Abstract
Background:
Amoebiasis, the cause of dysentery and extra-intestinal abscesses, now becomes second fatal parasitic disease in the world. As routine microscopic diagnosis cannot differentiate causative Entamoeba histolytica from non-pathogenic E. dispar and E. moshkovskii, better diagnosis has to be searched.
Materials and Methods:
Multiplex single round PCR was tested and compared with results of microscopy of wet preparation on 30 samples of diarrheic stools and extra intestinal lesions from amoebiasis suspected patients.
Results:
Microscopy examination showed that 21 (70%) of the samples were positive for E. histolytica/E. dispar/E. moshkovskii complex and 18 (86%) of them contained hematophagous trophozoites. Multiplex single round PCR showed 12 positive results, from which seven were positive for E. histolytica, two were positive for E. moshkovskii, and three showed mixed of E. histolytica and E. moshkovskii. No samples were positive for E. dispar. High positive rate of microscopy might be related with highly suspected amoebiasis cases, while lower positive PCR might be caused by low parasite density and time-related trophozoite disintegration.
Conclusion:
The study showed that multiplex single-round PCR is a valuable diagnostic tool for species differentiation, but cannot replace microscopy in the diagnosis of amoebiasis because of its low sensitivity and impossibility to discriminate the form of E. histolytica and whether it is in the disease-causing stage, while microscopic examination is capable to demonstrate the presence of hematophagous trophozoites that indicates it is invasive and at the disease-causing stage of E. histolytica.
Keywords: E.histolytica/E.dispar/E.moshkovskii, multiplex single round PCR, microscopic diagnosis, hematophagous, invasive trophozoite
Introduction
Amoebiasis shows itself as diarrhea and dysentery in the bowel, while abscesses in intestinal wall and extraintestinal organs may cause amoebomas, fistula, perforations and bleedings (Davidson et al., 1988; Stanley, 2003), makes it the second parasitic disease with highest fatality rate after malaria. Therefore, rapid diagnosis of intestinal amoebiasis is important to determine the right treatment to prevent the worsening of the disease, extraintestinal spread, and to avoid unnecessary use of antiamoebics that may cause drug resistance (Bansal et al., 2006).
However, routine microscopic examination cannot distinguish E. histolytica from non-pathogenic species E. dispar and E. moshkovskii that show similar morphology, while sometimes diagnostic report only write “amoeba positive” without mentioning the species and stage found in the sample. E. histolytica itself may appear as “commensal” lumen dweller and cysts that do not correlate with clinical symptoms. On the other hand, dysentery can be caused by bacteria and virus as well. It is therefore mandatory to consider whether the amoebas found are cyst or trophozoites and whether the trophozoites are at invasive state and disease-causing (Gonzales-Ruiz et al., 1994; WHO, 1997; Lucas and Upcroft, 2001; Ackers, 2002; Shimokawa et al., 2012).
Among methods developed for better diagnostics, PCR is considered the leading one, as it is highly sensitive and specific. However, PCR is technically complex and time-consuming (Tanyuksel and Petri, 2003; Fotedar, 2007b,). Considering that multiplex single round PCR offers a much easier and time saving method, Zulhainan Hamzah, et al. in 2006 developed this technique for diagnosis of amoebiasis with the use of a single genus-specific forward primer that is run together with two or three species-specific reverse primers (specific for E. histolytica, E. dispar and E. moshkovskii) in a single mixture using one microtube and run them in a single round (Zulhainan et al., 2006; 2011).
In this study, we compared multiplex single round PCR, which has never been done in Indonesia, with routine microscopy of wet preparations in the diagnosis of amoebiasis and discussed the benefits and disadvantages of the two methods and also among other similar technics.
Materials and Methods
Sample collection
Informed consent was approved by Medical Ethics Committee under the no. 191/Panke.KKE/VII/2013 issued on 9 July 2013. Thirty samples of stools, abscess fluid, pleural effusion and pus were collected from children and adults suspected to be amoebiasis from Dr. Soetomo Hospital, Surabaya, Indonesia, from August 2013 to June 2014. Macroscopic and microscopic examination was done in the Laboratory of the Department of Parasitology, Faculty of Medicine, Universitas Airlangga while PCR work was performed at the Tropical Disease Institute of Universitas Airlangga, Surabaya.
