Abstract
Understanding of cellular transdifferentiation is limited by the technical inability to track multiple lineages in vivo. To overcome this we developed a new tool to simultaneously fate map two distinct cell types in the kidney, and genetically test whether cells of renin lineage (CoRL) can transdifferentiate to a podocyte fate. Ren1cCreER/tdTomato/Nphs1-FLPo/FRT-EGFP mice (CoRL-PODO mice) were generated by crossing Ren1c-CreER/tdTomato CoRL reporter mice with Nphs1-FLPo/FRT-EGFP podocyte reporter mice. Following tamoxifen administration in these animals, CoRL were labeled with red fluorescence (tdTomato) and co-localized with renin. Podocytes were labeled green (enhanced green fluorescent protein) and co-localized with nephrin. Following podocyte loss by nephrotoxic antibody and subsequent enalapril-enhanced partial replacement, tdTomato-EGFP-labeled CoRL were detected as yellow-colored cells in a subset of glomerular tufts, without the use of antibodies. Co-localization with podocin indicated that these cells are podocytes, derived from CoRL origin. Thus, our novel study shows that two distinct cell types can be simultaneously labeled in the mouse kidney and provide strong genetic evidence in vivo that lost podocytes can be replaced in part by CoRL.
Key words not in title: reporter mouse, glomerulus, migration, podocin, enalapril, lineage tracing, Cre-recombination, FLP-recombination
INTRODUCTION
Site-specific recombinase (SSR) is an essential tool for lineage tracing cells in mice1. DNA recombinases from the P1 bacteriophage (Cre) and Saccharomyces cerevisiae (flippase, FLP), combined with cell-specific promoters and reporters silenced by loxP- or FRT-flanked STOP sequences, allow for permanent expression of either fluorescent or enzymatic reporters in specific cell populations2. Combined with ligand-dependent activation, such as the tamoxifen-responsive CreERT2 recombinase, SSR can be achieved within specific temporal periods 3, 4. This provides a non-invasive method of mapping cell fate throughout development or tracing cell lineages during disease/regeneration.
SSR techniques have been used extensively in the study of kidney regeneration, and has led to several significant findings regarding the ability of distinct cell populations to undergo repair 4. Yet, despite these genetic advances, simultaneously lineage tracing of two distinct cell types has been a challenge, in part because the majority of SSR in the kidney utilizes only Cre-loxP recombination5, 6. Expression of Cre recombinase, even under the control of distinct promoters and utilizing distinct reporters, cannot uniquely label two cell types. This has posed a barrier in our ability to observe transdifferentiation events, in which one cell type adopts the characteristics of another. The limited numbers of studies that have been performed that utilize Cre and FLP systems simultaneously, outside of the kidney, are intersectional or subtractive genetic fate mapping, that do not allow for the continuous tracing of two distinct populations, and require enzymatic or immunohistochemical staining of the β-gal reporter 1, 7.
Here, we have succeeded in simultaneously labeling two distinct kidney cell populations by combining Cre-lox and FLP-FRT recombination strategies in the same mouse, regardless of sex, with the use of directly observable fluorescent reporters, that we are calling dual lineage tracing. We utilized this methodology to genetically demonstrate that cells of renin lineage (CoRL) transdifferentiate towards a podocyte fate following podocyte depletion.
