Abstract
DNA replication requires separation of genomic duplex DNA strands that is performed by hexameric ring shaped helicase machines in all domains of life. The structures and chemo-mechanical actions of these fascinating machines are coming into focus. There is no evolutionary relationship between the hexameric helicases of bacteria and archaea/eukaryotes, yet they share many fundamental features. We review recent studies on these two groups of hexameric helicases that have unveiled some very surprising distinctions.
Keywords: hexameric helicase, replisome, origin initiation, DNA polymerase, DNA replication
Introduction
The elegant simplicity of the double helix intuited by Watson and Crick suggested that its replication would also be simple 1, but years of research have shown that nothing could be further from the truth. We now know that a whole army of enzymes are needed for DNA duplication. There remain numerous questions about how each enzyme works and how they function together as a “replisome” machine 2,3. At a minimum replisomes in all domains of life contain a helicase, DNA polymerase, primase, and they also require sliding clamps and clamp loaders identified not long ago4,5. This review focuses on the helicase that operates within the context of a replisome machine.
Most helicases are monomers that act in pathways of DNA repair, transcription and translation 6,7. But the helicase at the replication fork of all cells is a ring-shaped hexamer 8–10. Even some bacteriophage and viruses use hexameric helicase rings. Why did helicases evolve a ring shape for replication? Perhaps the ring enhances the grip on DNA to unwind long genomes. Regulation is another possible reason for a ring shape, as replicative helicases usually require other proteins to load them onto DNA, the key objective of origin initiation 1–3,5. For example, in eukaryotes the hexameric helicase is loaded onto double strand (ds) DNA in G1 phase (called licensing), but it remains inactive until S phase when accessory factors activate the hexamer and reposition it onto single strand (ss) DNA 1,11–14. This cell cycle staging of an active helicase underlies why origins fire only once per cell cycle in eukaryotes.
Central life processes in the Last Universal Common Ancestor (LUCA) are typically conserved across all domains of life 15,16. For example, all cells use a universal genetic code and the core elements of the translation and transcription machinery are homologous between bacteria, archaea and eukaryotes. Surprisingly, this is not the case for replication. The bacterial helicase, polymerase and primase are not homologous to their counterparts in eukaryotes/archaea, implying these enzymes evolved twice, independently 17. Presumably LUCA did not evolve a replication process with the precision and efficiency to survive natural selection.
In this review, we compare general features of the bacterial and eukaryotic/archaeal hexameric helicases, and then discuss the main hexameric helicase systems individually. We sincerely apologize for the many studies that are not cited for lack of space.
Comparison of bacterial and eukaryotic hexameric replicative helicases
Helicases can be sorted by sequence homology into six superfamilies (SF) 7. The SF1 and SF2 helicases are typically monomers involved in repair, recombination, transcription and other processes. The hexameric helicases involved in DNA replication include members of SF3, SF4 and SF6. These assort into two main groups, bacterial (SF4) and eukaryotic (SF3,6). The E. coli Rho factor that terminates particular RNA transcripts is a SF5 hexamer that has shed considerable light on replicative helicase function; it translocates on RNA not DNA. Except for eukaryotic CMG, these hexameric helicases are homohexamers. The bacterial SF4 and SF5 helicases contain ATP sites based on the RecA motif and translocate 5′-3′, while the ATP site of eukaryotic SF3 (bovine papilloma virus (BPV) E1 helicase and SV40 T-antigen) and SF6 (eukaryotic CMG and archaeal MCM) helicases are based on the AAA+ fold (ATPases Associated with a variety of Activities) and translocate 3′-5′ 8,9. The RecA and AAA+ motifs are within the larger class of the ASCE fold (The Additional Strand Catalytic glutamatE (ASCE) P-loop family) 18. However, the relationship is distant and the RecA and AAA+ based helicases not only track on DNA in opposite directions but also contain DNA binding elements located at different positions in the secondary structure of the ATP fold, and the orientation of the ATP fold is different within the higher-order quaternary structure of the RecA based and AAA+ based hexamers 9.
Despite their different evolutionary lineages, the bacterial and eukaryotic helicases have many similarities. For example, the individual subunits are dumbbell shaped, composed of N-terminal (NTD) and C-terminal (CTD) domains that give the hexamer an N-tier and C-tier that look like two stacked rings (Fig. 1A,B) 8–10. In all hexameric replicative helicases, the ATP sites are in the CTD at subunit interfaces, with active site residues donated by both subunits (Fig. 1C). This bipartite ATP site construction enables coordination among the different subunits during ATP binding and hydrolysis. All replicative hexameric helicases also contain DNA binding loops that extend into the central pore for DNA interaction. Besides replicative helicases, there are other types of ring shaped oligomeric machines that utilize the RecA fold or AAA+ fold for ATPase-based mechanical activities and also contain substrate binding loops in the central pore 8,9. Examples include the F1 ATPase hexamer ring that rotates a shaft protein to generate a membrane potential, hexameric peptide translocases like ClpX that unfold and feed proteins into cylindrical protease chambers, the NSF/p97 hexamers that function in vesicle fusion, pentameric DNA packaging motors that spool DNA into viral capsids, and pentameric clamp loaders that assemble processivity clamps onto DNA.
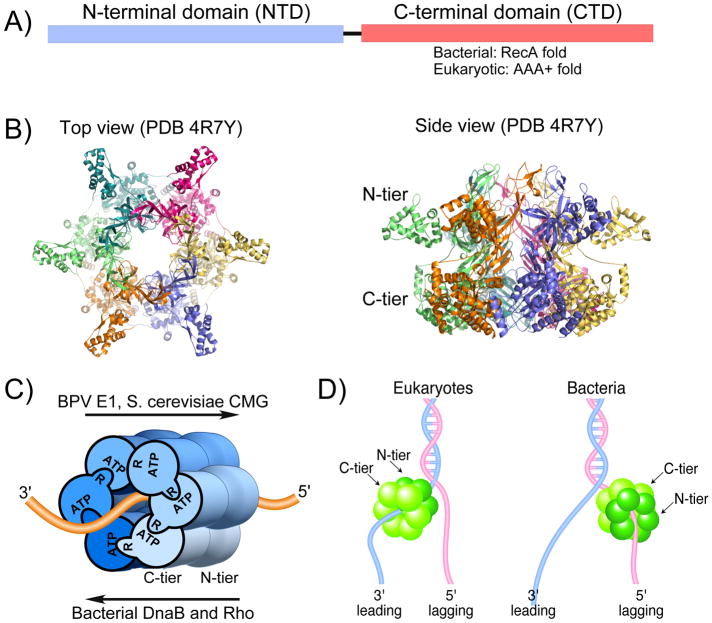
Figure 1. Overview of replicative hexameric helicases.
