Abstract
Runt-related transcription factor 1 (RUNX1), a member of the RUNX family, is one of the key regulatory proteins in vertebrates. RUNX1 is involved in embryonic development, hematopoiesis, angio-genesis, tumorigenesis and immune response. In the past few decades, studies mainly focused on the effect of RUNX1 on acute leukemia and cancer. Only few studies about the function of RUNX1 in the patholog-ical process of pulmonary diseases have been reported. Recent studies have demonstrated that RUNX1 is highly expressed in both mesenchymal and epithelial compartments of the developing and postnatal lung and that it plays a critical role in the lipopolysaccharide induced lung inflammation by regulating the NF-κB pathway. RUNX1 participates in the regulation of the NF-κB signaling pathway through interaction with IkB kinase complex in the cytoplasm or interaction with the NF-κB subunit P50. NF-κB is well-known signaling pathway necessary for inflammatory response in the lung. This review is to highlight the RUNX1 structure, isoforms and to present the mechanism that RUNX1 regulates NF-κB. This will illus-trate the great potential role of RUNX1 in the inflammation signaling pathway in pulmonary diseases.
Keywords: RUNX1, NF-κB, IKK, P50, lung, pulmonary inflammation
1. Introduction
The Runt-related transcription factor (RUNX) gene family are also known as the acute myeloid leukemia (AML), core-binding factor-α (CBFα) and polyoma enhancer-binding protein-2α (PEBP2α) family, encodes the DNA-binding protein α-chain partners of the heterodimeric CBF complex. The RUNX protein family includes the RUNX1, RUNX2, and RUNX3 transcription factors, which are also known as AML1, AML2 and AML3, respectively [1]. The Runt-related transcription factor 1 (RUNX1/AML1/CBFA2/ PEBP2aB) was first identified in 1991 as a gene involved in the chromosome rearrangement t (8; 21), which is associated with acute myeloid leukemia [2]. Functional disruption of RUNX1 by chromosomal translocation and somatic point mutation occurs frequently in myeloid leukemia. There are over 30 different translocations on chromosome which have been implicated in acute myeloid leukemia and RUNX1 mutations linked with familial predisposition to acute leukemia have also been uncovered [1, 3]. The indispensable role of RUNX1 in hematopoiesis was discovered by transgenic mice in 1996 [4, 5]. In the following decades, a series of studies suggested that in addition to participating in hematopoiesis or angiogenesis, RUNX1, an important transcription factor, is also involved in embryonic development, tumorigenesis, immune response, and especially the inflammatory response [6-9].
2. The Structure of RUNX1
2.1. The Main Domains of RUNX1
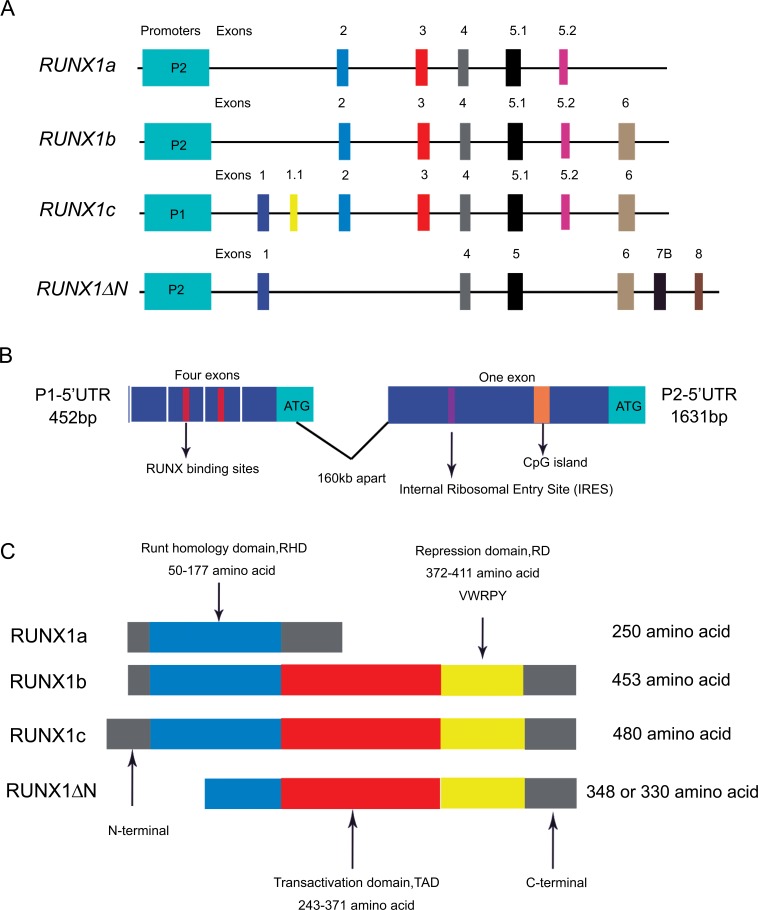
The human RUNX1 gene is located on chromosome 21q22.3 and contains 12 exons with a total length of more than 260kb [10]. The RUNX1 protein consists of three domains, including the runt homology domain(RHD)within the N-terminal region, C-terminal transactivation domain (TAD) and the repression domain (RD) (Fig. 1C). The RHD is coded by exons 2, 3, and 4 of RUNX1 and located in the N-terminal part (amino acids 50-177), while exon 6 codes for the TAD (amino acids 243-371), and part of exon 7 and exon 8 codes for the RD (amino acids 371-411 or 208-243) [11-14]. Significantly, to maintain