RESULTS
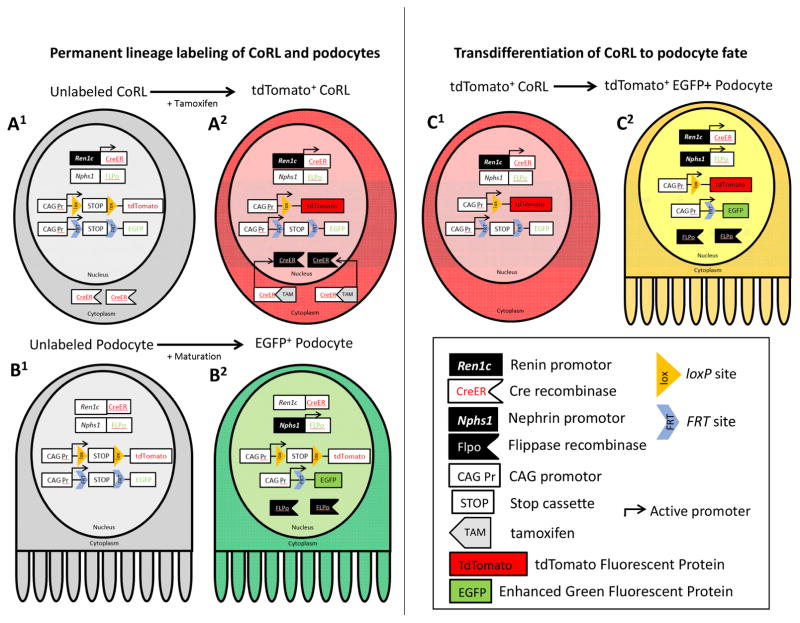
Figure 1 shows a schema of our dual transgenic approach. Unlabeled CoRL activate the Ren1c promoter inducing CreER expression, however CreER remains sequestered in the cytoplasm and unable bind loxP (Figure 1A1). Following tamoxifen, CreER translocates to the nucleus and recombines the loxP sites to remove the STOP cassette, thus inducing permanent tdTomato expression (Figure 1A2). As podocytes mature (Figure 1B1), the Nphs1 promoter is activated to drive FLP expression, which recombines the FRT sites to remove the STOP cassette, thus inducing permanent EGFP expression (Figure 1B2). During transdifferentiation to a podocyte fate, tdTomato+ CoRL (Figure 1C1) activate the Nphs1 promoter, inducing FLP expression. FLP removes the FRT-flanked STOP cassette, inducing permanent EGFP expression and turning the originally red cell, yellow (Figure 1C2).
Figure 1. Schema of dual lineage tracing transgenic approach.
(A1) Unlabeled CoRL activate the Ren1c promoter (black box, white type) inducing CreER (white flag, red type). CreER remains sequestered in the cytoplasm and unable to bind loxP. (A2) Tamoxifen (gray flag, black type) binds to the ligand-binding domain of the Cre-estrogen receptor fusion protein, resulting in translocation to the nucleus (black flag, white type) and recombination of loxP sites (orange triangle, black type) and the intervening STOP cassette (white box, black type), inducing permanent tdTomato (red box, black type) expression. (B1) Immature unlabeled podocytes have not yet activated the Nphs1 promoter (white box, black type) (B2) During podocyte maturation, the Nphs1 promoter is activated (black box, white type), driving FLPase (black flag, white type) expression, which recombines the FLPase recognition targets (blue chevron, black type) and STOP cassette (white box, black type) to induce permanent enhanced green fluorescent protein (green box, black type) expression. (C1) tdTomato+ CoRL have not yet activated the Nphs1 promoter (white box, black type). (C2) Upon transdifferentiation to a podocyte fate, the Nphs1 promoter is activated (black box, white type), driving FLPase (black flag, white type) expression; FLPase recombines the FRT sites (blue chevron, black type) and STOP cassette (white box, black type) inducing permanent enhanced green fluorescent protein (green box, black type), resulting in a yellow color.
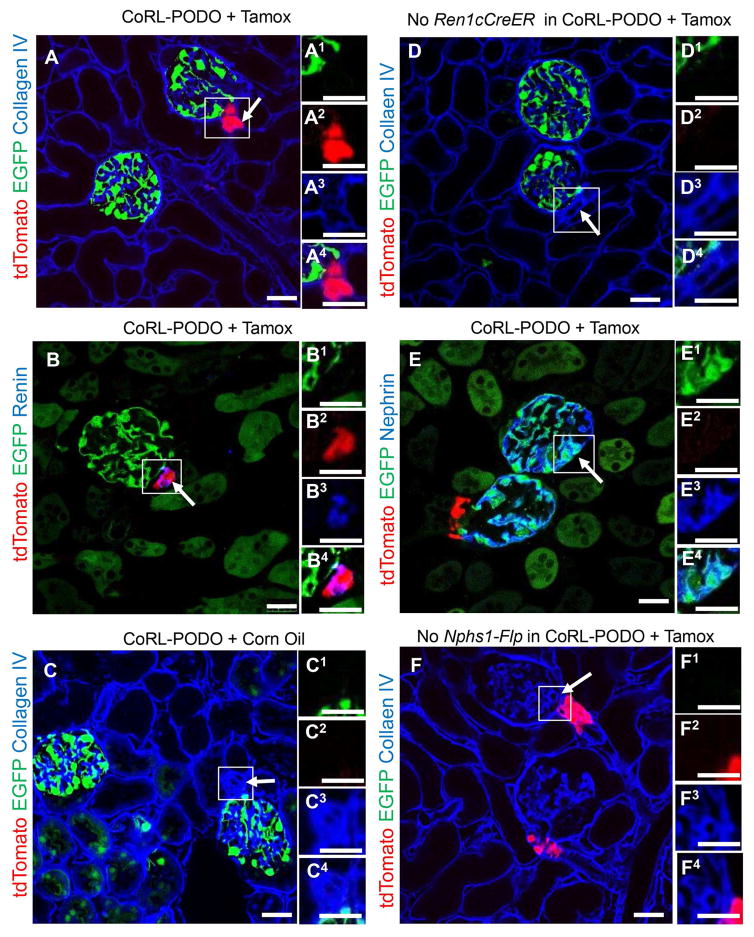