A) Subunits of hexameric helicases contain NTD and CTD domains; the motors are in the CTD. B) Top and side views of an archaeal MCM homohexamer (PDB 4R7Y), illustrating the two stacked ring appearance. C) ATP sites at subunit interfaces require residues of both subunits. One subunit binds ATP and the other subunit contributes residues, such as a catalytic arginine finger. High resolution structures of 3 replicative helicase-DNA complexes (and E. coli Rho-RNA) reveal that ssDNA passes through the motors similarly, but translocation direction depends on whether the motor is constructed from the RecA fold (bacterial DnaB and Rho) or the AAA+ fold (eukaryotic BPV E1 and S. cerevisiae CMG). Adapted from Fig. 1 of 23. D) Eukaryotic AAA+ fold helicases encircle the leading strand and travel 3′-5′ and bacterial RecA fold helicases encircle the lagging strand and travel 5′-3′.
Hexameric helicases are thought to unwind DNA by encircling and translocating along one strand and partitioning the other strand to the outside of the ring (Fig. 1D) 10,19–21. As the helicase translocates on the strand it encircles, the front tier acts as a moving wedge that peels off the complementary strand of duplex DNA. This process is referred to as “steric exclusion”. The main evidence for the steric exclusion model lies in biochemical studies that test the ability of a hexameric helicase to bypass a bulky substituent placed on one strand (i.e. excluded from the central channel), but not on the strand it tracks upon. However, the structure of replicative hexameric helicases are generally with ssDNA, not forked DNA as discussed later.
There exists only three high resolution structures of replicative helicases bound to nucleic acid in the motors, one each for SF3 (BPV E1), SF4 (DnaB) and SF6 (S. cerevisiae CMG), and also a crystal structure of the SF5 Rho RNA translocase 22–25. The four structures reveal that they bind the nucleic acid with the same polarity through their motors, regardless of RecA (bacterial) or AAA+ (eukaryotic) based architecture22–25. Considering that bacterial and eukaryotic replicative helicases track in opposite directions, yet nucleic acid threads through the motors the same (Fig. 1C) led to the proposal that they fire ATP in opposite directions around the ring to achieve 5′-3′ and 3′-5′ translocation 25, placing the bacterial DnaB SF4 on the lagging strand and eukaryotic SF3 and SF6 helicases on the leading strand (Fig. 1D).
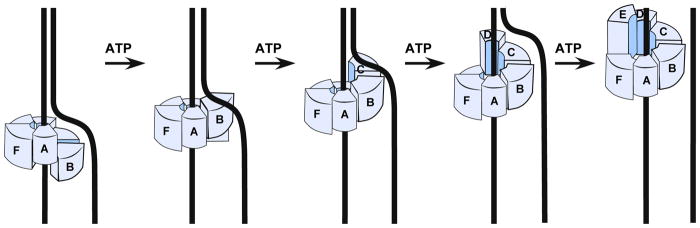
The process by which hexameric helicases translocate on nucleic acid, derived from crystal structures of homohexameric BPV E1 and E. coli Rho, have been proposed 8,9. However, this issue awaits further study, especially for eukaryotic CMG as discussed later in this review. Initial insight into translocation was obtained from T7 gp4 without DNA 22, and then expanded upon from the structure of the eukaryotic AAA+ BPV E1 bound to ssDNA and ADP 22. The E1 subunits bind the ssDNA as a spiral inside the central pore. DNA binding loop(s) in the central channel are on a domain that changes orientation around the ring, correlating with changes in the ADP binding site geometry (dissussed further, below). The domains containing the loops progressively rotate from up (proposed ATP state) to down (proposed apo state) around the ring. The structure is interpreted as sequential ATP hydrolysis around the ring that drives domain rotations to escort DNA down the pore. In this “escort” or “staircase” model, neither helicase nor DNA actually rotate. Instead, DNA binding elements stay attached to the same phosphodiester bond during one round of ATP turnover events, but when a subunit reaches the end of the spiral staircase, hydrolysis detaches that subunit from DNA and its DNA binding loops leapfrog over all the other DNA-binding loops and rebind DNA 6 bases away (see Fig. 2). Progression of these actions six times around the ring results in moving six bases. Structures of RecA based bacterial hexameric Rho RNA translocase (SF5) and DnaB helicase (SF4) with ssDNA suggest a similar mechanism 24,25. Individual subunits of E1 and Rho bind one phosphodiester bond, and the opposing directions of translocation are proposed to be driven by hydrolysis clockwise or counterclockwise around the ring 25. DnaB binds two phosphodiester bonds per subunit and entire subunits are proposed to move rather than rotation of domains 24.
Figure 2. Rotary staircase model of translocation by hexameric helicases.
Each segment of the ring represents a DNA binding element (i.e. DNA binding loop). When one subunit hydrolyzes ATP the DNA binding element lets go of DNA and leapfrogs all the other elements, then rebinds the DNA. In this fashion each DNA binding element of a subunit escorts the same DNA nucleotide(s) through the central pore. There is no rotation of DNA or helicase.
It is not clear to what extent helicases harness the energy of ATP to melt DNA, or to translocate on ssDNA following spontaneous thermal fraying of DNA. Harnessing ATP to unwind DNA is referred to as active unwinding; taking advantage of spontaneous fraying is referred to as passive unwinding. There are reports that helicases may employ both process 27,28. However, if spontaneous fraying were sufficiently rapid, DNA polymerases would not need helicases because polymerases also translocate on DNA and should take advantage of thermal fraying (i.e. would not need a fully passive helicase).
Individual Replicative Hexameric Helicases
Having reviewed general features we now focus on unique features of the six main model replicative helicases. The helicases are divided into two main groups, bacterial (RecA based SF4) and eukaryotic (AAA+ based SF3 and SF6). Structural studies of the bacterial Rho RNA translocase (RecA based SF5) is a also often referred to 25.