the normal function of RUNX1, RHD and TAD are both simultaneously required [15]. The RHD in the N-terminal region of RUNX1 protein harbors a conserved domain of 128 residues, which is homologous to the runt transcription factor of Drosophila Melanogaster [16, 17]. The RHD is responsible for DNA-binding and protein-protein interaction. The RHD is able to combine with the TG(T/C)GGT motif, which is known as the runt domain-binding element. In addition to binding to DNA, the RHD is sufficient for interacting with CBFβ, which is coded by a single gene in mammals. CBFβ does not bind to DNA directly, although it confers high-affinity DNA binding and stabilizes the interaction between DNA and the runt domain [18, 19]. The third domain, RD, mediating the transcription of RUNX1 gene function, is divided into different regions. For instance, RD1 is located at the C-terminus of the RHD, which can raise co-arrest factors such as EAR-2 and SIN3A to inhibit transcription of target genes [20, 21]; RD2 is located at the C-terminus of the TAD and plays a role in transcriptional repression or even gene silencing by contacting SUV39H1, a histone methyltransferase [22]. RD3 is located at the C-terminus of the entire RUNX1 protein structure, containing VWRPY motifs in this region, and plays a role in inhibiting the transcription of target genes [23]. In addition to main domains, RUNX1 also contains a nuclear matrix targeting sequence [23, 24]. Taken together, RUNX1 can serve as a transcriptionally repressive or active factor, as well as the nucleus of a trans activator.
Fig. (1).
The structure of the RUNX1 gene and protein. (A) Expression of RUNX1 is initiated by the following two promoters: distal P1 and proximal P2. Different mRNAs of RUNX1 are translated by different exons. (B)Alternative promoters and elaborate splicing alternatives result in generating different 5’-untranslated regions (5’UTRs). (C)Four subtypes of the RUNX1 protein are composed of different combinations of domains that give rise to different features and functions.
2.2. Promoters of RUNX1
In vertebrates, the expression of RUNX1 is regulated by two distantly located promoter regions, distal P1 and proximal P2, which code at least 12 different alternatively spliced isoforms with distinct amino-terminal sequence. The distal P1 and proximal P2 are 160 kb apart. The proximal P1 is located at upstream of the distal P2 [25, 26]. The P1 and P2 promoter regions contain several dispersed binding sites for the RUNX proteins, suggesting an auto-regulation and raising the possibility of cross-regulations between the RUNX genes [10, 16, 27-29]. It is important that in the human and mouse RUNX1 gene, two RUNX sites are identically located at the beginning of the P1 5’-untranslated region (5’UTR) within a conserved sequence of 18bp (Fig. 1B) [30]. Some in vitro studies have attributed the role of positive or negative transcriptional regulation of these dispersed binding sites to bind RUNX protein [31, 32], while using various RUNX knock out (KO) mice in vivo demonstrated that these RUNX-binding sites located at the beginning of the P1 5’UTR do not always function in cross-regulation [33, 34]. The P1 and P2 regions of the RUNX1 gene possess promoter activity [27, 28, 35], that is to say, cells normally expressing RUNX1 have higher promoter-reporter readouts [36].