Figure 2 shows validation of our dual lineage tracing approach. Following tamoxifen, young adult Ren1cCreER/tdTomato/Nphs1-FLPo/FRT-EGFP mice (herein called CoRL-PODO mice) expressed tdTomato specifically in cells in the juxta-glomerular compartment (JGC) (Figure 2A, arrow). As shown previously, tdTomato overlapped with 95% of renin+ cells (Figure 2B, arrow). When CoRL-PODO mice were given the tamoxifen vehicle corn oil, tdTomato was not detected in the JGC (Figure 2C, arrow). Similarly, in mice lacking the Ren1cCreER transgene, tdTomato was not detected following tamoxifen administration (Figure 2D, arrow).
Figure 2. CoRL-PODO mice specifically and inducibly label renin expressing cells, and constitutively label nephrin expressing cells (podocytes).
Ren1cCreER/tdTomato mice were crossed with Nphs1-FLPo/FRT-EGFP mice to generate dual-reporting Ren1cCreER/tdTomato/Nphs1-FLPo/FRT-EGFP mice. Immunohistochemistry for Collagen IV is used to easily identify glomeruli in panels A, C, D and F. Higher magnification images of the areas of interest outlined by the white boxes are shown to the right in superscript 1–4 for green, red, far red and merged fluorescence channels respectively.
A) Confocal direct fluorescence of representative CoRL-PODO mice administered tamoxifen, express tdTomato in CoRL in the JGC (arrow) and EGFP in podocytes in the glomerular tuft. B) Immunohistochemistry for renin in tamoxifen-induced CoRL-PODO mice, shows co-labeling of renin (blue) and tdTomato in CoRL in the JGC (purple color, arrow). C) When CoRL-PODO mice are given the vehicle corn oil, there is no induced expression of tdTomato (arrow). The constitutive Nphs1-FLPo/FRT-EGFP transgene allows for the expression of EGFP. D) In CoRL-PODO mice given tamoxifen, that lack the Ren1c-CreER transgene, there is no inducible labeling of tdTomato CoRL (arrow) but the constitutive Nphs1-FLPo/FRT-EGFP transgene allows for the expression of EGFP. E) Immunohistochemistry for the podocyte protein nephrin in tamoxifen induced CoRL-PODO mice shows co-labeling of nephrin and EGFP in podocytes (arrow). F) In CoRL-PODO mice given tamoxifen which do not have the Nphs1-FLPo transgene, there is no labeling of EGFP in podocytes, but the induction of the Ren1c-CreER transgene allows for the expression of tdTomato in CoRL in the JGC (arrow). Scale bars = 25μm.
In the same mouse, EGFP (green) was detected within the glomerular tuft in a podocyte distribution pattern (Figure 2A) and overlapped with staining for the podocyte marker nephrin in 98% of EGFP+ cells (Figure 2E, arrow). However, in CoRL-PODO mice lacking the Nphs1-FLPo transgene, EGFP was not detected (Figure 2F, arrow). There was no overlap between tdTomato and EGFP under normal non-stressed conditions (Figure 2A). These results show distinct inducible tdTomato labeling of CoRL in the JGC and constitutive EGFP labeling of podocytes in the glomerular tuft, in the same kidney. As this approach enabled simultaneous tracking of two cell types in vivo, we called it dual lineage tracing.