Bacterial RecA based 5′-3′ hexameric helicases
Phage T7 gp4 helicase
The phage T7 gene protein 4 (gp4) contains a primase domain attached to the N-terminal domain of the helicase 29. The first insight into a sequential process of translocation on DNA was derived from the structure of T7 gp4 bound to AMPPNP (no DNA) 26. Four subunits bind AMPPNP yet appear structurally asymmetric, possibly resembling ATP, ADP and apo states. Further, the position of loops protruding into the central pore (presumed to bind DNA) were also asymmetric, interpreted as movements in response to sequential hydrolysis around the ring and moving DNA through the pore 26. Biochemical studies support strict sequential hydrolysis, as mixing with mutant subunits demonstrate that a single inactive subunit poisons the entire T7 gp4 hexamer 30. Cocrystals of T7 gp4 with ssDNA have not been obtained as yet to confirm this proposal.
Single-molecule studies demonstrate that gp4 often backtracks, letting DNA reanneal and then resuming forward progression 31. T7 gp4 interacts directly with the T7 Pol 29, and the Pol stimulates helicase unwinding 32. Recent cryoEM studies reveal the organization of a T7 gp4 helicase/primase-dipolymerase replisome (Box 1) 33.
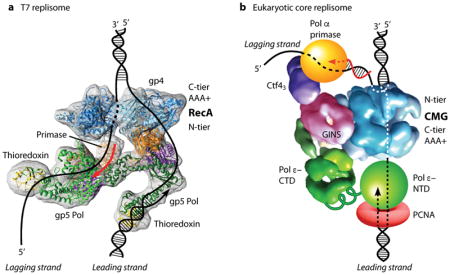
Box 1. . Hexameric helicases are organizing centers of replisomes.
The structure of replisome machines are only recently coming into focus due to the advent of direct electron detection technology. Replicative helicases bind replisome proteins. Phage T7 gp4 helicase contains the primase in the gp4 helicase NTD, and binds the T7 DNA polymerase-thioredoxin complex 29,32. EM 3D reconstruction of the T7 replisome (see figure, panel A) indicates that two T7 Pols bind the same face of the gp4 helicase 33. A crystal structure of another study of the T7 replisome shows a heptamer gp4 bound to 3 Pols, proposed as an intermediate in replisome assembly 89. Eukaryotic CMG binds the leading strand Pol ε 90 and Ctf4 that binds to Pol α-primase 91, forming a core replisome 76. 3D EM studies of CMG-Pol ε reveal Pol ε is located on the C-tier side of CMG, and the Ctf4-Pol α-primase is on the N-tier side of CMG (see figure, panel B, adapted with permission from Fig. 8A of 23) 76. Thus eukaryotic Pols appear to reside above and below the helicase. Chemical cross-link-mass spectrometry and EM studies indicate that the catalytic domain of Pol ε is flexibly attached to the body of Pol ε 23,92,93. Thus the catalytic Pol domain may periodically vacate the leading 3′ terminus while staying attached to CMG, providing access of the leading strand to other factors as needed to continue leading synthesis under various conditions (see 23).
Cellular replicative helicase DnaB
DnaB (SF4 like T7 gp4) is the bacterial chromosomal helicase 7, and like gp4, DnaB functions in the steric exclusion mode 34 and couples to the replicative Pol, stimulating helicase rate 35. Unique to DnaB, the enzyme can use any rNTP, the N-tier is a trimer of dimers, and the C-tier motor is a spiral hexamer (Fig. 3A) 24,36,37.
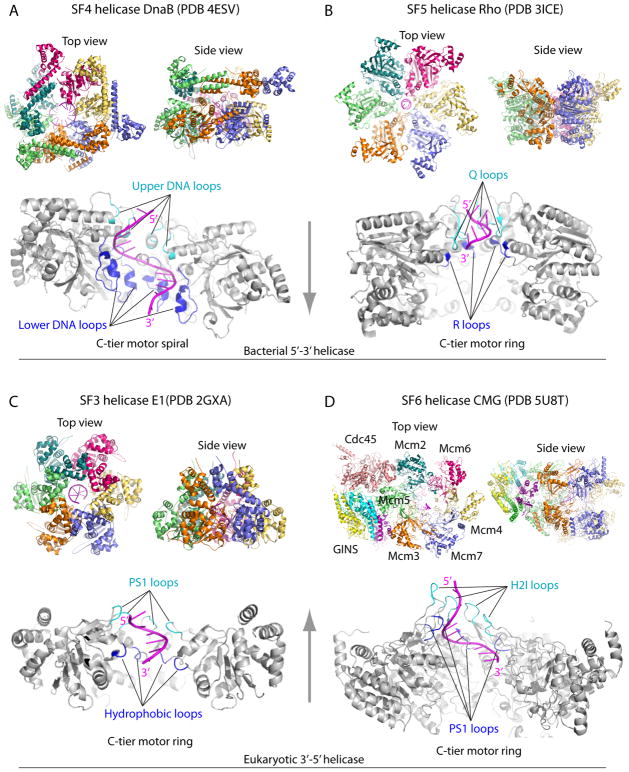
Figure 3. Structures of hexameric helicases bound to a single stranded nucleic acid.
A) Bacterial SF4 DnaB-ssDNA, B) bacterial SF5 Rho transcription terminator-RNA, C) eukaryotic SF3 BPV E1-ssDNA, D) eukaryotic SF6 S. cerevisiae CMG-ssDNA. The helicases are not drawn to the same scale. The vertical arrows between Panels A and B and between panels C and D indicate helicase translocation directions. In each panel, the upper two sub-panels show the N-tier and side views of the hexamers (color), and the lower sub-panel shows the side view of the C-tier motor ring (gray), with the front two subunits removed to better visualize the nucleic acid (magenta)-binding loops (cyan and blue) inside the motor ring. In all cases, the nucleic acids form a righ-handed spiral, bind an upper loop and a lower loop in each of the six protein subunits, with the exception of Mcm2-7 in which the H2I and PS1 loops of the removed Mcm4 and Mcm7 in the front do not bind DNA.
Co-crystals of Bacillus stearothermophilus DnaB with ssDNA and GDP-AlF4 show the ssDNA has a pitch of A form duplex with 11 bases in the C-tier motors, held by loops that each bind 2 phosphodiester bonds (Fig. 3A) 24,38. This 2 base/subunit structure predicts a step size of 2bp/ATP, consistent with biochemical studies 39, twice the 1 bp/ATP predicted for E. coli Rho and BPV E1 22,25. The DnaB central pore is much wider than Rho and E1 (compare Fig. 3A,B,C) and biochemical studies demonstrate that DnaB can encircle and track on dsDNA, but encircles ssDNA at the replication fork 34,40.