Fresh unpreserved samples were processed as soon as possible for macroscopic and microscopic examination. The rest were kept frozen in minus 200C for PCR that performed at the earliest convenience. Specimen for all microscopy and PCR procedures were taken from several parts of stool especialy parts suspected to contain trophozoites. For instance, bloody or mucous discharge and sloughed tissue. For liver abscess, the clinician was advised to include specimen from the edge of the abscess where amoebas were expected to recide (Faust et al, 1970). Method to collect the specimen of stool or other specimen was written on the leaflet supplied for nurses and medical analyst.
Microscopy examination was performed using Olympus Student Microscope CX 21 under high power or immersion oil objectives on samples of wet preparation with normal saline to find motile trophozoites and Lugol’s iodine solution to confirm the morphology. Microscopic slides were prepared from feces and other part of stool as mentioned before (mucous or bloody part and sloughed tissue) (Faust et al, 1970;1987; W.H.O, 1997; Tannich, 2004; Carrol, 2007; Crede, 2007).
E. histolytica was considered as hematophagous or at invasive forms if the trophozoites showing specific morphology of E. histolytica containing one or more erythrocytes in the cytoplasm (Gonzalez-Ruiz et al., 1994; C.D.C, 2016) as seen in Figure 2. Positive diagnosis was considered by at least two examiners.
Figure 2.
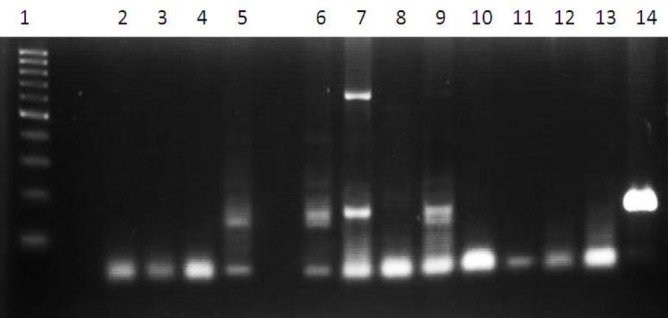
Example of electrophoresis image of PCR products of samples from patients with suspected amoebiasis. Lane 1. Marker with 100 bp leader. Lane 2. Negative control. Lane 3, 4, 8, 10, 11, 12, 13. Negative results.. Lane 5, 9. Positive results from stool samples. Lane 6, 7. Positive results from samples with subcutaneous absces. Lane 14. Positive result showing intensive band. This specimen was taken from an empyema fluid of a patient that was used as positive control after had been checked to the gene bank. Lane 7 showed two bands of mixed infection of E. histolytica and E. moshkovskii.
Polymerase Chain Reaction
DNA was extracted directly from samples using QIAamp stool DNA extraction mini kit (QIAGEN, Hilden, Germany Cat. No 51504) according to the manufacturer’s protocol (Tanyuksel and Petri, 2003; Zulhainan et al., 2006; Fotedar et al., 2007a). The DNA was then stored in -20 0C until use.
The single round PCR method as developed by Hamzah Zulhainan et al. in 2006 was applied (Zulhainan, 2006; 2011). Three (“multiplex”) reverse primers were used to detect different species of the complex E. histolytica, E. dispar and E. moshkovskii. The technics will be informed briefly here. As the forward primer, the conserved sequence of all three species were taken from the central region of the small subunit rRNA gene, i.e., 5’ –ATG CAC GAG AGC GAA AGC AT-3’. Three reverse primers were specific for each of 3 species, i.e., EhR : 5’- GAT CTA GAA ACA ATG CTT CTC T-3’ specific for E. histolytica, EdR: 5’–CAC CAC TTA CTA TCC CTA CC- 3’ for E. dispar, and EmR, 5’-TGA CCG GAG CCA GAG ACA T- 3’ for E. moshkovskii. These primers produced bands on 166, 752 and 580 bp for E. histolytica, E. dispar and E. moshkovski, respectively (Zulhainan, 2006; 2011). PCR amplification was done in a Perkin Elmer thermocycler and the product was electrophoresized and photographed.
Statistical analysis
The results were statistically analyzed by chi-square and McNemar tests in an SPSS version 16 with the level of significance on p = 0.05. Accuracy was proved by the assessment of sensitivity, specificity, positive predictive value and negative predictive value (Fletcher and Fletcher, 2005).
Results
Complete process could be performed to totally 30 samples, consisting of 26 stool samples from children with diarrhea, one sample from a child with empyema, two samples of abscess fluid from a subcutaneous abscess on the lower back, and one sample of lung empyema as an extension of a liver abscess in an adult male. Eighteen samples were from males and 12 from females.