To genetically demonstrate that a subset of CoRL can replace lost podocytes in disease, experimental FSGS, characterized by podocyte loss and subsequent partial replacement, was induced in CoRL-PODO mice with a nephrotoxic antibody8, 9. Mice were treated with enalapril, which we have previously shown increases podocyte repletion 8, 9. Dual lineage tracing was performed to follow migration and transdifferentiation (Figure 3). At baseline, tdTomato+ CoRL were restricted to the JGC. However, following podocyte depletion and enalapril treatment, tdTomato+ CoRL were visualized in the glomerular tuft. While a subset of CoRL that migrated remained only red, another subset expressed both tdTomato and EGFP, indicating transdifferentiation to podocytes occurred (Figure 3).
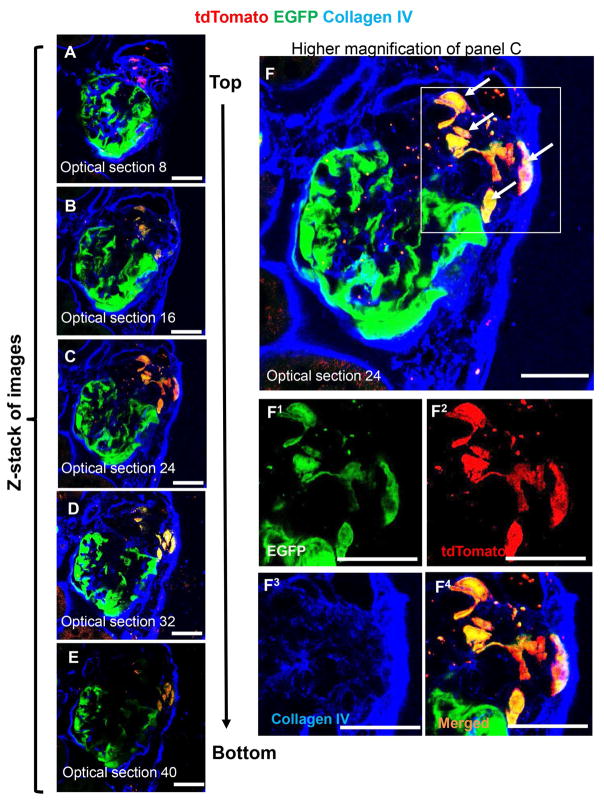
Figure 3. A subset of tdTomato+ CoRL migrate onto the glomerular tuft and begin to co-express EGFP.
Immunohistochemistry for Collagen IV is used to easily identify glomeruli.
(A–E) Fifty 0.4 μm optical sections were taken by confocal microscopy through a 20μm thick section of a glomerulus from CoRL-PODO mice with FSGS. Five equally distributed representative optical sections are shown. F) Higher magnification of optical section 32 (panel D) showing tdTomato+ EGFP+ co-labeled cells on the glomerular tuft, which appears yellow in merged images (arrows). Images of the areas of interest outlined by the white boxes are shown below in superscript 1–4 for green, red, far red and merged fluorescence channels respectively. Scale bars = 25μm.
Collagen IV staining was performed to better visualize and define the boundaries of glomeruli (Figure 3). Again, co-localization was observed between the tdTomato and EGFP reporters. Noteworthy was that optical sectioning by confocal microscopy through a 20 μm kidney section identified tdTomato-EGFP-labeled CoRL in the glomerular tuft that would have been missed by simply looking at a single 4 μm section (Figure 3). EGFP+ cells were not detected in the JGC.
