryos were imaged using confocal microscopy at 1, 8, 24, and 48 h post-injection (hpi) (Figure 1a), and confocal micrographs were generated for the entire embryo (whole organism level) as well as from a region caudal to the cloaca (tissue level) (Figure 1b,d and Figure S1). We developed a quantification method to compare levels of circulating liposomes, extravasation, and accumulation in different blood vessel types between formulations (Figure 1c,e–h and Figure S2).
Figure 1.
A zebrafish model for liposome biodistribution. (a) Schematic of liposome injection and quantification in zebrafish. Fluorescently labeled liposomes (1 mM total lipids containing 1 mol % Rhod-PE) were injected into the duct of Cuvier at 54 hpf. Confocal microscopy is performed in a defined region (boxed) caudal to the yolk extension at 1, 8, 24, and 48 h after injection. (b) Whole-embryo view of liposome distribution in kdrl:GFP transgenic embryos, 1 hpi with three different liposome formulations (AmBisome, EndoTAG-1, and Myocet). (c) High-resolution imaging allows quantification of liposomes in circulation (measured in the lumen of the dorsal aorta (white box)) and liposome association with different blood vessel types (see Supporting Information). CHT-EC: caudal hematopoietic tissue endothelial cells, DLAV: dorsal longitudinal anastomotic vessel. ISV: intersegmental vessel. (d) Tissue level view of liposome distribution in kdrl:gfp transgenic embryos, 1 h and 8 h after injection with three different liposome formulations and a single confocal section through the dorsal aorta (DA) at 1 h after injection. (e) Quantification of liposome levels in circulation based on mean rhodamine fluorescence intensity in the lumen of the dorsal aorta at 1, 8, 24, and 48 h after injection (error bars: standard deviation.) n = 6 individually injected embryos per formulation per time point (in two experiments). (f) Quantification of liposome levels associated with venous vs arterial endothelial cells based on rhodamine fluorescence intensity associated with caudal vein (CV) vs DA at 8 h after injection. (g) Quantification of extravascular liposome levels based on rhodamine fluorescence intensity outside of the vasculature between the DLAV and DA at 8 h after injection. (h) Quantification of liposome levels associated with the vessel wall based on rhodamine fluorescence intensity associated with all endothelial cells relative to rhodamine fluorescence intensity in circulation at 1h after injection. (f–h) Bar height represents median values, dots represent individual data points, brackets indicate significantly different values (*: p < 0.05, **: p < 0.01, ***: p < 0.001) based on Kruskal–Wallis and Dunn’s tests with Bonferroni correction for multiple testing. n = 12 individually injected embryos per group (in 2 experiments). (i) Whole-embryo view of liposome distribution in kdrl:GFP transgenic embryos, 1 h after injection with DOPG and DSPC liposomes. Liposome accumulation for both formulations is observed in the primitive head sinus (PHS), common cardinal vein (CCV), posterior cardinal vein (PCV), and caudal vein (CV). (j) Tissue level view of liposome distribution in kdrl:GFP transgenic embryos, 1 h after injection with DOPG and DSPC liposomes at 102 hpf. Liposome accumulation is observed in the entire caudal vein (CV), but only on the dorsal side of the PCV (dPCV, arrows).
At 1 hpi, on a whole organism level, all three liposome formulations were found associated with the blood vasculature and over time, the fluorescence associated with freely circulating liposomes within the lumen of the dorsal aorta, decayed exponentially (Figure 1b,e). At the tissue level however, clear differences in liposome biodistribution were observed (Figure 1d). Consistent with their behavior in mammals, neutral Myocet liposomes were mostly seen circulating within the blood vessel lumen. At 1 hpi, liposome translocation through the vessel wall (extravasation) was already evident, and between 1 and 8 hpi, co-localization with plasma-exposed macrophages was observed (Figure 1d,g, Figure S3). Increasing the size of Myocet liposomes resulted in enhanced uptake by macrophages, whereas surface PEGylation—a strategy widely employed to limit nanoparticle clearance in vivo(30)—effectively inhibited phagocytotic uptake as described previously (Figure S3).23,26
For EndoTAG-1 and AmBisome, a large fraction of the injected dose was removed from circulation by 1 hpi and 8 hpi respectively, and these formulations were found associated with the vessel wall (Figure 1e,h). Strikingly however, anionic AmBisome liposomes associated only with ECs of a subset of blood vessels, namely the caudal vein (CV), the posterior and common cardinal veins (PCV and CCV), and the primary head sinus (PHS) as well as ECs within the caudal hematopoietic tissue (CHT-ECs) (Figure 1d,f–h).31 These comprise the majority of venous ECs within the zebrafish embryo at this developmental stage.32 Cationic EndoTAG-1 liposomes at 1 hpi associated with all ECs as expected33 but at later time points remain associated only with venous ECs.
AmBisome, EndoTAG-1, and Myocet are each composed of various mixtures of (phospho)lipids and cholesterol. In these cases, lipid headgroup chemistries, fatty acid chain saturation and cholesterol content, will together combine to affect the overall physicochemical character of the formulated liposomes and consequently their in vivo fate. To limit potential variation in liposome membrane composition, we next formulated and injected ∼100 nm liposomes composed of the individual (phospho)lipids constituting AmBisome, EndoTAG-1, and Myocet (Figure S4 and Table S1). We also included liposomes composed of 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG) and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). In these experiments, injected cationic liposomes (measured zeta potential; >30 mV) initially associated with both arterial and venous ECs of the embryonic fish. All anionic liposomes (<−30 mV) associated with venous ECs alone, and the behavior of neutral liposomes was dependent on lipid fatty acid chain saturation, whereby “fluid” liposome membranes (for example, DOPC), rich in unsaturated lipids, are freely circulating, whereas those composed of ‘rigid’, saturated lipids (for example, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)) associated with venous ECs. Of these, liposomes composed of DSPC and DOPG associated with venous ECs of the CCV, PHS, PCV, CHT, and CV most strongly (Figure 1i, Figure S4a,d). Both these liposomes also accumulated in macrophages within the CHT and along the CCV (Figure S5).
Differential distribution of nanoparticles over blood vessel networks has previously been attributed to differences in flow patterns.7,25 However, when injections were performed in 4 day-old zebrafish embryos, both DOPG and DSPC liposomes preferentially associated with only a subset of venous ECs along the dorsal side of the PCV (dPCV) (Figure 1j). Liposome association with a subset of ECs in a single, straight blood vessel (where flow patterns are expected to be similar throughout) indicated dPCV ECs are a cell type distinct from ventral PCV (vPCV) ECs. Indeed, differentiation of dPCV and vPCV ECs has previously been observed during the induction of lymphatic differentiation and subintestinal vein angiogenesis,34,35 suggesting dPCV differentiation may lead to the expression of specific receptors by these ECs which in turn could mediate the selective binding of DOPG and DSPC liposomes.
Identification of a Zebrafish EC Type Homologous to Mammalian LSECs
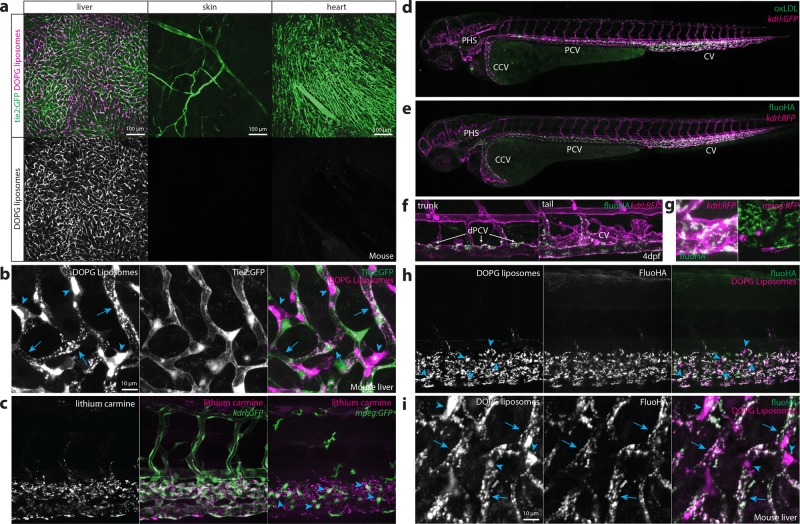
Selective association of liposomes with most venous ECs has not been observed in adult mammals. However, we hypothesized a more restricted subset of ECs in mammals could be functionally related to venous ECs of the embryonic zebrafish. To test this hypothesis, DOPG liposomes were injected intravenously into Tie2:GFP+ adult mice. In these mice, liposomes were removed from circulation within 1 hpi, and a striking accumulation was observed in the liver (Figure 2a). Within the liver, liposomes associated with Tie2:GFP+ sinusoidal ECs and with cells identified as KCs based on cell shape and intravascular localization (Figure 2b). No liposome accumulation was observed in hepatocytes or other analyzed organs. This suggested venous ECs and macrophages within the CHT and CV of the embryonic zebrafish were functionally homologous to LSECs and KCs of the mammalian liver and comprise the RES in zebrafish embryos. To confirm this, we injected colloidal lithium carmine (Li-Car), the most prominent vital stain originally used to define the mammalian RES, into zebrafish embryos. Making use of the inherent fluorescence of carminic acid,36 we observed accumulation of this colloid in the same blood vessels (CV, CHT, PCV, and PHS) and subcellular structures within venous ECs and macrophages, in which DOPG and DSPC liposomes also accumulate (Figure 2c).