Archaeal AAA+ based MCM 3′-5′ homohexameric helicase
The archaeal replicative helicase is a homohexamer of an MCM subunit (SF6 family), homologous to each of the eukaryotic Mcm2–7 subunits. The homohexamer status of archaeal MCM enables mutational studies of all 6 subunits at once (i.e. compared to individual mutations in Mcm2-7), and the lessons learned likely generalize to eukaryotic Mcm2-7 41.
The N-tier of archaeal MCM (and eukaryotic Mcm2-7) is quite large and probably has certain functions beyond processivity (compare archaeal MCM in Fig. 1B to CMG and other helicases in Fig. 3A–D). For example, in Sulfolobus solfataricus (Sso) MCM and Methanothermobacter thermautotrophicus (Mt) MCM, the N-tier binds ssDNA tighter than the C-tier 42. Crystal structures show the NTD has three domains 43–45. The A domain is an α helical region and deletion reduces DNA binding 42. The B domain contains cysteine residues that coordinate zinc 43–45 and replacement of one cysteine with a serine in Mt MCM eliminates helicase, DNA binding and ATPase activity 46. The C domain is an oligonucleotide/oligosaccharide binding (OB) fold, important for ssDNA binding and hexamerization 44,47,48. The crystal structure of an isolated archaeal hexameric N-tier shows it binds ssDNA perpendicular to the central channel via an OB fold loop 44. The perpendicular arrangement of the DNA suggests it is not in a helicase tracking mode, and is proposed to function in origin initiation 44.
The structure of an archaeal MCM C-tier motor domain bound to ssDNA is not yet known. The C-tier AAA+ motor domains of the SF6 family, to which MCMs belong, have two β hairpin loops referred to as presensor 1 (PS1) and helix 2 insert (H2I); both are required for helicase activity 47,48. The structure of an archaeal MCM monomer was originally modeled to form a hexamer, and indicated that the PS1 and H2I loops point into the central pore 47,49. Biochemical studies indicate the loops move with ATP state 50. Interestingly, H2I deletion mutants bind DNA 100-fold tighter than wt, implying the H2I loop may help translocation by destabilizing contacts with DNA 50. A very important step forward in understanding MCM structure/function came from a chimeric MCM hexamer composed of N- and C-tiers spliced together from different archaea, which is active as a helicase and it crystalized with ADP; indeed the PS1 and H2I loops point into the central pore 51. An archaeal MCM doped with three inactive subunits still contains demonstrable activity, casting doubt on strict sequential hydrolysis 52. FRET experiments that used ADP to load archaeal MCM onto a DNA fork having 44 ntd 3′ and 5′ tails concluded that MCM moves C-tier first during unwinding 53. However, ADP may not support translocation to the fork as envisioned, and MCM orientation on DNA may require reinterpretation of the FRET observations. High resolution structures of the archaeal MCM C-tier motors bound to DNA are needed to confirm or determine MCM orientation on DNA.
Biochemical studies indicate that the MCM functions by steric exclusion and make the interesting conclusion that the excluded strand may wrap around the outside perimeter of the ring 54. Furthermore, archaea have homologues to the Cdc45 and GINS and might be needed to correctly orient MCM on DNA 55. Interestingly, isolation of the CTD from the NTD hexameric tiers demonstrate that the C-tier alone has weak catalytic activity, and the N-tier enhances processivity 42,55.
The AAA+ motor domains of archaeal MCM are followed by 60–70 residues that form a winged helix domain (WHD) connected to the CTD by a flexible linker 41,49. A C-terminal WHD is conserved from archaea to most eukaryotic Mcm2-7 subunits. Deletion of the WHD in Sso MCM and Mt MCM enhance their activity 42,50, indicating the WHD in not required for helicase activity. The WHD mediates loading of the MCM hexamer by initiators in eukaryotes 56.
Eukaryotic AAA+ based 3′-5′ hexameric helicases
Viral hexameric helicases
The BPV E1 and SV40 T antigen model eukaryotic viral helicases are AAA+ proteins in the SF3 family. An important difference in SF3 AAA+ and SF6 AAA+ helicases, is that the SF3 AAA+ motor region lacks the H2I loop of SF6 AAA+ (i.e. MCM and CMG) helicases (compare Fig. 3C,D) 8,9. Both E1 and T antigen bind their respective origins in sequence specific fashion for which they have an extra N-terminal origin binding domain that is not essential for helicase activity.
The cocrystal structure of the E1 hexamer with ssDNA and ADP provided insight into helicase translocation, discussed earlier in this review 22, 57. BPV E1 translocates on ssDNA with the N-tier ahead of the C-tier, shown both structurally and biochemically 22,57, opposite the C-tier ahead of N-tier translocation of bacterial DnaB and Rho. No DNA binding elements have been identified in the E1 N-tier, unlike SF6 MCM and CMG helicases. The N-tier of E1 is quite small and is proposed to hexamerize the AAA+ domains and enhance processivity (Fig. 3C) 22. Single-molecule studies indicate that E1 undergoes backslippage 57, as observed in T7 gp4, proposed to prevent extensive unwinding until replisome components have assembled with the helicase.
SV40 T antigen has been crystalized in a variety of nucleotide states without DNA. Unlike other hexameric helicases, T antigen structures show a 6-fold symmetric ring 58–60. Thus the DNA binding loops of T antigen move in unison according to the nucleotide state, suggesting T antigen may function by concerted ATP hydrolysis with all subunits firing at once rather than sequential hydrolysis 61.
Recently T-antigen has been crystalized with origin dsDNA, and the central channel has constriction points that compress and partially melt the origin dsDNA 58,62,63. These observations suggest a squeeze-pump model of initial origin duplex DNA unwinding in which concerted ATP hydrolysis squeezes dsDNA making it melt 61. However, during unwinding at a replication fork T antigen is documented to act by steric exclusion 64.
CMG, the AAA+ helicase for eukaryotic chromosome replication
The eukaryotic nuclear helicase is a heterohexamer of Mcm2-7 65–68 in the sequence Mcm5-3-7-4-6-2 69. A subassembly of Mcm467 contains robust helicase activity 70, but S. cerevisiae Mcm2-7 displays weak activity that requires specialized conditions 71. Whole cell pull-outs using epitope tagged GINS in S. cerevisiae established that Mcm2-7 is tightly associated with Cdc45 and the GINS tetramer at the replication fork along with other factors 72. Purification from Drosophila by the Botchan lab proved the 11-subunit complex was a stable entity with substantial 3′-5′ helicase activity, coining the term CMG (Cdc45, Mcm2-7, GINS) 67,68.