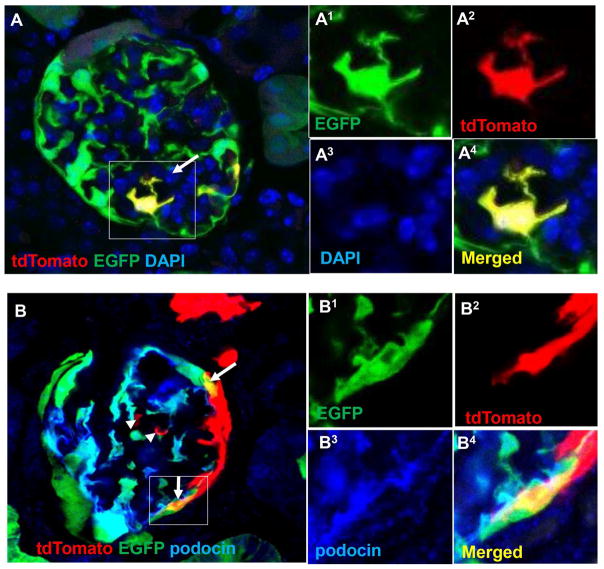
To further confirm that a subset of tdTomato+ EGFP+ CoRL (Figure 4A) transdifferentiated towards a podocyte fate, staining for podocin was performed. Indeed, a subset of CoRL in the glomerular tuft co-expressed tdTomato (red), EGFP (green) and podocin (blue) (Figure 4B, arrows). Quantitation showed that after induction with tamoxifen, there were no RFP+ cells on the glomerular tuft. However, at D28 of FSGS, 26.3±4.2% of glomeruli per kidney section still showed signs of podocyte injury, evidenced by a decrease in podocyte density, similar to what we have previously reported in this model 8, 10, 11. Of these glomeruli, 45.1±3.1% contained at least one RFP+ cell. Furthermore, in glomeruli with EGFP+ CoRL, there was an average of 6.7±1.2 individual RFP+ CoRL, and 53.8±0.7% were EGFP+.
Figure 4. tdTomato+ CoRL on the glomerular tuft co-express EGFP and stain for podocyte marker Podocin.
A) Another clear example of a single tdTomato+ EGFP+ CoRL on the glomerular tuft (arrow). B) Immunohistochemistry for the podocyte protein podocin (blue) shows co-labeling with tdTomato+ EGFP+ CoRL on the glomerular tuft (arrows). Images of the areas of interest outlined by the white boxes are shown to the right in superscript 1–4 for green, red, far red and merged fluorescence channels, respectively. It should be noted that the tdTomato and EGFP reporters are not targeted to any particular cellular compartment, whereas staining for native proteins such as podocin are localized to specific structures such as the slit daphragms of foot processes in normal podocytes. 24 Furthermore, not all tdTomato+ CoRL that migrate onto the tuft (arrow heads) express podocyte markers9. Scale bars = 25μm.
DISCUSSION
We set out to lineage trace two distinct cell types in the kidney and use dual SSR as genetic proof that tdTomato+ CoRL transdifferentiate to the fate of podocytes. By combining inducible Cre- and constitutive FLP-mediated recombinase systems with separate loxP-flanked STOP tdTomato and FRT-flanked STOP EGFP reporter systems, we could use tamoxifen to induce permanent labelling of CoRL with tdTomato using the Ren1c promoter and constitutively and permanently label podocytes with EGFP using the Nphs1 promoter. The two reporters did not overlap under normal (no disease) conditions. These results support dual lineage tracing is feasible in the kidney. The possibility of any off-target effects of Cre expression is highly unlikely given the extended washout period used in the current studies.
A similar dual SSR approach utilized dual loxP-flanked STOP and FRT-flanked STOP cassettes upstream of the EGFP gene. After removal of the flanked STOP cassettes via Cre- and FLPo-mediated recombination, EGFP reporter was expressed12. This system utilized two unique promoter-driven SSR systems, but labeled the cell of interest with EGFP only when both promoters were active. Therefore, it did not label two distinct cell types with two distinct reporters.
The second finding was genetic evidence that a subset of lineage traced CoRL transdifferentiate towards an adult podocyte fate following podocyte depletion in experimental FSGS. Previously, our group11, 13, 14 and others15, 16 have relied on the de novo co-expression of cell-specific markers in reporter-labeled cells as evidence of transdifferentiation. For example, labeled CoRL co-expressing podocin suggests a podocyte fate, claudin-1 a parietal epithelial cell fate, α8 integrin a mesangial cell fate and PDGFβ-receptor/NG2 a pericyte fate11, 13–16. Here we show tdTomato+ CoRL track into the glomerulus following podocyte depletion, and express EGFP under the control of nephrin promoter activity, a podocyte specific gene. These findings are consistent with genetic evidence of CoRL transdifferentiation towards a podocyte fate. Furthermore, de novo co-expression of podocyte-specific markers were observed in dual reporter+ CoRL.