Figure 2.
Identification of scavenger endothelial cells (SECs) in zebrafish embryos. (a, b) Ex vivo imaging of adult Tie2:GFP transgenic mouse organs, 1 h after injection with DOPG liposomes. (a) Liposome accumulation is observed in liver, but not in the ear skin or heart muscle. (b) Within the liver, DOPG liposomes are observed as punctae within Tie2:GFP+ sinusoidal ECs (arrows) as well as sinusoid-associated cells which based on shape and position were identified as KCs (arrowheads). (c) Tissue level view of lithium carmine distribution in kdrl:GFP and mpeg:GFP transgenic zebrafish embryos, 1 h after injection. Lithium carmine (carminic acid) fluorescence co-localizes both with kdrl:GFP+ endothelial cells in the caudal vein and mpeg:GFP+ monocytes/macrophages (arrowheads) within the CHT. (d) Whole-embryo view of fluorescent oxLDL distribution in kdrl:GFP transgenic embryos, 1 h after injection. Accumulation of oxLDL is observed in the PHS, CCV, PCV, and CV. (e) Whole-embryo view of fluoHA distribution in kdrl:RFP transgenic embryos, 1 h after injection. Accumulation of fluoHA is observed in the PHS, CCV, PCV, and CV. (f) Tissue level view of fluoHA distribution in kdrl:RFP transgenic embryos, 1 h after injection at 102 hpf. FluoHA accumulation is observed in the entire caudal vein (CV), but only on the dorsal side of the PCV (dPCV, arrows). (g) Tissue level view of fluoHA in kdrl:RFP and mpeg:RFP transgenic embryos. Co-localization of RFP expression and fluoHA is observed only within kdrl:RFP endothelial cells, but not mpeg:RFP monocytes/macrophages. (h) Tissue level view of co-injected fluoHA and DOPG liposomes, 1 h after injection reveals co-localization in SECs. Monocytes/macrophages (arrowheads) take up DOPG but not fluoHA. (i) Ex vivo imaging of adult mouse liver, 1 h after injection with fluoHA and DOPG liposomes reveals widespread co-localization within sinusoidal ECs (arrows). KCs (arrowheads) take up DOPG liposomes only.
A small number of transmembrane receptors are selectively expressed in mammalian LSECs compared to other blood vascular ECs.11 These include the scavenger receptors Stabilin-1 and -237 and the mannose receptor Mrc1. Analysis of the expression patterns of their orthologs (stab1, stab2 and mrc1a) in zebrafish embryos confirmed their restricted expression in venous ECs of the PHS, PCV, CHT, and CV as described previously.38,39 Importantly, expression of these genes becomes enriched in the dPCV, matching observed EC binding specificities of both DOPG and DSPC liposomes (Figure S6).
LSECs mediate the scavenging of macromolecular waste including oxLDL and HA through receptor-mediated endocytosis.40 Therefore, we injected fluorescently labeled oxLDL and HA (fluoHA) and observed their rapid endocytosis, within the same subset of venous ECs (within the PHS, CCV, (d)PCV, and CV) (Figure 2d–f). Based on the conserved uptake of DOPG liposome, oxLDL, fluoHA, and Li-Car from circulation and expression of known LSEC markers by this venous EC subset in zebrafish embryos, we define them as SECs - homologous to mammalian LSECs.
In contrast to DSPC and DOPG liposomes and to oxLDL, fluoHA uptake was specific to SECs, and no uptake was observed in macrophages (Figure 2g). We next used fluoHA as a marker for endocytosis in SECs. Co-injection of fluoHA with DSPC or DOPG liposomes resulted in precise intracellular co-localization in all SECs of the embryonic fish, while in macrophages only liposome internalization was observed (Figure 2h, Figure S7). Intracellular co-localization in LSECs (but not KCs) of fluoHA and DOPG liposomes was conserved in the adult mouse liver (Figure 2i). These results demonstrated fluoHA endocytosis is a selective vital marker for SECs in vertebrates and offered a convenient method to study SEC differentiation in the developing zebrafish embryo (Figure S8). Importantly, we found SECs were present at the earliest time point at which intravenous injection is possible (28 hpf). During embryonic and larval stages, SECs were maintained within the CV, but starting at 52 hpf became gradually restricted to the dPCV. No fluoHA uptake was observed in embryonic veins that develop during later stages, such as in the brain and subintestinal vasculature. These results show that SECs are one of the first EC subtypes to emerge during embryonic development and provide the first analysis of early embryonic SEC differentiation in any vertebrate.
Stabilin-2 Is Required for Uptake of Liposomes and Other Nanoparticles by SECs
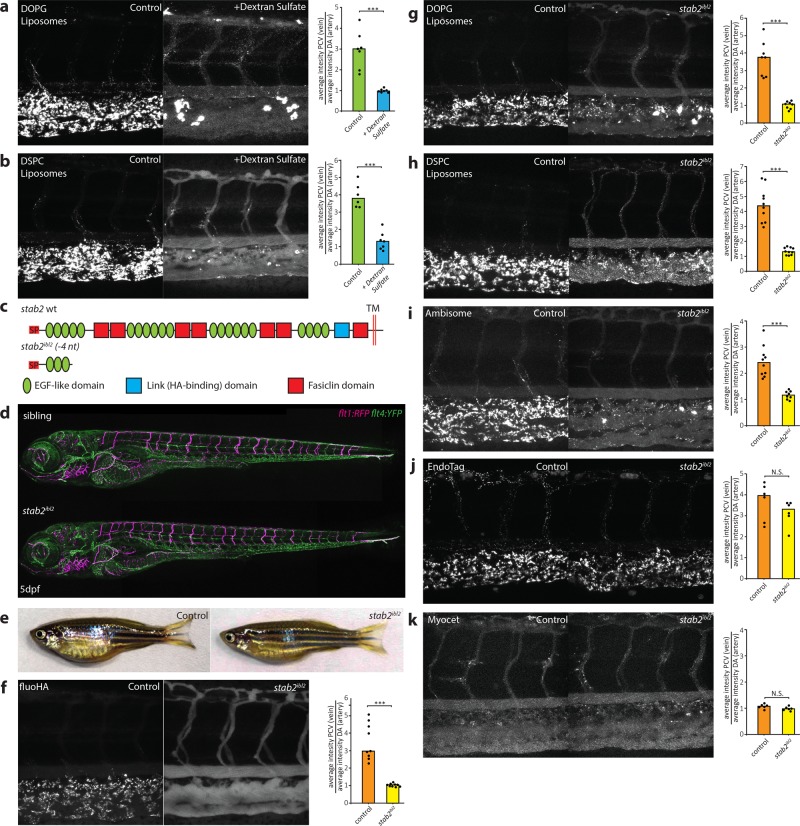
The precise intracellular co-localization of fluoHA with DOPG and DSPC liposomes in SECs indicated the use of a shared receptor for endocytosis. Importantly, one of the markers for SECs in zebrafish embryos and adult mammals, Stabilin-2, has been identified as the main HA clearance receptor in the mouse liver.40In vitro, Stabilin-2 and its paralog Stabilin-1 have been shown to bind to a large variety of endogenous (mostly anionic) macromolecules41 as well as phosphothiorate-modified antisense oligonucleotides (PS-ASO),42 apoptotic cell bodies,43 biotinylated albumin,44 and carbon nanotubes.45In vivo, Stabilin-1 and Stabilin-2 were shown to mediate sequestration (but not uptake) by LSECs of aged erythrocytes in a phosphatidylserine-dependent manner.46 Stabilin-1 and Stabilin-2 are both nonessential genes for development and normal physiology in mice, with mice lacking both Stabilin-1 and Stabilin-2 displaying deficient removal of nephrotoxic macromolecules from circulation.37 To test if stabilins were involved in liposome uptake by SECs, embryos were first pretreated with dextran sulfate - a competitive inhibitor of scavenger receptors, including stab1 and stab2.47,48 Subsequent liposome injection (or co-injection) resulted in a striking loss of liposome uptake by SECs, offset by an increase in circulating liposomes, and particularly in the case of DOPG liposomes, an increase in macrophage uptake (Figure 3a,b). In contrast, injection of mannan, a competitive inhibitor of mrc1a,49 did not inhibit liposome uptake by SECs (data not shown).