EM structures of Drosophila CMG 73–75 and S. cerevisiae CMG 23,76,77 show the two tiered ring of Mcm2-7 and that the 5 accessory factors bind the side of the Mcm ring (Fig. 3D). GINS and Cdc45 do not bind ATP and are thought to hold Mcm2-7 in an active conformation 67. A small 2nd channel is formed by the accessory proteins, suggesting that one DNA strand may go through it 74. But the high resolution structure of S. cerevisiae CMG reveals that the side chains fill the accessory channel 77. The accessory factors only bind the N-tier of Mcm2-7 77, and therefore the C-tier Mcm2-Mcm5 gate may open and close during catalytic cycles, risking loss of DNA from the central channel 74,77,78. Cross-linking studies demonstrate that Cdc45 helps to keep the leading strand in the central channel during the ATP catalytic cycle 78.
The CMG translocation mechanism is not clear. Two cryoEM studies of CMG-ssDNA reveal 2 phosphodiester bonds bound per subunit PS1 loop, but only 3–4 of the PS1 loops of the six Mcm2-7 subunits of CMG bind ssDNA in any one structure 23,73. Like archaeal MCM, ssDNA clearly interacts with the N-tier in CMG, as well as the motor C-tier. 23,73,77. S. cerevisiae CMG undergoes a large distance change between the N-and C-tiers that may couple ATP hydrolysis to inchworming along DNA 23. A cryoEM study of Drosophila CMG also suggests translocation via coordination between the N- and C- tiers, possibly involving twisting between them 73. A recent cryoEM of S. cerevisiae Mcm2-7 suggests an inchworm translocation process based on conformation changes in the C-tier alone, also suggested by S. cerevisiae CMG studies 23,79. Furthermore, mutation of any one of 4 ATP sites in Drosophila CMG have only a 2-fold, or less effect on Drosophila CMG helicase activity, inconsistent with staircasing translocation 67. Thus, the current evidence suggests CMG may translocate on DNA in a different way than staircasing, but clearly further studies are needed.
A recent cryoEM structure of ATP driven S. cerevisiae CMG engaged at a replication fork indicates that it translocates N-tier ahead of C-tier (Fig. 4) 23. This same orientation, N-tier before C-tier, is established for the SF3 AAA+ BPV E1 helicase 22,57. Low resolution EM studies of Drosophila CMG indicate the opposite orientation on DNA, but higher resolution is needed to unambiguosly determine how the motor domains bind DNA 75. A cryoEM study of Drosophila CMG-fork DNA-ATPγS did not address CMG orientation on DNA because the forked junction was not observed, possibly due to use of ATPγS not supporting translocation from the 3′ end of the leading strand 73. Considering the need for further structural information of metazoan CMG and archaeal MCM, the remainder of this review will discuss the orientation determined by high resolution for the S. cerevisiae CMG, which include numerous structural and biochemical data as presented below.
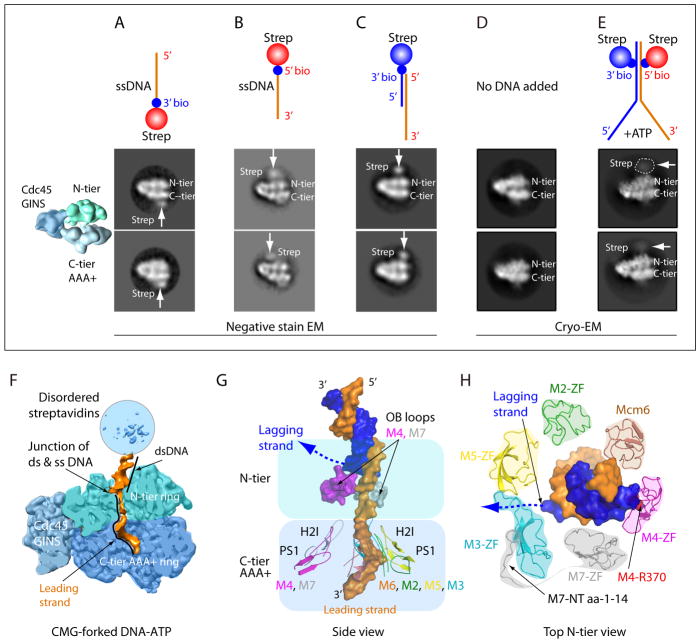
Figure 4. CMG-forked DNA structure with N-tier leading the C-tier during translocation.
A–C) Negative stain EM side views of CMG with streptavidin tipped oligos; colored diagram at left illustrates the tiers of Mcm2-7 and Cdc45/GINS. A) 20mer ssDNA 3′ streptavidin. B) 20mer ssDNA 5′ streptavidin. C) fork lacking a lagging strand and tipped with streptavidin on the ds end. D) CryoEM of CMG without DNA. Note the density of Mcm6 WHD protruding from the C-tier. E) CryoEM of CMG-forked DNA and ATP; CMG is stopped at the streptavidin blocks which are visible as a fuzzy (mobile) region. F) Cut away view through CMG-forked DNA illustrating dsDNA enters the top of the N-tier (light green) and leading ssDNA proceeds into the motor C-tier (blue). G) The DNA-interacting loops of Mcm2-7. Note that the upper OB loops of Mcm4 and Mcm7 block the lagging strand DNA. Inside the C-tier motor ring, the PS1 and H2I loops of Mcm 2, 3, 5, 6 spiral around and interact with the right-handed leading strand DNA coil; the corresponding loops in Mcm 4, 7 do not interact with DNA. Lagging ssDNA is not visible but a possible path is suggested by the dotted line. H) Top view of N-terminal ZF domains of Mcm2-7 binding the dsDNA. Note dsDNA tilts toward and interacts with ZFs of Mcm 7, 4, 6. The Mcm3 ZF has an unusually long loop that forms the base between ZFs of Mcm 3, 5. The ordered N-terminal peptide of Mcm7 warps around and pulls Mcm3 ZF from Mcm5 ZF, creating a gap through which the lagging strand may escape (dotted line). The EM images and illustrations in panels A- E are adapted with permission from Figs 5 and 6 of 23.