We acknowledge that there is some debate regarding the replenishment of adult podocytes following disease induced loss. Elegant studies by Wanner et al. used a diphtheria toxin podocyte injury model and showed a 38% renewal of ablated podocytes in that model 17. In the adriamycin nephropathy model of podocyte injury, Lasagni et al. showed a 5–10% regeneration of podocyte by renal progenitor cells 15. Our data showed that in a subset of individual glomeruli with reduced podocyte density, yellow cells were detected, which in the current study, is the genetic readout for CoRL transdifferentiating into podocytes. While CoRL themselves are clearly not adequate to fully replace podocytes, they together with other progenitor sources may be able to tip the balance enough to reduce scarring and improve proteinuria as was shown in these studies. In addition to the current model, we have shown CoRL migration in to the glomerulus following chronic podocyte depletion in the experimental remnant kidney model 14.
We considered using the mTmG system which also utilizes dual reporters, where membrane-targeted tandem dimer Tomato (mT) is expressed ubiquitously prior to Cre-mediated excision and membrane-targeted green fluorescent protein (mG) after excision. Such dual reporters allow for genetic evidence of transdifferentiation, however do not allow for active dual lineage tracing 18, 19.
We recognize that a limitation to the current study is the use of constitutive podocyte labeling, rather than inducible labeling. To our knowledge, ligand inducible FLP-FRT systems are limited at this time. Furthermore, the constitutive FLP-FRT system in this study proved helpful, as we were able to show strong genetic evidence of CoRL transdifferentiation to podocytes without requiring the continuous presence of ligand during the 28 days of study. Taken together, this is not a major weakness to the current study.
Others have shown that the use of 4 μm sections, particularly when observing glomeruli, is problematic due the relatively small sampling of the ≅80 μm glomerulus (4μm = 5%), and the inherent heterogeneity in glomerular size both normally and in disease20. In the current study, we utilized 20 μm sections and optical sectioning by confocal microscopy. We noted a marked increase in the ability to detect reporter+ CoRL compared to a single 4 μm section. We therefore recommend lineage tracing in glomerular studies be performed by optically sectioning thick sections or utilizing multiple physical thin sections.
In conclusion, we have successfully shown the effective use of dual lineage tracing in the kidney, which enables tracking two different cell types in vivo. We have furthermore utilized dual lineage tracing to demonstrate the transdifferentiation of a subset of CoRL into podocytes in the kidney glomerulus after injury. As dual lineage tracing is highly flexible and could be used for a variety of different promoters and genes, it provides a powerful new tool to monitor the cellular changes and transitions during homeostasis and regeneration.
METHODS
Animals
Dual reporting Ren1cCreER/tdTomato/Nphs1-FLPo/FRT-EGFP (CoRL-PODO) were bred by crossing our previously generated inducible Ren1cCreER/tdTomato mice, utilizing the tdTomato clone Ai14 (The Jackson Laboratory Stock # 007914)21,22 with Nphs1-FLPo/FRT-EGFP mice. The Nphs1-FLPo mouse expresses an optimized FLP recombinase (FLPo) under the control of a 4.2kb fragment of the mouse nephrin promoter and has been characterized and previously described by Goldberg et al 23. The FRT-EGFP mice, also known as RCE:FRT (JAX Stock#: 010812), were purchased from The Jackson Laboratory (Bar Harbor, ME). In Nphs1-FLPo/FRT-EGFP mice, the FRT-flanked STOP cassette is excised, allowing for specific and permanent EGFP reporter expression in podocytes. Control mice comprised triple transgenic mice without the Cre or FLP. Both male and female mice were utilized and no differences between sexes were observed in these studies. Mice were housed in the animal care facility of the University of Washington under specific pathogen-free conditions. Studies were reviewed and approved by the University of Washington Institutional Animal Care and Use Committee (2968-04).
Reporter induction
Permanent labeling of CoRL with td-Tomato was induced in young adult (8 week-old), male and female, CoRL-PODO mice by delivering 100 mg/kg Tamoxifen (Sigma-Aldrich, St Louis, MO), in corn oil (Sigma-Aldrich, St Louis, MO), 4 times intra-peritoneally, every other day. This was followed by a washout period of 4 weeks. Control for Tamoxifen was vehicle (corn oil) alone. No drugs were required for the induction of the EGFP reporter.
Podocyte depletion
Experimental focal segmental glomerulosclerosis (FSGS) was induced in CoRL-PODO mice by two intra-peritoneal injections, 24 hours apart, of sheep anti-glomerular antibody at 12mg/20g body weight9, 11, 14. Animals were given Enalapril (75 mg/L drinking water, Sigma-Aldrich, St Louis, MI) starting on day 3 of FSGS through day 28 to augment podocyte regeneration9.