Figure 3.
stab2 is required for anionic liposome uptake by SECs. (a, b) Tissue level view of DOPG (a) and DSPC (b) liposome distribution at 1 hpi in control and dextran sulfate injected embryos, with quantification of liposome levels associated with venous vs arterial endothelial cells based on rhodamine fluorescence intensity associated with CV vs DA. (c) stab2 domain structure predicted to be expressed from the wild-type stab2 and the stab2ibl2 allele. (d) Whole-embryo view of flt1:RFP, flt4:YFP double transgenic embryos at 5 dpf to visualize blood vascular and lymphatic development. No defects were identified during (lymph)angiogenesis and vascular patterning in stab2ibl2 homozygous embryos compared to sibling controls. (e) Fertile adult females (stab2ibl2 homozygous and sibling controls) at 3 months post-fertilization. (f–k) Tissue level view of fluoHA (f) and DOPG (g), DSPC (h), AmBisome (i), EndoTAG-1 (j), and Myocet (k) liposome distribution at 1 hpi in stab2ibl2 and sibling control embryos, with quantification of liposome levels associated with venous vs arterial endothelial cells based on rhodamine fluorescence intensity associated with CV vs DA. (a, b, f–k) Bar height represents median values, dots represent individual data points, and brackets indicate significantly different values (*: p < 0.05, **: p < 0.01, ***: p < 0.001, N.S.: not significant) based on Mann–Whitney test. n = 6–10 per group (in two experiments).
To identify the specific role of stab1 and stab2 in liposome uptake, mutants for both genes were generated through CRISPR/Cas9-mediated mutagenesis. Here, we report the analysis of a stab2 mutant line, in which we identified a 4nt deletion (stab2ibl2), leading to a frameshift in the stab2 coding sequence and a premature stop codon (C233X) (Figure 3c, Figure S9). This mutation is predicted to remove most conserved stab2 domains including all fasiclin domains, the HA binding Link domain, and the transmembrane and cytoplasmic segments. Homozygous stab2ibl2 mutants displayed a strong reduction of stab2, but not of stab1 or mrc1a, mRNA expression indicating normal SEC differentiation and nonsense-mediated decay of stab2ibl2 mRNA (Figure S10). Stab2ibl2 mutants survived throughout embryonic development without defects in either blood or lymphatic vascular systems, which were described previously for stab2 morphants,50,51 and fertile adults were identified in normal Mendelian ratios (Figure 3d,e). Consistent with the increase in circulating HA levels observed in mouse Stab2 knockouts,52 a complete loss of fluoHA uptake by SECs was observed in zebrafish stab2ibl2 mutants, showing a conserved role for stab2 in HA clearance in vertebrates (Figure 3f). Importantly, when either DOPG or DSPC liposomes were injected in stab2ibl2 mutants, a strong reduction of liposome endocytosis by SECs was observed, offset by an increase in circulating liposome levels and an increase in macrophage uptake (Figure 3g,h). Differential liposome uptake in neighboring venous ECs of embryos with a mosaic loss of stab2 function indicated a cell-autonomous role of stab2 function in liposome uptake by SECs (Figure S11). For the original three liposome formulations screened, loss of stab2 function affected AmBisome, but not Myocet or EndoTAG-1 biodistribution (Figure 3i–k). Since both AmBisome and EndoTAG-1 accumulated within SECs of wild-type embryos, stab2-mediated uptake by SECs appears dependent on specific physicochemical properties of liposomes and stab2 does not function in the clearance of cationic liposomes.
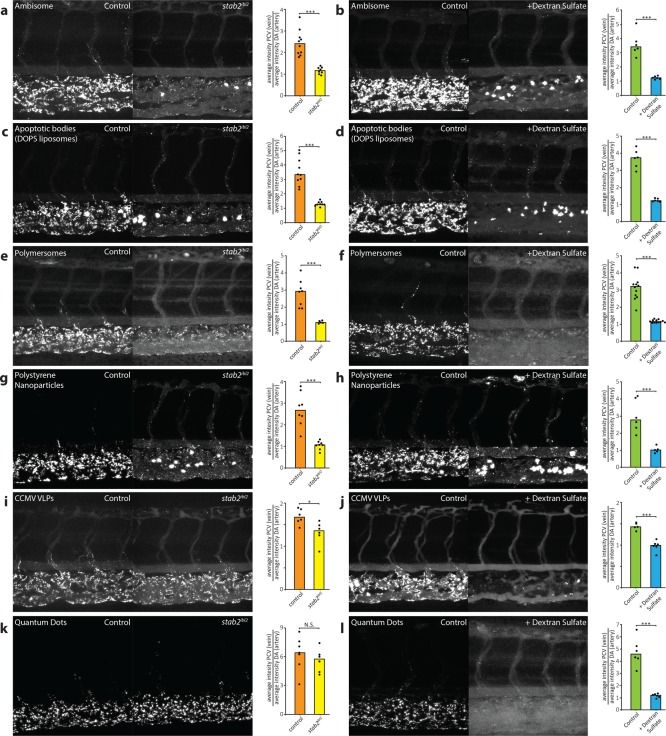
In vivo, several other scavenger receptors with similar binding profiles to stab2 are expressed,11 not only on SECs but also on other endothelial cells and macrophages. Given the significant increase in circulating DOPG, DSPC, and AmBisome liposomes in stab2ibl2 mutants, stab2 clearly plays a dominant role in removal of these liposomes from circulation compared to other scavenger receptors (including the structurally related stab1). Similarly, clearance of PS-ASOs was recently shown to be dominated by Stab2 in the mouse liver.42 To test the generality of stab2 function, several other polyanionic nanoparticles were injected in wild-type and stab2ibl2 mutant embryos as well as following dextran sulfate injection (Figure 4a–l). These included endogenous (DOPS liposomes, a model for apoptotic cell fragments), viral (Cowpea Chlorotic Mottle Virus-like particles, CCMV VLPs),53 polymeric (polymersomes54 and polystyrene beads), and inorganic (quantum dots, QDs) nanoparticles. All of these particles were endocytosed selectively by SECs in zebrafish embryos, and in all cases SEC endocytosis could be inhibited by dextran sulfate. However, not all nanoparticles were dependent on stab2 for SEC endocytosis. Although uptake by SECs of DOPS liposomes, polymersomes, and polystyrene nanoparticles was strongly decreased in stab2ibl2 mutants, uptake of CCMV VLPs was only partly dependent on stab2 and QD uptake appeared stab2-independent. Alternatively, QD uptake by SECs is also mediated in part by stab2, but its function is masked in stab2ibl2 mutants through redundancy with other scavenger receptors (such as stab1) that can be inhibited by dextran sulfate. CCMV VLPs (28 nm) and QDs (<10 nm) were the smallest nanoparticles screened in this study, suggesting size may be an important determinant of scavenger receptor–nanoparticle interactions.
Figure 4.
stab2-mediated scavenging of anionic nanoparticles in vivo. (a–i) Tissue level view of DOPS liposome (a, b), PIB-PEG polymersome (c, d), carboxylated polystyrene nanoparticle (e, f), CCMV virus-like particle (g, h), and carboxylated quantum dot (i, j) distribution at 1 hpi in stab2ibl2 and sibling control embryos (a, c, e, g, i) or control and dextran sulfate injected embryos (b, d, f, h, j). Quantification of nanoparticle levels associated with venous vs arterial endothelial cells based on rhodamine fluorescence intensity associated with caudal vein vs DA. (a–j) Bar height represents median values, dots represent individual data points, and brackets indicate significantly different values (*: p < 0.05, **: p < 0.01, ***: p < 0.001, N.S.: not significant) based on Mann–Whitney test. n = 5–12 per group (in two experiments).