The N-tier ahead of C-tier orientation of S. cerevisiae CMG was initially suggested by EM studies using either 3′ or 5′ streptavidin (SA) tipped ssDNA 20mers that were observed to bind opposite sides of CMG, indicating N-tier leads the C-tier during 3′-5′ translocation (Fig. 4A,B). Use of a SA tipped ds/ss hybrid lacking a lagging strand tail, identical to that used in earlier Drosophila studies was also consistent with an N-tier first orientation (Fig. 4C) 23. However, high resolution cryoEM studies directly observed the N-tier first orientation. Initially forked DNA was used with AMPPNP which showed only ssDNA in both the N-tier and C-tier, but failed to show CMG at the fork, probably because ATP hydrolysis is needed for CMG to translocate to the forked junction after threading onto the 3′ tail 23. Thus an experimental strategy was devised to use ATP and block CMG unwinding using SA-biotinylated nucleotide blocks on the duplex stem (Fig. 4D,E) 23. CryoEM 2D averages of side views reveal SA at the N-tier face of CMG (Fig. 4E). The negative control lacking DNA shows a density on the C-tier (bottom) that protrudes and looks like DNA (Fig. 4D), but this is the WHD of Mcm6 79. The 3D reconstruction of CMG-blocked fork DNA-ATP revealed a short stretch of dsDNA with both duplex stands contacted by protruding zinc fingers (ZF) of the N-tier, and the leading ssDNA proceeds into the PS1 loops of the C-tier motor domains (Fig. 3D and Fig. 4F,G,H). Thus unwinding occurs past the point of dsDNA binding by the zinc fingers (ZF), also supported by biochemical studies described below.
The ZFs bind the dsDNA at the fork 23. However the ZF domain of Mcm3 is unusual: (1) It lacks the four Zn-coordinating cysteines found in other Mcm proteins, (2) the Mcm3 ZF is pulled away from the neighboring Mcm5 ZF by the N-terminal 14-residues of Mcm7 that is fully ordered starting from Met1, leaving a sizable gap between the Mcm5 ZF and Mcm3 ZF, and (3) the Mcm3 ZF has a very long loop extending from the second β-strand and the loop forms the base of the gap between ZFs of Mcm 3, 5 (Fig. 4H). Furthermore, the dsDNA is tilted about 28° relative to the axis of the central channel, largely coordinated by the ZFs of Mcm7, Mcm4, Mcm6. Interestingly, the lagging strand ends at a position in the gap between the ZFs of Mcm5 and Mcm3 just described (Fig. 4H). Thus the unwound lagging strand, which was not visualized, perhaps being too mobile, may glance out of CMG via the gap (Fig 4H). Additionally, OB fold loops of Mcm 7 and Mcm 4 reach up to the unwinding point, but their possible involvement in unwinding is not yet determined (Fig. 4G) 23. In hindsight, the orientation that dsDNA enters the helicase from the N-tier makes structural sense, because the ZF domains at the entry of the N-tier ring are evolved to handle the dsDNA and the OB folds below the ZF’s are evolved to handle ssDNA (i.e. the lagging ssDNA).
Biochemical studies support dsDNA entry into S. cerevisiae CMG during normal helicase action 82. Specifically, CMG is inhibited by streptavidin bound to either strand, while homohexameric helicases are only inhibited by obstructions on the tracking strand. However, given time, S. cerevisie CMG pushes past lagging strand blocks mostly without displacing them, and thus ultimately acts by steric exclusion, as demonstrated for replisome action in Xenopus extracts 19,82. Presumably, the CMG-dsDNA connection is eventually lost, enabling bypass of a lagging strand block, although other scenarious are possible (discussed in 82). A recent study demonstrates that Mcm10 enables CMG to rapidly bypass lagging strand blocks and thus possibly disrupts the ZF-dsDNA interaction for bypass of DNA blocks 83. Neither RPA, Mrc1, Tof1, Csm3 or Pol ε-PCNA were capable of rapid block bypass withtout Mcm10 83.
In the S. cerevisiae CMG-forked DNA structure, only four PS1 loops and four H2I loops in the C-tier motor bind ssDNA (Mcm 2,3,5,6). It is fascinating that Drosophila cryoEM CMG-DNA-ATPγS showed 6 ntds of DNA bound to the PS1 loops of Mcm 7,4 and 6 73, while the S. cerevisiae CMG-forked DNA-ATP showed 8 ntds bound to the opposite side of the channel on the PS1 loops of Mcm 2,3,5,6 and no interaction with the PS1 loops of Mcm4 and 7 73. These two structures may resemble two intermediate states during unwinding. Alternatively, since the DNA orientation could not be assigned in the Drosophila cryoEM CMG study 73, and given that CMG encircles dsDNA at an origin, the DNA bound to PS1 loops 7,4,6 in Drosophila CMG might bind the opposite (i.e. lagging) strand, thus explaining the early conclusion of C-tier first polarity 75, and this may be involved in initial duplex binding and/or unwinding at an origin.
It is interesting that in the S. cerevisiae CMG helicase-fork DNA structure, Mcm 2,3,5,6 proteins engage the leading strand with their respective PS1 and H2I loops in the C-tier motor ring, while the PS1 and H2I loops of the two remaining Mcm proteins, Mcm 4 and 7 do not bind DNA. Instead, Mcm 4, 7 engage the ds/ssDNA junction point with their respective upper OB loops in the N-tier ring (Fig. 4G) 23. This architecture raises the possibility that there is a division of labor among the six Mcm proteins, with Mcm 4,7 functioning to split apart the dsDNA, and Mcm 2,3,5,6 motoring on the leading strand. One prediction of such scenario is that the PS1 loops – which are primarily responsibe for DNA translocation – are more important in Mcm 2,3,5,6 than in Mcm4 and 7. Consistent with this prediction, a Mcm3 PS1 mutant is lethal 84, a Mcm5 PS1 mutation has a growth defect at both 18° and 37° 85, but Mcm4 PS1 mutation exhibited no growth defect 85, and Mcm7 PS1 mutation exhibited slow growth but was not lethal 84.
Many replication proteins bind CMG, and EM analysis reveals the organization of a minimal S. cerevisiae replisome containing CMG with DNA polymerases (Box 1). Studies of origin initiation in S. cerevisiae reveal a head-to-head (i.e. N-tier-to-N-tier) double hexamer of Mcm2-7 at origins in G1 phase 11,80,81. A CMG that translocates with the N-tier ahead of the C-tier implies the individual Mcm2-7s of the N-to-N double Mcm2-7 hexamer are directed toward one another23, contrary to a decade of expectation that they are directed outward. This suggests a new paradigm for origin unwinding in which CMGs pass one another, and act as a quality control mechanism to ensure each origin results in two forks (see Box 2). Inward directed CMG motors has also been proposed to create torsional strain that destabilizes dsDNA for initial origin unwinding 23. A recent report that two Mcm2-7 are loaded at two separate origins of opposing polarity also implies that they translocate on DNA N-tier ahead of C-tier to form a head-to-head Mcm dimer 86. CMG requires Mcm10 to unwind the origin, and thus Mcm10 may either help activate CMG helicase or transition CMG onto ssDNA for bypass 2,12,14,83,87,.