Tissue collection
Mice were sacrificed with an overdose of Ketamine/Xylazine, cardiac perfused with 10–15ml of ice cold phosphate buffered saline (PBS) at 25 inches of gravity pressure through a 21G butterfly infusion set. Kidneys were removed when blanched and split lengthwise or “butterflied”. Kidneys were then placed into 4% paraformaldehyde in PBS (PFA, Affymetrix, Santa Clara, CA) for 45 minutes, washed briefly in PBS, placed in 30% sucrose in PBS (Sigma-Aldrich, St Louis, MO) overnight, blotted dry, embedded in Tissue-Tek® O.C.T. Compound (VWR, Radnor, PA), and frozen in a 100% ethanol/dry ice bath.
Visualization of dual (tdTomato/EGFP) reporter
In order to visualize tdTomato and EGFP, 20 μm cryosections were rinsed in PBS (pH 7.4) to remove OCT compound (VWR, Radnor, PA) and mounted with Vectashield with DAPI (Vector Labs, Burlingame, CA). No antibodies were required for visualization of the dual tdTomato and EGFP reporters. Confocal images were acquired with a Leica TCS SPE II laser scanning confocal microscope (Solms, Germany) with X40 (1.3 NA) oil objective, at 1,024 × 1,024 pixel format with 12 bit intensity resolution. Sets of 50 serial images were collected at 0.4 μm step size. The acquisition wavelengths were set as follows; EGFP 488 nm excitation, 497–510 nm emission, and tdTomato 561 nm excitation, 575–654 nm emission.
Multicolor immunofluorescence staining
Indirect immunofluorescence staining was performed on 20 μm sections from 4% PFA-fixed frozen tissue processed as described above, and as previously performed 11, 14. Frozen sections were warmed from −80°C to room temperature and allowed to air-dry. All sections were equilibrated in PBS (pH7.4) then blocked with Background Buster (Accurate Chemical & Scientific Corporation, Westbury, NY) for 30 minutes to minimize nonspecific protein interactions. Endogenous biotin activity was quenched with the avidin/biotin blocking kit (Vector Laboratories, Burlingame, CA). After blocking, sections were incubated overnight at 4°C with the appropriate primary antibodies: to identify podocytes, guinea pig antibody to nephrin, dilution 1:500 (Fitzgerald Industries International. Inc., 20R-NP002, Concord, MA) and rabbit antibody to podocin, dilution 1:4000 (Abcam, ab 50339, Cambridge, MA) were used. The appropriate biotinylated secondary antibodies (Vector Laboratories) were applied and followed by Streptavidin-AlexaFluor 647 (Life Technologies - Molecular Probes, Grand Island, NY). To demarcate the glomerular compartment, biotinylated collagen IV antibody, dilution 1:100 (Southern Biotechnology, 1340-08, Birmingham, AL) and Streptavidin-AlexaFluor 647 were used. Biotinylated anti-renin antibody, dilution 1:100 (Innovative Research, IASMPREN-GF-HT-BIO, Novi, MI) and Streptavidin-AlexaFluor 647 (Life Technologies) were used to detect renin. All immunofluorescence samples were mounted using Vectashield with DAPI. As a negative control, all staining was performed without primary antibodies.
Acknowledgments
Grant Support: 5 R01 DK 056799-10, 5 R01 DK 056799-12, 1 R01 DK097598-01A1, 5UH2 DK107343 02, R01DK058366, HL48459, P30CA016056
Abbreviations
- CoRL
Cell of Renin Lineage
- EGFP
enhanced green fluorescent protein
- PODO
Nphs1-FLPo/FRT-EGFP podocyte reporter
- SSR
site specific recombinase
- tdTomato
tdTomato (red) fluorescent protein
Footnotes
COI: None of the authors have any financial or other conflicts of interest. The results presented in this paper have not been published previously, in whole or part.