Targeted Liposomal Drug Delivery to SECs
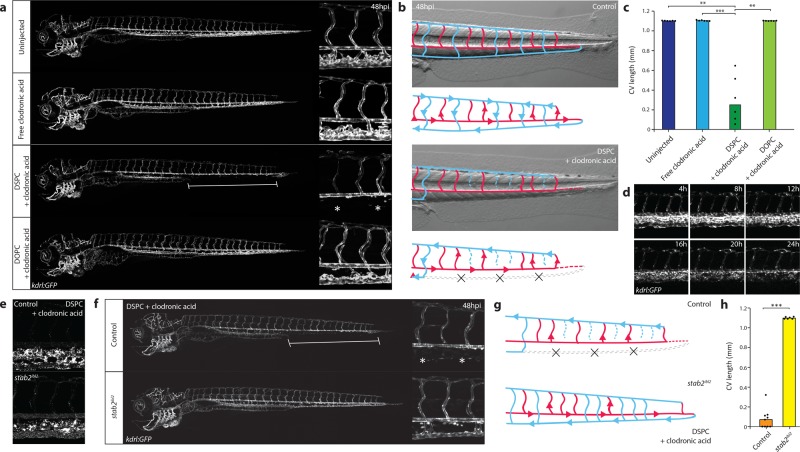
Finally, to demonstrate we could extend the observed interaction of nanoparticles with SECs to cell-selective drug delivery, we encapsulated a model drug, clodronic acid, within DSPC liposomes (Table S2). Clodronic acid requires active transport (endocytosis or phagocytosis) across the target cell membrane to illicit a cytotoxic effect.55 Liposome-mediated intracellular delivery of clodronic acid into monocytes/macrophages is used extensively as a research tool to selectively remove these cell populations in vivo.56 After 12–24 hpi, synchronous changes in the morphology of the CHT and caudal vein ECs were observed, followed by a gradual loss of kdrl:GFP+ endothelial cells or cell fragments and ultimately leading to the complete disappearance of the caudal vein between 24 and 48 hpi (Figure 5a–d, Movie S1 and S2). The PCV and other cell types within the CHT, including mpeg:GFP+ macrophages (most of which are not exposed to circulating nanoparticles) as well as mpx:GFP+ neutrophils, were largely unaffected (Figure S12). Injection of free clodronic acid (a control demonstrating the requirement of liposomal encapsulation) did not result in any observable changes to the venous endothelium. Similarly, injection of freely circulating DOPC-clodronic acid liposomes (a control demonstrating the requirement of selective nanoparticle uptake by SECs) did not affect the venous endothelium. The development of the dorsal aorta was unaffected by deletion of the CV and CHT, and blood supply to the caudal parts of the embryo was maintained through a rerouting of blood cells into the intersegmental vessels and dorsal longitudinal anastomotic vessel (DLAV). Embryos with a complete loss of the CV and CHT endothelial cells were agile and could survive at least until 6 dpf. Imaging of fluorescent DSPC-clodronic acid liposomes revealed selective stab2-dependent uptake by SECs analogous to empty DSPC liposomes (Figure 5e). Importantly, loss of stab2 function as observed in stab2ibl2 mutant embryos rescued the CV phenotype induced by injection of DSPC-clodronic acid liposomes (Figure 5 f-h, Movie S3). These results identify stab2-mediated uptake of liposomes by SECs as a simple strategy for intracellular compound delivery to this cell type in zebrafish embryos.
Figure 5.
Nanoparticle-mediated SEC deletion. (a) Whole-embryo and tissue level views at 48 hpi of the blood vasculature in kdrl:GFP transgenic control embryos, embryos injected with 1 mg/mL clodronic acid, or embryos injected with liposomes containing 1 mg/mL clodronic acid (DSPC or DOPC liposomes). Complete deletion of the caudal vein is observed in embryos injected with DSPC liposomes containing clodronic acid (brackets and asterisks). (b) Schematic representation of blood flow in control embryos or embryos injected with DSPC liposomes containing 1 mg/mL clodronic acid. Blue indicates venous or capillary blood vessels, and red indicates arterial blood vessels. Arrowheads indicate direction of blood flow (based on observations from Movie S1). The removal of the CV (dashed lines) leads to a rerouting of blood flow through the DLAV. (c) Quantification of PCV length in injected embryos. Bar height represents median values, dots represent individual data points, and brackets indicate significant values (**: p < 0.01, ***: p < 0.001) based on Kruskal–Wallis and Dunn’s tests with Bonferroni correction for multiple testing. n = 6 individually injected embryos per group (in two experiments). (d) Progression of SEC deletion. Individual frames from Movie S2 at indicated time points after injection of DSPC liposomes containing 1 mg/mL clodronic acid, injected into kdrl:GFP transgenic embryos. SEC fragmentation in this case is observed mostly between 12 hpi and 16 hpi, followed by a gradual loss of fluorescence or removal of cellular debris. (e) Tissue level view of distribution of DSPC liposomes containing 1 mg/mL clodronic acid at 1 hpi in stab2ibl2 and sibling control embryos. (f) Whole-embryo and tissue level views at 48 hpi of the blood vasculature in kdrl:GFP transgenic stab2ibl2 and sibling embryos. Embryos were injected with DSPC liposomes containing 1 mg/mL clodronic acid. Complete deletion of the caudal vein is observed in sibling control (brackets and asterisks), but not stab2ibl2 mutant embryos. (g) Schematic representation of blood flow in sibling control embryos or stab2ibl2 homozygous mutants, both injected with DSPC liposomes containing approximately 1 mg/mL clodronic acid. Blue indicates venous or capillary blood vessels, and red indicates arterial blood vessels. Arrowheads indicate direction of blood flow (based on observations from Movie S3). The removal of the CV (dashed lines) leads to a rerouting of blood flow through the DLAV in control embryos but not in stab2ibl2 homozygous mutants. (h) Quantification of PCV length in injected embryos. Bar height represents median values, dots represent individual data points, and brackets indicate significant values (***: p < 0.001) based on Mann–Whitney test.
Conclusion
In summary, we show stab2 is an important (scavenger) receptor mediating the uptake of circulating nanoparticles by SECs. In particular, anionic nanoparticles, between 50 and 250 nm in size, are avidly taken up by SECs in a stab2-dependent fashion. Here, binding and uptake appear independent of material and functional properties of nanoparticles and are solely dependent on surface charge. Given the comparable sizes and surface charge of many blood-borne viruses,16−18,57 clearance of these circulating pathogens by LSECs is also potentially mediated by stab2. These findings, combined with the high expression of stab2 by LSECs within the mammalian liver,11 implicate SECs as an important cell-type in the binding, uptake, and clearance of administered nanoparticles. As such, we support the re-adoption of the RES, over the MPS, as the most accurate term to describe the specialized cellular components involved in nanoparticle clearance from circulation.58
The ultimate goal of many nanoparticle-based technologies is cell-type-specific targeting. Yet reported targeting efficiencies rarely surpass 1% of the total injected nanoparticle dose.1 A major contributing factor has been off-target nanoparticle interactions within the mammalian liver.5 By revealing the molecular basis of nanoparticle interactions with specific cells of the embryonic zebrafish, we have been able to demonstrate nanoparticle targeting of, and drug delivery to, specific cell types with homologues in the mammalian liver. In addition, we show these interactions can be effectively inhibited by dextran sulfate. As stab2 is not essential for normal adult physiology,37 this offers a simple method to extend circulation lifetimes of nanoparticles by minimizing potential off-target liver interactions.59 This will likely be particularly beneficial in instances where active targeting of nanoparticles to cell types beyond the liver (for example, cancer cells) is desired.
Importantly, the SEC/selective drug delivery we describe has not resulted from adding further complexity to nanoparticle designs. Instead, through systematic screening of “simple” nanoparticles (i.e., liposomes composed of a single phospholipid), we have established what general properties and molecular mechanisms direct nanoparticles to specific cell types. The use of the embryonic zebrafish as a model organism, and the ability to visualize nanoparticle–cell interactions at high resolution in living organisms, has been essential in this process. We therefore propose that the embryonic zebrafish, with its established extensive genetic toolkit, is a valuable preclinical in vivo model allowing screening, optimization, and mechanistic understanding of nanoparticle biodistribution, predictive of their behavior in mammals.26
Materials and Methods
Reagents
Fluorescein-labeled hyaluronic acid (fluoHA) was prepared through conjugation of hyaluronic acid (100 kDa) with fluorescein isothiocyanate (Isomer I, Sigma-Aldrich) as previously described.60 Additional fluoHA was provided as a kind gift from W. Jiskoot (Leiden University, The Netherlands). Colloidal Li-Car was prepared as previously described.61 Rhodamine-loaded polymersomes on polyisobutadiene/polyethylene glycol (PIB/PEG) block copolymers54 were a kind gift from S. Askes and S. Bonnet (Leiden University, The Netherlands). Atto-647 labeled CCMV-virus-like particles (t = 3, 28 nm)53 were a kind gift from M. de Ruiter and J. Cornelissen (Twente University, The Netherlands). Purchased reagents are described in the Supporting Information.