As mentioned above, transcription of the RUNX1 gene is regulated at the transcriptional level by two promoters that give rise to mRNAs with distinct 5’UTRs and 3’UTRs. The two 5’UTRs have different sizes and structures and each has characteristic features (Fig. 1B) [37]. The P1 5’UTR is usually shorter than the P2 5’UTR. The P1 5’UTR contains four exons and the P2 5’UTR has only one. In RUNX1, the P1 5’UTR is 452bp and P2 5’UTR is exceptionally longer (1631bp). Functional analysis indicated that the P1 5’UTR controls translation via a cap-dependent mechanism, whereas the P2 5’UTR regulates translation by an internal ribosomal entry site (IRES) mechanism [38].
The expression of RUNX1 is highly and distinctly tissue-specific. Previously, it was thought that promoters of RUNX1 exhibit tissue specificity. Isoforms of RUNX1 derived from the P1 promoter are mainly distributed in hematopoietic stem cells, macrophages and T lymphocytes in thymus and spleen [39, 40]. P2 initiates RUNX1 isoforms mainly in non-hematopoietic tissues such as the brain, kidney, pancreas, heart and liver [29]. However, one study proved that none of the distal P1 or proximal P2 exhibited tissue specificity [27], indicating that additional tissue-specific genomic elements are required for regulating the RUNX1 expression, and RUNX1 is more likely to act as an organizer of transcription by recruiting tissue-specific regulatory elements to their binding sites in promoters [27, 41]. The regulatory element 1 (RE1) located in intron 1 of the RUNX1 gene and the regulatory element 2 (RE2) locating in intron 5.2 of the human RUNX1 gene have recently been identified as the element regulating promoter activity. The study of the spatial organization of the RUNX1 gene in different cell types showed that RE1 exhibited an enhancer activity [42, 43], while ER2 was neither an enhancer nor a silencer to distal P1 and the proximal P2 [44]. In addition, a highly conserved sequence named CpG islands, nested within P2 promoter and the last exon of RUNX1 gene, is reported to be responsible for the tissue-specific expression of RUNX1 [45, 46]. Notably, CpG islands are particularly nested in the P2 promoter and at the 3’ end of the gene, but are not found in the P1 promoter [14, 30], suggesting that CpG islands are not absolutely necessary for tissue-specific expression of RUNX1. As tissue-specific expression of RUNX1 is regulated by diverse specific elements or complexes, the mechanism of positive or negative regulation of RUNX1 gene in specific tissues requires further research.
3. RUNX1 Isoforms
RUNX1 exhibits a complex pattern of expression, regulated at the levels of transcription, splicing and translation [14]. The P1 and P2 derived RUNX1 transcripts are processed into a variety of alternatively spliced mRNA isoforms that are differentially expressed in different cell types and at different developmental stages. Therefore, the combination of alternative promoters and elaborate splicing alternatives results in generating different 5’UTRs and thus distinct N-terminal coding regions which leads to production of a large number of isoforms with a variety of biological functions [25, 47-52].
At present, four variants are known for RUNX1: RUNX1a, RUNX1b, RUNX1c, and RUNX1∆N. In general, these different isoforms ranges in size between 20-52 kDa [14]. RUNX1a is translated by exons 2, 3, 4, and 5.1, its synthesis is terminated within exon 5.2, and the protein has 250 amino acids. RUNX1b contains 453 amino acids, which is translated by exons 2, 3, 4, 5.1, and 5.2, and the termination of synthesis occurs within exon 6. The RUNX1c isoform, a 480-amino acid protein, is similar to the RUNX1b but has a longer 5’-terminal regions because its translation starts within exons 1 and 1.1. In the case of the RUNX1∆N isoform, exons 3, 4, 5.1, and 5.3 are involved and its translation is terminated within exon 6. The above mentioned different RUNX1 isoforms coded by the different exons is referenced to the description by E.N. Markova [53]. While Zhang et al. [54] described that RUNX1a is translated by exons 3, 4, 5, 6 and 7A, RUNX1b is coded by exons 3, 4, 5, 6, 7B and 8, RUNX1c is coded by exons 1, 2, 3, 4, 5, 6, 7B and 8, and RUNX1∆N is translated by exons 1, 4, 5, 6, 7B and 8. The reason for this difference in exons description is that exons are not all identified at the same time. Exons 1-6 were the first to be described. New exons, such as 1.1 and 5.1, which were identified between the known ones, were named recently. Therefore, the description made by E.N. Markova et al. was referenced in the structures of RUNX1gene in Fig. (1A) in this review.