DISCLOSURES:
All the authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jensen P, Dymecki SM. Essentials of recombinase-based genetic fate mapping in mice. Methods in molecular biology (Clifton, NJ) 2014;1092:437–454. doi: 10.1007/978-1-60327-292-6_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Feil R, Brocard J, Mascrez B, et al. Ligand-activated site-specific recombination in mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romagnani P, Rinkevich Y, Dekel B. The use of lineage tracing to study kidney injury and regeneration. Nature reviews Nephrology. 2015;11:420–431. doi: 10.1038/nrneph.2015.67. [DOI] [PubMed] [Google Scholar]
- 5.Chai OH, Song CH, Park SK, et al. Molecular regulation of kidney development. Anatomy & cell biology. 2013;46:19–31. doi: 10.5115/acb.2013.46.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F. Conditional targeting in the kidney. Nephron Physiology. 2007;107:p10–16. doi: 10.1159/000106483. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Shook NA, Kanisicak O, et al. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis (New York, NY: 2000) 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaverina NV, Eng DG, Schneider RR, et al. Partial podocyte replenishment in experimental FSGS derives from nonpodocyte sources. American journal of physiology Renal physiology. 2016;310:F1397–1413. doi: 10.1152/ajprenal.00369.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichtnekert J, Kaverina NV, Eng DG, et al. Renin-Angiotensin-Aldosterone System Inhibition Increases Podocyte Derivation from Cells of Renin Lineage. Journal of the American Society of Nephrology: JASN. 2016;27:3611–3627. doi: 10.1681/ASN.2015080877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng DG, Sunseri MW, Kaverina NV, et al. Glomerular parietal epithelial cells contribute to adult podocyte regeneration in experimental focal segmental glomerulosclerosis. Kidney international. 2015;88:999–1012. doi: 10.1038/ki.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaverina NV, Kadoya H, Eng DG, et al. Tracking the stochastic fate of cells of the renin lineage after podocyte depletion using multicolor reporters and intravital imaging. PloS one. 2017;12:e0173891. doi: 10.1371/journal.pone.0173891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sousa VH, Miyoshi G, Hjerling-Leffler J, et al. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebral cortex (New York, NY: 1991) 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharib SA, Pippin JW, Ohse T, et al. Transcriptional landscape of glomerular parietal epithelial cells. PloS one. 2014;9:e105289. doi: 10.1371/journal.pone.0105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pippin JW, Kaverina NV, Eng DG, et al. Cells of renin lineage are adult pluripotent progenitors in experimental glomerular disease. American journal of physiology Renal physiology. 2015;309:F341–358. doi: 10.1152/ajprenal.00438.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasagni L, Angelotti ML, Ronconi E, et al. Podocyte Regeneration Driven by Renal Progenitors Determines Glomerular Disease Remission and Can Be Pharmacologically Enhanced. Stem cell reports. 2015;5:248–263. doi: 10.1016/j.stemcr.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starke C, Betz H, Hickmann L, et al. Renin lineage cells repopulate the glomerular mesangium after injury. Journal of the American Society of Nephrology: JASN. 2015;26:48–54. doi: 10.1681/ASN.2014030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanner N, Hartleben B, Herbach N, et al. Unraveling the role of podocyte turnover in glomerular aging and injury. Journal of the American Society of Nephrology: JASN. 2014;25:707–716. doi: 10.1681/ASN.2013050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzumdar MD, Tasic B, Miyamichi K, et al. A global double-fluorescent Cre reporter mouse. Genesis (New York, NY: 2000) 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 19.Hickmann L, Steglich A, Gerlach M, et al. Persistent and inducible neogenesis repopulates progenitor renin lineage cells in the kidney. Kidney international. 2017 doi: 10.1016/j.kint.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puelles VG, Bertram JF, Moeller MJ. Quantifying podocyte depletion: theoretical and practical considerations. Cell and tissue research. 2017;369:229–236. doi: 10.1007/s00441-017-2630-z. [DOI] [PubMed] [Google Scholar]
- 21.Pippin JW, Sparks MA, Glenn ST, et al. Cells of renin lineage are progenitors of podocytes and parietal epithelial cells in experimental glomerular disease. The American journal of pathology. 2013;183:542–557. doi: 10.1016/j.ajpath.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg S, Adair-Kirk TL, Senior RM, et al. Maintenance of glomerular filtration barrier integrity requires laminin alpha5. Journal of the American Society of Nephrology: JASN. 2010;21:579–586. doi: 10.1681/ASN.2009091004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suleiman HY, Roth R, Jain S, et al. Injury-induced actin cytoskeleton reorganization in podocytes revealed by super-resolution microscopy. JCI insight. 2017:2. doi: 10.1172/jci.insight.94137. [DOI] [PMC free article] [PubMed] [Google Scholar]