Liposome Preparation and Characterization
All liposomes (without encapsulated drugs) were formulated in ddH2O at a total lipid concentration of 1 mM. Individual lipids, as stock solutions (1–10 mM) in chloroform, were combined at the desired molar ratios and dried to a film, first under a stream of N2 and then >1h under vacuum. With the exception of Myocet 325 and 464 nm, lipid films were hydrated in 1 mL ddH2O at >65 °C (with gentle vortexing if necessary) to form large/giant multilamellar vesicles. Large unilamellar vesicles were formed through extrusion above the Tm of all lipids (>65 °C, Mini-extruder with heating block, Avanti Polar Lipids, Alabaster, US). Hydrated lipids were passed 11 times through 2 × 400 nm polycarbonate (PC) membranes (Nucleopore Track-Etch membranes, Whatman), followed by 11 times through 2 × 100 nm PC pores. All liposomes were stored at 4 °C. With the exception of DSPC liposomes (significant aggregation after 1 week storage), all liposomes were stable for at least 1 month. Myocet 325 and 464 nm liposomes were formulated by gentle hydration of lipid films at 35 °C (without vortexing). In the case of 464 nm Myocet liposomes, hydrated lipids were passed through a 800 nm PC membrane 7 times at 35 °C. In the case of 325 nm Myocet liposomes, hydrated lipids were passed through a 400 nm PC membrane 7 times at 35 °C. See Supporting Information for nanoparticle characterization methods and Table S1 for all lipid compositions, size, and zeta potentials of nanoparticles used in this study.
Clodronic Acid Encapsulation and Quantification
Lipid films (10 mM total lipids) were hydrated with ddH2O containing 200 mgmL–1 clodronic acid (1 mL) and formulated through extrusion as described for the corresponding “empty” liposomes. Unencapsulated clodronic acid was removed by size exclusion chromatography (illustra NAP Sephadex G-25 DNA grade premade columns (GE Healthcare) used according to the supplier’s instructions). Eluted clodronic acid-encapsulated liposomes were diluted 2.5× during SEC and injected without further dilution. Quantification of encapsulated clodronic acid was determined by UV absorbance as previously reported.62 Briefly, liposomes were first destroyed through a 1:1 dilution with 1% v/v Triton X-100 solution before further dilution into an acidic CuSO4 solution (1:2.25:2.25; Liposome-Triton X-100 mix: 3 mM HNO3: 4 mM CuSO4). The concentration of clodronic acid was determined by UV absorbance (Cary 3 Bio UV–vis spectrometer) at 240 nm and quantified against a predetermined calibration curve (50 μM to 2.5 mM clodronic acid). All UV–vis absorbance measurements were taken at room temperature. Blanks were made using liposome solutions without encapsulated clodronic acid but prepared otherwise identically (including SEC procedure). The final encapsulated clodronic acid concentration varied between 0.9 and 1.7 mg mL–1 (see Supporting Information Table S2).
Zebrafish Strains, in Situ Hybridization, and CRISPR/Cas9 Mutagenesis
Zebrafish (Danio rerio, strain AB/TL) were maintained and handled according to the guidelines from the Zebrafish Model Organism Database (http://zfin.org) and in compliance with the directives of the local animal welfare committee of Leiden University. Fertilization was performed by natural spawning at the beginning of the light period, and eggs were raised at 28.5 °C in egg water (60 ug/mL Instant Ocean sea salts). The following previously established zebrafish lines were used Tg(kdrl:GFP)s843,63Tg(kdrl:RFP-CAAX)s916,38Tg(mpeg:GFP)gl22,64Tg(mpeg:RFP-CAAX)ump2,65Tg(flt1enh:RFP)hu5333,66Tg(flt4BAC:YFP)hu7135,67 and Tg(mpx:GFP)uwm1.68 Whole-mount in situ hybridization was performed as described.69Supporting Information Table S3 lists primers for probe generation. Cloning-free sgRNAs for CRISPR/Cas9 mutagenesis were designed and synthesized as described.70 sgRNAs (125 pg) and cas9 mRNA (300 pg) were co-injected into single-cell wild-type, albino or flt4:YFP; flt1:RFP transgenic embryos. Mutagenesis efficacy, founder identification, and genotyping were performed using CRISPR-STAT.71 The nucleotide sequences and predicted stab2 amino acid sequences in the stab2ibl2 line are shown in Figure S11. Table S3 lists guide RNA sequences and genotyping primers. For mosaic analysis, heterozygous embryos (stab2ibl/+) obtained from a cross between a stab2ibl2 homozygous parent and a kdrl:GFP (stab2+/+) parent were co-injected with sgRNAs (125 pg) and cas9 mRNA (300 pg) to create second-hit mutations in the wild-type allele.
Zebrafish Intravenous Injections
Liposomal formulations were injected into 2 day old zebrafish embryos (52–56 hpf) using a modified microangraphy protocol.72 Embryos were anesthetized in 0.01% tricaine and embedded in 0.4% agarose containing tricaine before injection. To improve reproducibility of microangiography experiments, 1 nL volumes were calibrated and injected into the sinus venosus/duct of Cuvier. We created a small injection space by penetrating the skin with the injection needle and gently pulling the needle back, thereby creating a small pyramidal space in which the liposomes and polymers were injected. Successfully injected embryos were identified through the backward translocation of venous erythrocytes and the absence of damage to the yolk ball, which would reduce the amount of liposomes in circulation. For injections at later stages (>80 hpf), 0.5 nL volumes were injected into the CCV. The following concentrations were injected: dextran sulfate (20 mg/mL), FluoHA (0.2 mg/mL), oxLDL (1 mg/mL), CCMV-VLP (1 mg/mL), QDs (1:25 dilution), lithium carmine (1:50 dilution), polymersomes (1 mg/mL), latex beads (1:10 dilution). Dextran sulfate was injected 20 min prior to nanoparticle injection.
Zebrafish Imaging and Quantification
For each treatment or time point, at least six individual embryos (biological replicates) using at minimum two independently formulated liposome preparations were imaged using confocal microscopy. Embryos were randomly picked from a dish of 20–60 successfully injected embryos (exclusion criteria were: no backward translocation of erythrocytes after injection and/or damage to the yolk ball). Confocal z-stacks were captured on a Leica TCS SPE confocal microscope, using a 10× air objective (HCX PL FLUOTAR) or a 40× water-immersion objective (HCX APO L). For whole-embryo views, 3–5 overlapping z-stacks were captured to cover the complete embryo. Laser intensity, gain, and offset settings were identical between stacks and sessions. Images were processed and quantified using the Fiji distribution of ImageJ.73,74 Quantification (not blinded) of liposome biodistribution was performed on 40× confocal z-stacks (with an optical thickness of 2 μm/slice) as described in the Supporting Information.
Mouse Injections and Imaging
All experiments were performed in accordance with the guidelines of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences, The Netherlands. Tg(TIE2GFP)287Sato/J mice were sedated using isoflurane inhalation anesthesia (1.5–2% isoflurane/O2 mixture). 100 μL of DOPG liposomes (10 mM DOPG + 1% Rhod-PE) diluted 1:5 in PBS were injected retro-orbitally with an insulin syringe (BD). After 1 h, mice were sacrificed, and organs were harvested and imaged ex vivo on glass bottom dishes. Images were taken with a Leica SP8 multiphoton microscope with a chameleon Vision-S (Coherent Inc.), equipped with four HyD detectors: HyD1 (<455 nm), HyD2 (455–490 nm), HyD3 (500–550 nm), and HyD4 (560–650 nm). Different wavelengths between 700 nm and 1150 nm were used for excitation; HA and Rhod-PE were excited with a wavelength of 960/1050 nm and detected in HyD3 and HyD4. All images were in 12 bit and acquired with a 25× (HCX IRAPO N.A. 0.95 WD 2.5 mm) water objective.