Macroscopic examination showed that all 26 fecal samples consisted of loose stools, with liquid or somewhat thicker consistency, with various colors. Only nine showed typical colour of dysentery, i.e. red or brown, while the others were yellowish or greenish. Microscopic examination showed the presence of free erythrocytes in all nine dysenteric and in five non-dysenteric samples.
Direct microscopy of wet preparations revealed 21 (70%) of 30 samples were positive for E. histolytica/E. dispar/E. moshkovskii complex and 18 (86%) of them contained hematophagous trophozoites that indicated invasive stage, while three samples only showed non-hematophagous trophozoites, typical lumen dwellers of E. histolytica (Table 1, Table 2 and figure 1).
Table 1.
The results of PCR and microscopy in detection of E. histolytica, E. dispar and E. moshkovskii complex
| PCR* | Microscopy | Total | |
|---|---|---|---|
| Positive (%) | Negative (%) | ||
| Positive | 10 (83.3) | 2(16.7) | 12 (100.0) |
| Negative | 11 (61.1) | 7 (38.9) | 18 (100) |
| Total | 21 (70.0) | 9 (30.0) | 30 (100) |
Sensitivity 47.6%, specificity 77.7%, PPV 83.3%, NPV 38.9%, p=0.022 (McNemar test)
Table 2.
Results of species identification by PCR on positive and negative microscopic samples
| Microscopy | PCR | |
|---|---|---|
| Species | Number positive (n, %) | |
| Positive (n = 21) |
E. histolytica | 6 (28.6) |
| E. dispar | 0 (0.0) | |
| E. moshkovskii | 1 (4.8) | |
| Mixed (E. histolytica&E. moshkovskii) | 3 (14.3) | |
| Negative (n = 9) |
E. histolytica | 1 (11.1) |
| E. moshkovskii | 1 (11.1) | |
Figure 1.

Photomicrographs of Entamoeba histolytica trophozoites having one (A, B, C) or more (D) erythrocytes inside the cytoplasm (hematophagous trophozoites). Prepared from fresh dysenteric stool in normal saline (B and D) and lugol iodine solution (A and C).
Multiplex single round PCR revealed 12 (40%) positive test results. From the positive ones, seven (58%) were positive for E. histolytica, two (17%) for E. moshkovskii, and three (25%) showed mixed infections of E. histolytica and E. moshkovskii. From three samples of extraintestinal lesions, two were positive for E. histolytica and one, which was the subcutaneous abscess, was a mixed infection of E. histolytica and E. moshkovskii. No samples were positive for E. dispar (Table 2). Statistical analysis showed that multiplex single round PCR was less sensitive (42,9%) and less specific (90%) than microscopy (Table 3). Compared with detection of hematophagous trophozoites, multiplex single round PCR was also less sensitive (44.4%) and less specific (83.3%) (Table 4). An example of the results of multiplex single round PCR was visualized on electrophoresis image in Figure 2.
Table 3.
Microscopic detection of E. histolytica trophozoites among PCR E. Histolytica positive and negative samples.
| PCR* E. histolytica |
Microscopy positive (n, %) | Microscopy negative (n, %) | Total (n, %) |
|---|---|---|---|
| Positive | 9 (90) | 1 (10) | 10 (100) |
| Negative | 12 (60) | 8 (40) | 20 (100) |
| Total | 21 (70) | 9 (30) | 30 (100) |
Sensitivity 42.9%, specifity 90.0%, PPV 90.0%, NPV 45% p value 0.003 (McNemar test).
Table 4.
The presence of hematophagous and non-hematophagous trophozoites of E. histolytica in microscopy among PCR E. histolytica positive and negative samples
| PCR* E. histolytica | Microscopy | ||
|---|---|---|---|
| Hematophagous trophozoites (n,%) | Non hematophagous trophozoites (n,%) | Total (n, %) | |
| Positive | 8 (88.9) | 1 (11.1) | 9 (100) |
| Negative | 10 (83.3) | 2 (16.7) | 12 (100) |
| Total | 18 (85.7) | 3 (14.3) | 21 (100) |
Sensitivity 44.4%, specifity 83.3%, PPV 80.0%, NPV 50%, p value 0.039 (McNemar test)
Discussion
Multiplex single round PCR detected the presence of two of three species of the Entamoeba complex, but compared to routine microscopy (70% positive), statistically, this PCR (40% positive) was less sensitive and less specific, although two of the 12 PCR-positive cases were not diagnosed by microscopy.