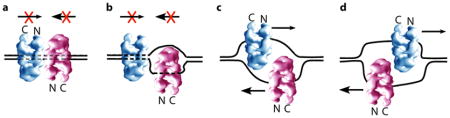
Box 2. A quality control mechanism to ensure bidirectional forks are produced at origins.
In SV40 and eukaryotes the helicase encircles dsDNA at the origin as a head-to-head double hexamer 11,13,62,63. Given the N-first direction of yeast CMG 23, each CMG heads into the other and must transition from dsDNA to encircle opposite strands of ssDNA before they can pass one another (see Figure). If only one CMG makes this transition it will remain blocked by the CMG that encircles dsDNA. Hence, a head-to-head double helicase that encircles dsDNA will ensure that both helicases transition to encircle ssDNA to move out of the origin, thereby ensuring that origins produce two forks and not only one. S. cerevisiae CMG has ZF elements that bind dsDNA and prevent CMG bypass of obsticals on the excluded strand 23,82. This is solved by Mcm10 83, a protein that is required as a last step in origin activation 2,12,14. Mcm10 facilitates CMG bypass of obsticals on the opposite (excluded) strand, and may reflect its function at an origin and also at termination when two forks must again pass one another.
How far the replicative helicase field has come
Intially DNA was thought to wrap on the outside of ring shaped hexameric helicases. But now many structures of hexameric helicases have been solved, and those with DNA show that ssDNA adopts a helical path within the central channel. Studies in the past 10 years show the replicative hexameric helicases ultimately function by steric exclusion. Exact translocation mechanisms are still in play, although a staircasing process with sequential ATP hydrolysis around the ring is the most popular. Despite these large advances, there is clearly a need for much more information to truly understand how these complex machines function.
Important questions within reach
It is still a mystery why bacterial and eukaryotic helicases translocate on opposite strands of a replication fork. Perhaps it is a fluke of evolution, but future studies may solve this riddle. A related question is why bacterial helicases translocate with the C-tier ahead of the N-tier, while eukaryotic S. cerevisiae CMG and BPV E1 helicases travel with the N-tier ahead of the C-tier. It is important in the near term to obtain high resolution structures of archaeal MCM and metazoan CMG bound to DNA to confirm whether the orientation of E1 and S. cerevisiae CMG generalize. The sequential hydrolysis staircase mechanism of translocation is appealing, but different processes have been proposed for CMG and future studies should determine if a re-thinking of this process is needed, or whether different helicases act in various ways. Besides unwinding DNA, replicative helicases must resolve collisions with nucleosomes, transcribing RNA polymerase, recombination intermediates, DNA specific transcription regulators, fork barrier proteins, DNA cross-links, nicks and other lesions. Initial studies have begun to address these encounters, but little is currently known, and these mechanisms are important to genomic instability and disease. How double hexamers unwind dsDNA at origins and transition to encircle ssDNA is also unknown. A detailed understanding of how helicases are loaded onto origins is also central to helicase action. Assembly of CMG requires numerous proteins and the entire process has been recapitulated by pure proteins in vitro 14,87,88, yet numerous questions remain about the detailed process. Longer term questions include knowledge about helicase action in DNA damage checkpoints, programmed fork arrest, retention of epigenetic programing during replication, and cohesion of sister chromosomes. The stage is set for new landmark discoveries and no shortage of important questions to answer about these central hexameric machines.
Acknowledgments
We are grateful to Nina Yao (Rockefeller University) and Lin Bai (Van Andel Research Institute) for making some of the illustrations in this review. This work was supported by grants from the NIH (GM111472 and GM124170 (to H.L.) and GM115809 (to M.O.D.)), the Van Andel Research Institute (H.L.); and the Howard Hughes Medical Institute (M.O.D.). We thank members of the O’Donnell Lab and the Li Lab for their contribution to some of the studies described here.
References
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–7. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Bell SP, Labib K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–67. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–37. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg A, Baker TA. DNA Replication. W.H. Freeman; New York: 1992. [Google Scholar]
- 6.Eoff RL, Raney KD. Helicase-catalysed translocation and strand separation. Biochem Soc Trans. 2005;33:1474–8. doi: 10.1042/BST0331474. [DOI] [PubMed] [Google Scholar]
- 7.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 8.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–57. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–8. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–97. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 11.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–5. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samel SA, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–66. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–5. doi: 10.1038/nature14285. This report reconstitues replication from an origin using pure proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steel M, Penny D. Origins of life: Common ancestry put to the test. Nature. 2010;465:168–9. doi: 10.1038/465168a. [DOI] [PubMed] [Google Scholar]
- 16.Theobald DL. A formal test of the theory of universal common ancestry. Nature. 2010;465:219–22. doi: 10.1038/nature09014. [DOI] [PubMed] [Google Scholar]
- 17.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leipe DD, Koonin EV, Aravind L. Evolution and classificationof P-loop kinases and related protiens. J Mol Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 19.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–41. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker KJ, Johnson KA. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry. 1997;36:14080–7. doi: 10.1021/bi971644v. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem. 2003;278:49171–82. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 22.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 23.Georgescu R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1620500114. This report determined the high resolution structure of S. cerevisiae CMG at a replication fork, indicating CMG is oriented on DNA a direction opposite to long held conventional beliefs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–77. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 27.Donmez I, Rajagopal V, Jeong YJ, Patel SS. Nucleic acid unwinding by hepatitis C virus and bacteriophage t7 helicases is sensitive to base pair stability. J Biol Chem. 2007;282:21116–23. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulczyk AW, Richardson CC. The Replication System of Bacteriophage T7. Enzymes. 2016;39:89–136. doi: 10.1016/bs.enz.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Patel SS, Hingorani MM, Ng WM. The K318A mutant of bacteriophage T7 DNA primase-helicase protein is deficient in helicase but not primase activity and inhibits primase-helicase protein wild–type activities by heterooligomer formation. Biochemistry. 1994;33:7857–68. doi: 10.1021/bi00191a013. [DOI] [PubMed] [Google Scholar]
- 31.Sun B, et al. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–5. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stano NM, et al. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–3. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulczyk AW, Moeller A, Meyer P, Sliz P, Richardson CC. Cryo-EM structure of the replisome reveals multiple interactions coordinating DNA synthesis. Proc Natl Acad Sci U S A. 2017;114:E1848–E1856. doi: 10.1073/pnas.1701252114. This report provides a cryoEM organization of the T7 phage polymerases relative to the helicase-primase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan DL, O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–57. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–50. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 36.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–63. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 37.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–48. [PubMed] [Google Scholar]