Statistical Analysis and Data Availability
Because of small sample sizes, nonparametric tests were used exclusively. For comparisons between two groups, two-tailed Mann–Whitney tests were performed. For comparisons between multiple groups, we used Kruskal–Wallis tests followed by two-tailed Dunn’s tests with Bonferroni correction using the PMCMR package in R.75 No statistical methods were used to predetermine sample size, but group sizes were >5 in order for the null distribution of the Kruskal–Wallis statistic to approximate the X2 distribution (with k–1 degrees of freedom). With the exception of Figure 1e, graphs show all individual data points and the median. Confocal image stacks (raw data) are available from the corresponding authors upon reasonable request.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO, Dutch Technology Foundation STW, project no. 12520 (J.B.), and NWO-VICI, project no. 724.014.001, F.C., G.A.-A., J.B., A.K.), “Stiftung zur Förderung des pharmazeutischen Nachwuchses in Basel” and “Freiwilligen Akademischen Gesellschaft Basel” (S.S.) and the Dutch Cancer Society (KWF, project no. 6660, F.L.B.)
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b06995.
Supporting information, methods, tables and figures (PDF)
Movie S1: An uninjected control embryo and three DSPC-clodronic acid (10mM total lipids) liposome injected embryos showing blood flow dynamics in the tail region and normal embryonic development 48 h after injection. Black arrows indicate the most caudal end of the PCV that contains bloodflow, and white arrows indicate the most caudal perfused ISV (AVI)
Movie S2: Time lapse confocal imaging of a kdrl:GFP transgenic embryo injected with DSPC-clodronic acid (10 mM total lipids) liposome. Imaging started 6 hpi. Confocal z-stacks were captured every 20 minutes for 24 h (AVI)
Movie S3: Three sibling control embryo and three stab2ibl2 homozygous mutants DSPC-clodronic acid (10 mM total lipids) liposome injected embryos showing blood flow dynamics in the tail region and normal embryonic development 48 h after injection (AVI)
Author Present Address
# Princess Máxima Center for Pediatric Oncology, Utrecht 3584CT, The Netherlands
Author Contributions
⊥ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. W. Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater. 2016, 1, 16014. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- Blanco E.; Shen H.; Ferrari M. Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol. 2015, 33, 941–951. 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawidczyk C. M.; Kim C.; Park J. H.; Russell L. M.; Lee K. H.; Pomper M. G.; Searson P. C. State-of-the-Art in Design Rules for Drug Delivery Platforms: Lessons Learned from FDA-Approved Nanomedicines. J. Controlled Release 2014, 187, 133–144. 10.1016/j.jconrel.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Z.; Langer R.; Farokhzad O. C. Nanoparticle Delivery of Cancer Drugs. Annu. Rev. Med. 2012, 63, 185–198. 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- Zhang Y. N.; Poon W.; Tavares A. J.; McGilvray I. D.; Chan W. C. Nanoparticle-Liver Interactions: Cellular Uptake and Hepatobiliary Elimination. J. Controlled Release 2016, 240, 332–348. 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Bertrand N.; Leroux J. C. The Journey of a Drug-Carrier in the Body: An Anatomo-Physiological Perspective. J. Controlled Release 2012, 161, 152–163. 10.1016/j.jconrel.2011.09.098. [DOI] [PubMed] [Google Scholar]
- Tsoi K. M.; MacParland S. A.; Ma X. Z.; Spetzler V. N.; Echeverri J.; Ouyang B.; Fadel S. M.; Sykes E. A.; Goldaracena N.; Kaths J. M.; Conneely J. B.; Alman B. A.; Selzner M.; Ostrowski M. A.; Adeyi O. A.; Zilman A.; McGilvray I. D.; Chan W. C. Mechanism of Hard-Nanomaterial Clearance by the Liver. Nat. Mater. 2016, 15, 1212–1221. 10.1038/nmat4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkopf C.; Fahr A.; Fricker G.; Scherphof G. L.; Kamps J. A. Uptake of Phosphatidylserine-Containing Liposomes by Liver Sinusoidal Endothelial Cells in the Serum-Free Perfused Rat Liver. Biochim. Biophys. Acta, Biomembr. 2005, 1668, 10–16. 10.1016/j.bbamem.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Kamps J. A. A. M.; Morselt H. W. M.; Swart P. J.; Meijer D. K. F.; Scherphof G. L. Massive Targeting of Liposomes, Surface-Modified with Anionized Albumins, to Hepatic Endothelial Cells. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 11681–11685. 10.1073/pnas.94.21.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. K.; Utsumi T.; Seo Y. E.; Deng Y.; Satoh A.; Saltzman W. M.; Iwakiri Y. Cellular Distribution of Injected PLGA-Nanoparticles in the Liver. Nanomedicine 2016, 12, 1365–1374. 10.1016/j.nano.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen K. K.; Simon-Santamaria J.; McCuskey R. S.; Smedsrod B. Liver Sinusoidal Endothelial Cells. Compr. Physiol. 2015, 5, 1751–1774. 10.1002/cphy.c140078. [DOI] [PubMed] [Google Scholar]
- MacPhee P. J.; Schmidt E. E.; Groom A. C. Intermittence of Blood Flow in Liver Sinusoids, Studied by High-Resolution in Vivo Microscopy. Am. J. Physiol. 1995, 269, G692–8. 10.1152/ajpgi.1995.269.5.G692. [DOI] [PubMed] [Google Scholar]
- Wisse E. An Electron Microscopic Study of the Fenestrated Endothelial Lining of Rat Liver Sinusoids. J. Ultrastruct. Res. 1970, 31, 125–150. 10.1016/S0022-5320(70)90150-4. [DOI] [PubMed] [Google Scholar]
- Aschoff L. Das Reticulo-Endotheliale System. Ergebnisse der Inneren Medizin und Kinderheilkunde 1924, 1–118. 10.1007/978-3-642-90639-8_1. [DOI] [Google Scholar]
- Seternes T.; Sorensen K.; Smedsrod B. Scavenger Endothelial Cells of Vertebrates: A Nonperipheral Leukocyte System for High-Capacity Elimination of Waste Macromolecules. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 7594–7597. 10.1073/pnas.102173299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan L. P.; Mohanty S.; Kim J.; Clark K. R.; Robinson J. M.; Anderson C. L. Rapid and Efficient Clearance of Blood-Borne Virus by Liver Sinusoidal Endothelium. PLoS Pathog. 2011, 7, e1002281. 10.1371/journal.ppat.1002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates J. M.; Yao Z. L.; Cheplowitz A. M.; Suer O.; Phillips G. S.; Kwiek J. J.; Rajaram M. V. S.; Kim J.; Robinson J. M.; Ganesan L. P.; Anderson C. L. Mouse Liver Sinusoidal Endothelium Eliminates HIV-Like Particles from Blood at a Rate of 100 Million per Minute by a Second-Order Kinetic Process. Front. Immunol. 2017, 8, 35. 10.3389/fimmu.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiner K. M.; Schaller H.; Knolle P. A. Endothelial Cell-Mediated Uptake of a Hepatitis B Virus: A New Concept of Liver Targeting of Hepatotropic Microorganisms. Hepatology 2001, 34, 803–808. 10.1053/jhep.2001.27810. [DOI] [PubMed] [Google Scholar]
- Knolle P. A.; Wohlleber D. Immunological Functions of Liver Sinusoidal Endothelial Cells. Cell. Mol. Immunol. 2016, 13, 347–353. 10.1038/cmi.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen K. K.; McCourt P.; Berg T.; Crossley C.; Le Couteur D.; Wake K.; Smedsrod B. The Scavenger Endothelial Cell: A New Player in Homeostasis and Immunity. Am. J. Physiol Regul Integr Comp Physiol 2012, 303, R1217–R1230. 10.1152/ajpregu.00686.2011. [DOI] [PubMed] [Google Scholar]