In the first trial of the method (multiplex single round PCR for amoebiasis) in Thailand, positive PCR was found in ten (33%) out of 30 samples with positive microscopy (Zulhainan et al., 2006). This result seems comparable, showing that PCR gave less positivity, but our research tested all samples, not only the positive one in microscopy like that in the first trial.
According to Fotedar (Fotedar et al., 2007a; 2007b), PCR could be negative when the density of parasites is low on microscopic examination. Both microscopy and PCR use small amounts of material that may not contain amoeba, especially for parasites inhabiting distal intestine because the parasites are not evenly distributed in stool. An Iranian study showed 100% PCR-positivity if samples were cultured before (Mojarad et al., 2010). This suggests that heightening the number of parasites by cultivation could increase positivity of single round PCR.
Problem that comes from the presence of DNA polymerase inhibitors in stool samples that normally could cause opaque and thinner bands on the electrophoretic patterns had been anticipated by the use of QIAamp DNA stool kit (from Qiagen); in this work and procedures following its use like concentration, etc. could minimize the effect of polymerase inhibitors (Fotedar, 2007 b). However, a sample coming from empyema patient produced much thicker and broader bands (lane 14 on fig.2) despite the use of the same DNA extraction kit. It could be caused by the presence of higher density of parasites and deposit of their remnants in the abscess. This becomes a factor that builds the opinion that hepatic amoebiasis is easier to diagnose than invasive intestinal amoebiasis (Haque et al, 1998; Petri and Singh, 1999; Ackers, 2002).
Unlike nested PCR, in which the second round can increase the amount of PCR product, this compensation for the effect of polymerase inhibitors in the stool is absent in single-round PCR (Khainar and Parija, 2007). However, using nested PCR, a group of researchers in the United Arab Emirates (El-Bakri, et al, 2013) still found less positive rate of PCR compared to microscopy (23 versus 36 out of 120 fecal samples). Lower positive rate of PCR compared to microscopy was also observed using a two-step PCR, i.e., 81% of microscopy positive samples, also after re-checking (Fotedar et al, 2007b).
As possible explanations for the lower positivity was that the number of parasites fell below detection level and (quick) degeneration of trophozoites during time lapse before starting the PCR (usually first work of PCR /DNA extraction only started after microscopic work finished). Lower positive rate of PCR than microscopy was also showed in a similar study in India (Khairnar and Parija, 2007) and Malaysia using nested PCR (Lau et al, 2013). The use of RT-PCR increased the positive rate but still lower than that of microscopy.
The lower positive rate of PCR in our study could also relate to the very high positivity of microscopic finding (70%) as the standard for comparison. Besides false positivity due to individual examiner, the samples also came from highly suspected cases. The clinicians sent samples that really suspected as amoebiasis intentionally and most patients also showed lowered immune responses like leukemia, human immunodeficiency virus infection, tuberculosis and malnutrition, that made them more vulnerable to amoebic infection. Samples also got special attention and intensive microscopic examination in the laboratory. Anyhow, there are also other amoebas, protozoa and leucocytes that resemble E. histolytica in the stool that results in false positivity (Tanyuksel and Petri, 2003; Zulhainan, 2006; Carroll and Crede, 2007).
In this study in Indonesia, E. dispar was not detected. This was different from studies in other countries where the prevalence of E. dispar was mostly higher than that of E. histolytica (Ali et al., 2003; Khairnar and Parija, 2007; and El-Sobky, 2011). Only Malaysian study among people with gastrointestinal complaints showed higher E. histolytica than E.dispar (Lau, 2013). As reviewed by WHO (1997), E. dispar may be ten times more common than E. histolytica worldwide, but the prevalence may vary significantly with location and therefore needs to be confirmed for different areas. Our findings in Surabaya push us to do further research to confirm whether the prevalence of E. dispar in the Indonesian community outside the hospital is really low as well.
Meanwhile, E. moshkovskii was found in five (16.7%) of 30 samples, two as single species and three as mixed infections with E. histolytica, while Zulhainan et al. (2006; 2011) in Thailand did not find E. moshkovskii in patient’s samples. But a comparable result from a study in Bangladesh using nested PCR among preschool children revealed 21% E. moshkovskii (Ali, et al., 2003). E. moshkovskii, formerly called E. histolytica Laredo strain and was reported as a free-living amoeba, has now been noted as a true human parasite that can be associated with diarrhea, fatigue and weight loss in human, as well as diarrhea and colitis in mice (Tanyuksel et al., 2007; Shimokawa et al, 2012).