- 38.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–6. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 39.Jezewska MJ, Rajendran S, Bujalowski W. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1998;273:9058–69. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–99. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 41.Miller JM, Enemark EJ. Archaeal MCM Proteins as an Analog for the Eukaryotic Mcm2-7 Helicase to Reveal Essential Features of Structure and Function. Archaea. 2015;2015:305497. doi: 10.1155/2015/305497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35:988–98. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fletcher RJ, et al. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10:160–7. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 44.Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. A conserved MCM single-stranded DNA binding element is essential for replication initiation. Elife. 2014;3:e01993. doi: 10.7554/eLife.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Pucci B, Rossi M, Pisani FM, Ladenstein R. Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain. Nucleic Acids Res. 2008;36:3235–43. doi: 10.1093/nar/gkn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poplawski A, Grabowski B, Long SE, Kelman Z. The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J Biol Chem. 2001;276:49371–7. doi: 10.1074/jbc.M108519200. [DOI] [PubMed] [Google Scholar]
- 47.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–6. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasiviswanathan R, Shin JH, Melamud E, Kelman Z. Biochemical characterization of the Methanothermobacter thermautotrophicus minichromosome maintenance (MCM) helicase N-terminal domains. J Biol Chem. 2004;279:28358–66. doi: 10.1074/jbc.M403202200. [DOI] [PubMed] [Google Scholar]
- 49.Bae B, et al. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–22. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Jenkinson ER, Chong JP. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci U S A. 2006;103:7613–8. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JM, Arachea BT, Epling LB, Enemark EJ. Analysis of the crystal structure of an active MCM hexamer. Elife. 2014;3:e03433. doi: 10.7554/eLife.03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–14. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 53.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–62. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 54.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem. 2007;282:34229–34. doi: 10.1074/jbc.M706300200. This report makes the interesting observation that the excluded ssDNA strand bends around the nexus of the tiers in replicative hexameric helicases. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y, Gristwood T, Hodgson B, Trinidad JC, Albers SV, Bell SD. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc Natl Acad Sci U S A. 2016;113:13390–13395. doi: 10.1073/pnas.1613825113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Z, et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee SJ, et al. Dynamic look at DNA unwinding by a replicative helicase. Proc Natl Acad Sci U S A. 2014;111:E827–35. doi: 10.1073/pnas.1322254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gai D, Wang D, Li SX, Chen XS. The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA. Elife. 2016;5 doi: 10.7554/eLife.18129. A crystal structure of SV40 T antigen with origin DNA is observed to squeeze dsDNA for initial melting. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 60.Li D, et al. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–8. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 61.Gai D, Chang YP, Chen XS. Origin DNA melting and unwinding in DNA replication. Curr Opin Struct Biol. 2010;20:756–62. doi: 10.1016/j.sbi.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomez-Lorenzo MG, et al. Large T antigen on the simian virus 40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 2003;22:6205–13. doi: 10.1093/emboj/cdg612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuck S, Stenlund A. Assembly of a double hexameric helicase. Mol Cell. 2005;20:377–89. doi: 10.1016/j.molcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 64.Yardimci H, et al. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–9. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bell SD, Botchan MR. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol. 2013;5:a012807. doi: 10.1101/cshperspect.a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–73. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–58. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. This report proved that the eukaryotic CMG helicase as a tight isolable 11-subunit complex with demonstrable helicase activity, a holy grail landmark finding for the eukaryotic field. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278:4491–9. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 70.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–13. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 71.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–93. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 72.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 73.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. This report infers a cooperation, possibly due to twisting between the C-tier and N-tier for translocation rather than a staircase process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–7. doi: 10.1038/nsmb.2004. This initial negative stain EM study revealed the position of GINS/Cdc45 within CMG for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–82. doi: 10.1038/nsmb.3113. The overall organization of a minimal replisome containing the Pol epsilon and Pol alpha-primase with Ctf4 on CMG is determined by negative stain EM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–24. doi: 10.1038/nsmb.3170. This cryoEM study of CMG suggests an inchworm model of translocation along DNA, either via movements of the N- and C-tiers, or within the C-tier alone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petojevic T, et al. Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc Natl Acad Sci U S A. 2015;112:E249–58. doi: 10.1073/pnas.1422003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhai Y, et al. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol. 2017;24:300–308. doi: 10.1038/nsmb.3374. This cryoEM analysis of yeast Mcm2-7-Cdt1 suggests an inchworm process of translocation occuring within the C-tier alone. [DOI] [PubMed] [Google Scholar]
- 80.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524:186–91. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 81.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langston L, O’Donnell M. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. Elife. 2017;6:e23449. doi: 10.7554/eLife.23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langston L, et al. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. Elife. 2017;6:e29118. doi: 10.7554/eLife.29118. This report finds that Mcm10 activates CMG helicase and enables it to bypass lagging strand blocks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lam SK, Ma X, Sing TL, Shilton BH, Brandl CJ, Davey MJ. The PS1 hairpin of Mcm3 is essential for viability and for DNA unwinding in vitro. PLoS One. 2013;8:e82177. doi: 10.1371/journal.pone.0082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramey CJ, Sclafani RA. Functional Conservation of the Pre-Sensor One Beta-Finger Hairpin (PS1-hp) Structures in Mini-Chromosome Maintenance Proteins of Saccharomyces cerevisiae and Archaea. G3: genes, genomes, genetics. 2014;4:1319–1326. doi: 10.1534/g3.114.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coster G, Diffley JFX. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 2017;357:314–318. doi: 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Looke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol Cell. 2017;65:131–141. doi: 10.1016/j.molcel.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wallen JR, et al. Hybrid Methods Reveal Multiple Flexibly Linked DNA Polymerases within the Bacteriophage T7 Replisome. Structure. 2017;25:157–166. doi: 10.1016/j.str.2016.11.019. A crystal structure is presented for three T7 polymerases bound to a gp4 hexamer, suggesting a structure that may preceed loading onto DNA or be involved during replisome function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langston LD, et al. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A. 2014;111:15390–5. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Asturias FJ, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 93.Zhou JC, et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1700530114. This cryoEM study reveals flexible positions of the catalytic domain of Pol epsilon bound to CMG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–94. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]