- Allen T. M.; Cullis P. R. Liposomal Drug Delivery Systems: From Concept to Clinical Applications. Adv. Drug Delivery Rev. 2013, 65, 36–48. 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Sercombe L.; Veerati T.; Moheimani F.; Wu S. Y.; Sood A. K.; Hua S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen L.; Johansen P. L.; Koster G.; Zhu K.; Herfindal L.; Speth M.; Fenaroli F.; Hildahl J.; Bagherifam S.; Tulotta C.; Prasmickaite L.; Mælandsmo G. M.; Snaar-Jagalska E.; Griffiths G. Zebrafish as a Model System for Characterization of Nanoparticles against Cancer. Nanoscale 2016, 8, 862–877. 10.1039/C5NR07289A. [DOI] [PubMed] [Google Scholar]
- Fenaroli F.; Westmoreland D.; Benjaminsen J.; Kolstad T.; Skjeldal F. M.; Meijer A. H.; Van Der Vaart M.; Ulanova L.; Roos N.; Nyström B.; Hildahl J.; Griffiths G. Nanoparticles as Drug Delivery System against Tuberculosis in Zebrafish Embryos: Direct Visualization and Treatment. ACS Nano 2014, 8, 7014–7026. 10.1021/nn5019126. [DOI] [PubMed] [Google Scholar]
- Jiang X. Y.; Sarsons C. D.; Gomez-Garcia M. J.; Cramb D. T.; Rinker K. D.; Childs S. J. Quantum Dot Interactions and Flow Effects in Angiogenic Zebrafish (Danio Rerio) Vessels and Human Endothelial Cells. Nanomedicine 2017, 13, 999–1010. 10.1016/j.nano.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Sieber S.; Grossen P.; Detampel P.; Siegfried S.; Witzigmann D.; Huwyler J. Zebrafish as an Early Stage Screening Tool to Study the Systemic Circulation of Nanoparticulate Drug Delivery Systems in Vivo. J. Controlled Release 2017, 264, 180–191. 10.1016/j.jconrel.2017.08.023. [DOI] [PubMed] [Google Scholar]
- Batist G.; Barton J.; Chaikin P.; Swenson C.; Welles L. Myocet (Liposome-Encapsulated Doxorubicin Citrate): A New Approach in Breast Cancer Therapy. Expert Opin. Pharmacother. 2002, 3, 1739–1751. 10.1517/14656566.3.12.1739. [DOI] [PubMed] [Google Scholar]
- Schmitt-Sody M.; Strieth S.; Krasnici S.; Sauer B.; Schulze B.; Teifel M.; Michaelis U.; Naujoks K.; Dellian M. Neovascular Targeting Therapy: Paclitaxel Encapsulated in Cationic Liposomes Improves Antitumoral Efficacy. Clin. Cancer Res. 2003, 9, 2335–2341. [PubMed] [Google Scholar]
- Cornely O. A.; Maertens J.; Bresnik M.; Ebrahimi R.; Ullmann A. J.; Bouza E.; Heussel C. P.; Lortholary O.; Rieger C.; Boehme A.; Aoun M.; Horst H. A.; Thiebaut A.; Ruhnke M.; Reichert D.; Vianelli N.; Krause S. W.; Olavarria E.; Herbrecht R. Liposomal Amphotericin B as Initial Therapy for Invasive Mold Infection: A Randomized Trial Comparing a High-Loading Dose Regimen with Standard Dosing (AmBiLoad Trial). Clin. Infect. Dis. 2007, 44, 1289–1297. 10.1086/514341. [DOI] [PubMed] [Google Scholar]
- Immordino M. L.; Dosio F.; Cattel L. Stealth Liposomes: Review of the Basic Science, Rationale, and Clinical Applications, Existing and Potential. Int. J. Nanomedicine 2006, 1, 297–315. [PMC free article] [PubMed] [Google Scholar]
- Murayama E.; Kissa K.; Zapata A.; Mordelet E.; Briolat V.; Lin H. F.; Handin R. I.; Herbomel P. Tracing Hematopoietic Precursor Migration to Successive Hematopoietic Organs during Zebrafish Development. Immunity 2006, 25, 963–975. 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Isogai S.; Horiguchi M.; Weinstein B. M. The Vascular Anatomy of the Developing Zebrafish: An Atlas of Embryonic and Early Larval Development. Dev. Biol. 2001, 230, 278–301. 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- Cho E. C.; Xie J. W.; Wurm P. A.; Xia Y. N. Understanding the Role of Surface Charges in Cellular Adsorption versus Internalization by Selectively Removing Gold Nanoparticles on the Cell Surface with a I-2/KI Etchant. Nano Lett. 2009, 9, 1080–1084. 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- Hen G.; Nicenboim J.; Mayseless O.; Asaf L.; Shin M.; Busolin G.; Hofi R.; Almog G.; Tiso N.; Lawson N. D.; Yaniv K. Venous-Derived Angioblasts Generate Organ-Specific Vessels during Zebrafish Embryonic Development. Development 2015, 142, 4266–4278. 10.1242/dev.129247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltowska K.; Paterson S.; Bower N. I.; Baillie G. J.; Lagendijk A. K.; Astin J. W.; Chen H. J.; Francois M.; Crosier P. S.; Taft R. J.; Simons C.; Smith K. A.; Hogan B. M. Mafba Is a Downstream Transcriptional Effector of Vegfc Signaling Essential for Embryonic Lymphangiogenesis in Zebrafish. Genes Dev. 2015, 29, 1618–1630. 10.1101/gad.263210.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapelfeldt H.; Jun H.; Skibsted L. H. Fluorescence Properties of Carminic Acid in Relation to Aggregation, Complex Formation and Oxygen Activation in Aqueous Food Models. Food Chem. 1993, 48, 1–11. 10.1016/0308-8146(93)90213-Y. [DOI] [Google Scholar]
- Schledzewski K.; Géraud C.; Arnold B.; Wang S.; Gröne H. J.; Kempf T.; Wollert K. C.; Straub B. K.; Schirmacher P.; Demory A.; Schönhaber H.; Gratchev A.; Dietz L.; Thierse H. J.; Kzhyshkowska J.; Goerdt S. Deficiency of Liver Sinusoidal Scavenger Receptors Stabilin-1 and −2 in Mice Causes Glomerulofibrotic Nephropathy via Impaired Hepatic Clearance of Noxious Blood Factors. J. Clin. Invest. 2011, 121, 703–714. 10.1172/JCI44740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. M.; Bos F. L.; Bussmann J.; Witte M.; Chi N. C.; Duckers H. J.; Schulte-Merker S. ccbe1 Is Required for Embryonic Lymphangiogenesis and Venous Sprouting. Nat. Genet. 2009, 41, 396–398. 10.1038/ng.321. [DOI] [PubMed] [Google Scholar]
- Wong K. S.; Proulx K.; Rost M. S.; Sumanas S. Identification of Vasculature-Specific Genes by Microarray Analysis of Etsrp/Etv2 Overexpressing Zebrafish Embryos. Dev. Dyn. 2009, 238, 1836–1850. 10.1002/dvdy.21990. [DOI] [PubMed] [Google Scholar]
- Zhou B.; Weigel J. A.; Fauss L.; Weigel P. H. Identification of the Hyaluronan Receptor for Endocytosis (HARE). J. Biol. Chem. 2000, 275, 37733–37741. 10.1074/jbc.M003030200. [DOI] [PubMed] [Google Scholar]
- Simon-Santamaria J.; Malovic I.; Warren A.; Oteiza A.; Le Couteur D.; Smedsrod B.; McCourt P.; Sorensen K. K. Age-Related Changes in Scavenger Receptor-Mediated Endocytosis in Rat Liver Sinusoidal Endothelial Cells. J. Gerontol., Ser. A 2010, 65, 951–960. 10.1093/gerona/glq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. M.; Donner A. J.; Blank E. E.; Egger A. W.; Kellar B. M.; Ostergaard M. E.; Seth P. P.; Harris E. N. Stabilin-1 and Stabilin-2 Are Specific Receptors for the Cellular Internalization of Phosphorothioate-Modified Antisense Oligonucleotides (ASOs) in the Liver. Nucleic Acids Res. 2016, 44, 2782–2794. 10.1093/nar/gkw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y.; Jung M. Y.; Kim H. J.; Lee S. J.; Kim S. Y.; Lee B. H.; Kwon T. H.; Park R. W.; Kim I. S. Rapid Cell Corpse Clearance by Stabilin-2, a Membrane Phosphatidylserine Receptor. Cell Death Differ. 2008, 15, 192–201. 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- Balogh P.; Petz A. Selective Binding of Biotinylated Albumin to the Lymphoid Microvasculature. Histochem. Cell Biol. 2005, 123, 357–363. 10.1007/s00418-005-0778-1. [DOI] [PubMed] [Google Scholar]