In our study, two pure E. moshkovskii cases showed diarrhea with greenish yellow stools, no macroscopic bleeding, no hematophagous trophozoites and no free erythrocytes in microscopy, unlike that in dysentery caused by E. histolytica. The three other cases were mixed with E. histolytica, two showed macroscopic bleeding and hematophagous trophozoites and free erythrocytes.
The third case was a subcutaneous abscess with sanguineous fluid and microscopic finding of hematophagous trophozoites of E. histolytica. The finding of E. moshkoskii from subcutaneous abscess is odd. We checked that E. moshkoskii was found in second specimen from the patient when the abscess had been excised and there was still an open wound that enabled contamination from outside. It was known that E. moshkoskii can be found living in various source of water (Fonseca et al, 2016). Domination of histolytic symptoms in mixed infection in this study made the pathogenic role of E. moshkovskii obscured.
Technically, putting the forward primer (EntaF) and all three reverse primers (EhR, EdR and EmR) together in a single tube like that in our study saves much reagent and time compared to methods which put each primer in three separate tubes and then complete the three tubes with similar amounts of reagents like that in nested PCR. Nested PCR needs additional time for further process to identify species after genus identification. The multiplex single round PCR system also has another advantage, i.e. the presence of mixed infection can be seen directly as double bands (or more) on one lane in the electrophoretic image as seen in Figure 2.
Apart from its disadvantages, microscopic examination is easy to do and remains the only way to demonstrate the presence of hematophagous trophozoites that indicates invasion of the intestinal wall or other tissues where erythrocytes are present to be engulfed. This provides a strong argument that the amoeba is indeed the cause of the disease, especially in cases of dysentery (Gonzalez-Ruiz et al., 1994). While a PCR positive for E. histolytica does not tell whether the parasite is in the trophozoite or cyst stage and whether the parasite is a harmless lumen dweller or an invasive pathogen.
Conclusion
In conclusion, multiplex single-round PCR is relatively simple that can distinguish E. histolytica from E. moskovskii, but cannot replace microscopy in the diagnosis of amoebiasis because of its low sensitivity and its inability to discriminate several forms of E. histolytica and whether it has been in the disease-causing stage. Whereas, microscopic examination remains the only way to demonstrate the presence of hematophagous trophozoites and indicates whether it is invasive and in the disease-causing stage.
Acknowledgement
The study was financed by Airlangga University Research Council (DIPA BOPTN No.7673/UN3/KR/2013). We also thank Budiono, MD, for statistical analysis, and technical assistance of Moh. Yasin, Rachmadany, Moh. Amin and Ms. Fitriah. We also thank and appreciate Prof. Dr. Peterhans van den Broek, MD of Leiden University Medical Center for his precious advice and corrections of the manuscript.
Footnotes
Conflict of Interest: The authors of this manuscript declare that there is no conflict of interest with this study.
References
- 1.Ackers J. P. The diagnostics implication of the separation of Entamoeba histolytica and E. dispar. J. Biosci. 2002;27:573–578. [Google Scholar]
- 2.Ali I. KM, Hossain MB, Roy S, Ayeh-Kumi PF, Petri WA, Jr, Haque R, Clark C. G. Entamoeba moshkovskii infection in children in Bangladesh. Emerg. Infect. Dis. 2003;9:580–584. doi: 10.3201/eid0905.020548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bansal D, Sehgal R, Chawla Y, Malla N, Mahajan R. C. Multidrug resistance in amoebiasis patients. Indian J. Med. Res. 2006;124:189–94. [PubMed] [Google Scholar]
- 4.Carrol M. J. Section 9.2. Collection and preservation of fecal specimens. In: Garcia L. S, Bruckner DA, editors. Diagnostic Medical Parasitology. 4th ed. Washington, DC: ASM Press; 2007. pp. 14–25. [Google Scholar]
- 5.Carrol M. J, Crede P. Section 9.3. Macroscopic and microscopic examination of fecal specimen. In: Garcia L. S, Bruckner DA, editors. Diagnostic Medical Parasitology. 4th ed. Washington, DC: ASM Press; 2007. pp. 28–37. [Google Scholar]
- 6.C.D.C. Laboratory identification of parasitic diseases of public heath concern. Amebiasis. 2016 [Google Scholar]