- Alidori S.; Bowman R. L.; Yarilin D.; Romin Y.; Barlas A.; Mulvey J. J.; Fujisawa S.; Xu K.; Ruggiero A.; Riabov V.; Thorek D. L.; Ulmert H. D.; Brea E. J.; Behling K.; Kzhyshkowska J.; Manova-Todorova K.; Scheinberg D. A.; McDevitt M. R. Deconvoluting Hepatic Processing of Carbon Nanotubes. Nat. Commun. 2016, 7, 12343. 10.1038/ncomms12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J.; Park S. Y.; Jung M. Y.; Bae S. M.; Kim I. S. Mechanism for Phosphatidylserine-Dependent Erythrophagocytosis in Mouse Liver. Blood 2011, 117, 5215–5223. 10.1182/blood-2010-10-313239. [DOI] [PubMed] [Google Scholar]
- Harris E. N.; Weigel P. H. The Ligand-Binding Profile of HARE: Hyaluronan and Chondroitin Sulfates A, C, and D Bind to Overlapping Sites Distinct from the Sites for Heparin, Acetylated Low-Density Lipoprotein, Dermatan Sulfate, and CS-E. Glycobiology 2008, 18, 638–648. 10.1093/glycob/cwn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y.; Adachi H.; Osuga J.; Ohashi K.; Yahagi N.; Sekiya M.; Okazaki H.; Tomita S.; Iizuka Y.; Shimano H.; Nagai R.; Kimura S.; Tsujimoto M.; Ishibashi S. FEEL-1 and FEEL-2 Are Endocytic Receptors for Advanced Glycation End Products. J. Biol. Chem. 2003, 278, 12613–12617. 10.1074/jbc.M210211200. [DOI] [PubMed] [Google Scholar]
- Kawasaki T.; Etoh R.; Yamashina I. Isolation and Characterization of a Mannan-Binding Protein from Rabbit Liver. Biochem. Biophys. Res. Commun. 1978, 81, 1018–1024. 10.1016/0006-291X(78)91452-3. [DOI] [PubMed] [Google Scholar]
- Rost M. S.; Sumanas S. Hyaluronic Acid Receptor Stabilin-2 Regulates Erk Phosphorylation and Arterial--Venous Differentiation in Zebrafish. PLoS One 2014, 9, e88614. 10.1371/journal.pone.0088614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S. J.; Bartsch S.; Kroll J. HOXC9 Regulates Formation of Parachordal Lymphangioplasts and the Thoracic Duct in Zebrafish via Stabilin 2. PLoS One 2013, 8, e58311. 10.1371/journal.pone.0058311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y.; Saijou E.; Sugano Y.; Takeshita F.; Nishimura S.; Nonaka H.; Chen Y.-R.; Sekine K.; Kido T.; Nakamura T.; Kato S.; Kanke T.; Nakamura K.; Nagai R.; Ochiya T.; Miyajima A. Inhibition of Stabilin-2 Elevates Circulating Hyaluronic Acid Levels and Prevents Tumor Metastasis. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 4263–4268. 10.1073/pnas.1117560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwegen M.; Cornelissen J. J. L. M. Clustered Nanocarriers: The Effect of Size on the Clustering of CCMV Virus-like Particles with Soft Macromolecules. Macromol. Biosci. 2015, 15, 98–110. 10.1002/mabi.201400326. [DOI] [PubMed] [Google Scholar]
- Askes S. H. C.; Pomp W.; Hopkins S. L.; Kros A.; Wu S.; Schmidt T.; Bonnet S. Imaging Upconverting Polymersomes in Cancer Cells: Biocompatible Antioxidants Brighten Triplet–Triplet Annihilation Upconversion. Small 2016, 12, 5579–5590. 10.1002/smll.201601708. [DOI] [PubMed] [Google Scholar]
- Plosker G. L.; Goa K. L. Clodronate. A Review of Its Pharmacological Properties and Therapeutic Efficacy in Resorptive Bone Disease. Drugs 1994, 47, 945–982. 10.2165/00003495-199447060-00007. [DOI] [PubMed] [Google Scholar]
- van Rooijen N.; Hendrikx E. Liposomes for Specific Depletion of Macrophages from Organs and Tissues. Methods Mol. Biol. 2010, 605, 189–203. 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
- Michen B.; Graule T. Isoelectric Points of Viruses. J. Appl. Microbiol. 2010, 109, 388–397. 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Elvevold K.; Smedsrod B.; Martinez I. The Liver Sinusoidal Endothelial Cell: A Cell Type of Controversial and Confusing Identity. Am. J. Physiol Gastrointest Liver Physiol 2008, 294, G391–400. 10.1152/ajpgi.00167.2007. [DOI] [PubMed] [Google Scholar]
- Patel K. R.; Li M. P.; Baldeschwieler J. D. Suppression of Liver Uptake of Liposomes by Dextran Sulfate 500. Proc. Natl. Acad. Sci. U. S. A. 1983, 80, 6518–6522. 10.1073/pnas.80.21.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belder A. N.; Wik K. O. Preparation and Properties of Fluorescein-Labelled Hyaluronate. Carbohydr. Res. 1975, 44, 251–257. 10.1016/S0008-6215(00)84168-3. [DOI] [PubMed] [Google Scholar]
- Kawai Y.; Smedsrod B.; Elvevold K.; Wake K. Uptake of Lithium Carmine by Sinusoidal Endothelial and Kupffer Cells of the Rat Liver: New Insights into the Classical Vital Staining and the Reticulo-Endothelial System. Cell Tissue Res. 1998, 292, 395–410. 10.1007/s004410051069. [DOI] [PubMed] [Google Scholar]
- Voloshin T.; Alishekevitz D.; Kaneti L.; Miller V.; Isakov E.; Kaplanov I.; Voronov E.; Fremder E.; Benhar M.; Machluf M.; Apte R. N.; Shaked Y. Blocking IL1 Pathway Following Paclitaxel Chemotherapy Slightly Inhibits Primary Tumor Growth but Promotes Spontaneous Metastasis. Mol. Cancer Ther. 2015, 14, 1385–1394. 10.1158/1535-7163.MCT-14-0969. [DOI] [PubMed] [Google Scholar]
- Jin S.-W. Cellular and Molecular Analyses of Vascular Tube and Lumen Formation in Zebrafish. Development 2005, 132, 5199–5209. 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Ellett F.; Pase L.; Hayman J. W.; Andrianopoulos A.; Lieschke G. J. mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood 2011, 117, e49. 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Chi M.; Phan Q. T.; Gonzalez C.; Dubremetz J.-F.; Levraud J.-P.; Lutfalla G. Transient Infection of the Zebrafish Notochord with E. Coli Induces Chronic Inflammation. Dis. Models & Mech. 2014, 7, 871–882. 10.1242/dmm.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J.; Bos F. L.; Urasaki A.; Kawakami K.; Duckers H. J.; Schulte-Merker S. Arteries Provide Essential Guidance Cues for Lymphatic Endothelial Cells in the Zebrafish Trunk. Development 2010, 137, 2653–2657. 10.1242/dev.048207. [DOI] [PubMed] [Google Scholar]
- Hogan B. M.; Herpers R.; Witte M.; Helotera H.; Alitalo K.; Duckers H. J.; Schulte-Merker S. Vegfc/Flt4 Signalling Is Suppressed by Dll4 in Developing Zebrafish Intersegmental Arteries. Development 2009, 136, 4001–4009. 10.1242/dev.039990. [DOI] [PubMed] [Google Scholar]
- Mathias J. R.; Perrin B. J.; Liu T.-X.; Kanki J.; Look A. T.; Huttenlocher A. Resolution of Inflammation by Retrograde Chemotaxis of Neutrophils in Transgenic Zebrafish. J. Leukocyte Biol. 2006, 80, 1281–1288. 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- Thisse C.; Thisse B. High-Resolution in Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat. Protoc. 2008, 3, 59–69. 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Varshney G. K.; Pei W.; Lafave M. C.; Idol J.; Xu L.; Gallardo V.; Carrington B.; Bishop K.; Jones M.; Li M.; Harper U.; Huang S. C.; Prakash A.; Chen W.; Sood R.; Ledin J.; Burgess S. M. High-Throughput Gene Targeting and Phenotyping in Zebrafish Using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington B.; Varshney G. K.; Burgess S. M.; Sood R. CRISPR-STAT: An Easy and Reliable PCR-Based Method to Evaluate Target-Specific sgRNA Activity. Nucleic Acids Res. 2015, 43, e157. 10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B. M.; Stemple D. L.; Driever W.; Fishman M. C. Gridlock, a Localized Heritable Vascular Patterning Defect in the Zebrafish. Nat. Med. 1995, 1, 1143–1147. 10.1038/nm1195-1143. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlert T.The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R Package; R Foundation: Vienna, Austria, 2014. https://cran.r-project.org/web/packages/PMCMR/index.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.