- 7.Davidson BR, Neoptolemos JP, Watkin D, Talbot I. C. Invasive amoebiasis: an unusual presentation. Gut. 1988;29:682–685. doi: 10.1136/gut.29.5.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Bakri A, Samie A, Ezzedine S, Odeh R. A. Differential detection of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii in fecal samples by nested PCR in the United Arab Emirates (UAE) Acta Parasitol. 2013;58:185–190. doi: 10.2478/s11686-013-0128-8. [DOI] [PubMed] [Google Scholar]
- 9.El-Sobky MM, El Melegi M. A, Abo-Khalil N. A. Use of multiplex PCR in the differential detection of Entamoeba histolytica and/or E. dispar in comparison to microscopic examination. P.U.J. 2011;4:193–200. [Google Scholar]
- 10.Faust EC, Russell P. F, Jung R. C. Craig and Faust's Clinical Parasitology. eighth ed. Chapter 46. hiladelphia: LEA & Febiger; 1970. pp. 783–806. [Google Scholar]
- 11.Fletcher R. H, Fletcher SW. Chapter 3. Diagnosis. In: Fletcher R. H, Fletcher S. W, editors. Clinical Epidemiology: The Essentials. 4th eds. Philadelphia: Lippincott Williams & Wilkins; 2005. pp. 35–58. [Google Scholar]
- 12.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 2007a;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. PCR detection of Entamoeba histolytica, E. dispar and E. moshkovskii in stool samples from Sydney, Australia. J. Clin. Microbiol. 2007b;45:1035–1037. doi: 10.1128/JCM.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Ruiz A, Haque R, Aguirre A, Castañón G, Hall A, Guhl F, Ruiz- Palacios G, Miles M. A, Warhurst D. C. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J. Clin. Pathol. 1994;47:236–239. doi: 10.1136/jcp.47.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque R, Ali IKM, Akther S, Petri W. A. Comparison of PCR, Isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J. Clin. Microbiol. 1998;36:449–452. doi: 10.1128/jcm.36.2.449-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khairnar K, Parija S. C. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. B.M.C. Microbiol. 2007;7:47. doi: 10.1186/1471-2180-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau YL, Anthony C, Fakhrurrazi SA, Ibrahim J, Ithoi I, Mahmud R. Real-Time PCR assay in differentiating E. histolytica, E. dispar and E. Moshkovskii infection in Orang Asli settlements in Malaysia. Parasites & Vectors. 2013;6:250–257. doi: 10.1186/1756-3305-6-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas R, Upcroft J. A. Clinical Significance of redefinition of the agent of amoebiasis. Rev. Latinoam. Microbiol. 2001;43:183–187. [PubMed] [Google Scholar]
- 19.Mojarad EN, Nochi Z, Sahebekhtiari N, Nejad MR, Dabiri H, Haghighi A. Characterization of Entamoeba histolytica and E. dispar in fresh stool by PCR. Gastroenterol. Hepatol. From bed to bench. 2010;3:37–41. [Google Scholar]
- 20.Shimokawa C, Kabir M, Taniuchi M, Mondal D, Kobayashi S, Ali IKM, Sobuz SU, Senba M, Houpt E, Haque R, Petri W.A, Hamano S. Entamoeba moshkovski is associated with diarrhea in infants and causes diarrhea and colitis in mice. J. Infect. Dis. 2012;206:744–751. doi: 10.1093/infdis/jis414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley K. Amoebiasis. The Lancet. 2003;361:1024–1034. [Google Scholar]
- 22.Tannich E. Zur Labordiagnostik von Entamoeba histolytica - infektionen. J. Lab. Med. 2004;28:491–497. [Google Scholar]
- 23.Tanyuksel M, Petri W. A. Laboratory diagnosis of amebiasis. Clin. Microbiol. Rev. 2003;16:713–729. doi: 10.1128/CMR.16.4.713-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanyuksel M, Ulukanliqil M, Guclu Z, Araz E, Koru O, Petri W. A. Two cases of rarely recognized infection with Entamoeba moshkovskii. Am. J. Trop. Med. Hyg. 2007;76:723–724. [PubMed] [Google Scholar]
- 25.World Health Organization. World Health Organization/Pan American Health Organization / UNESCO report of a consultation of experts on amoebiasis. Wkly. Epidemiol. Rev. 1997;72:97–99. [Google Scholar]
- 26.Zulhainan H, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR Assay. J. Clin. Microbiol. 2006;44:3196–3200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zulhainan H, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Development of duplex PCR for detection of Entamoeba moshkovskii and Entamoeba histolytica. [Retrieved on October 12, 2016];Conference Paper. Zulhainan H. 2011 https://www.researchgate.net/publication/215516902 .