Two promoters of the RUNX1 gene produce different isoforms. The P1 promoter encodes RUNX1c, and the P2 promoter encodes RUNX1a, RUNX1b and RUNX1∆N [25]. The RUNX1a isoform retains the DNA-binding runt domain but lacks a large part of the C-terminal region. The RUNX1b is a full-size transcription factor and contains the runt domain and the C-terminal transactivation domains. RUNX1c, the longest isoform, has 27 more N-terminal amino acid residues than RUNX1b. The difference between RUNX1b and RUNX1c is that N-terminal 5-amino-acid sequence of RUNX1b is replaced by a different 33-amino-acid sequence in RUNX1c. The RUNX1∆N isoform, 348 or 330 amino acids, is generated by an alternative splicing of RUNX1 mRNA. This isoform has only half of the runt domain because it is truncated at the N end but contains the complete C-terminal region [14, 45, 52, 54, 55].
The different RUNX1 isoforms are functionally different. For instance, RUNX1b and RUNX1c regulate transcription of target genes by recruiting transactivation factors and interacting with the runt domain-binding sites. RUNX1a and RUNX1∆N, the shorter isoforms of RUNX1, commonly regulate transcription of target genes by acting as negative regulatory factors and by competing with the full-size transcription factors. For example, the region between AML1∆N amino acids 209 and 340, a sequestration site for positive factors, is primarily responsible for the inhibition of AML1 activity [54]. In addition, RUNX1 acts differently in different cell lines, activating the transcription of a target gene in some cells while suppressing it in other cells. For instance, RUNX1 promotes PU.1 expression in myeloid cells and B lymphocytes, but inhibits PU.1 expression in T cells, erythroid cells, and megakaryocytes [56].
4. Expression of RUNX1 in the Lung
In the mouse, RUNX1 expression in the embryo was first detected in definitive hematopoietic stem cells (HSC) and in the endothelial cells at HSC emergence sites. Studies on mouse models have shown that homozygous disruption of RUNX1 between E11.5 and E12.5 results in death of embryos because of a complete absence of hematopoiesis [4, 19]. Conditional knockout of RUNX1 in mice demonstrated that RUNX1 is not absolutely required for hematopoiesis, although the results showed an 80% reduction in platelets and a decrease in megakaryocyte numbers [57]. However, over the past few decades, RUNX1 expression in non-hematopoietic tissues has not been thoroughly investigated and rare study linked the RUNX1 to respiratory system. In 2001, immunohistochemistry staining showed that RUNX1 was expressed in multi-organs, including liver, thymus, olfactory mucosa of the nasal cavity, palatal ridges, mucosa of the esophagus, stomach, as well as the bronchi and respiratory in murine at E14.5-E16.5 [10]. This study fully demonstrated RUNX1 was expressed in both epithelium and mesenchyme of mice during embryogenesis. Hereafter, in 2011, Haley, K. J found that RUNX1 is highly expressed in developing human lung. In this study, RT-qPCR and immunostaining were used to assess the expression of RUNX1 in 20 samples of developing human lung obtained from discarded surgical material. The findings demonstrated that RUNX1 is highly expressed in the human lung at both the pseudoglandular and canalicular stages of fetal lung. Double staining with an epithelial cell marker confirmed that the immunopositive cells were located predominantly in cartilage and epithelium [58], indicating that the pattern of RUNX1 expression in developing human lung is similar to that observed in murine. To further demonstrate the levels of RUNX1 in the respiratory epithelial cells, RUNX1 expression in lung tissues was assessed in both fetal and postnatal mice. We further observed that RUNX1 was expressed in both respiratory epithelial cells and non-epithelial cells at E18.5, postnatal 1 day and 8 weeks. RUNX1 protein was higher (4.6-fold) in the adult lung than that in E18.5 [59]. RUNX1 staining was located in both mesenchymal and epithelial cell compartments of the developing and postnatal lung [59]. According to the diverse functions of RUNX1, it is reasonable to speculate that RUNX1 in respiratory epithelial cells may have a certain role in pulmonary diseases.
5. RUNX1 and NF-ΚB Signaling Pathway in Pulmonary Inflammation
RUNX1 is a highly conserved transcription factor that modulates the embryological development, epithelial cells, angiogenesis, hematopoiesis, immune responses and the inflammatory response [6]. Although it is expressed in epithelial cells in both fetal and postnatal lung, the role of RUNX1 in lung morphogenesis and/or homeostasis are largely unknown at the present. The first research on the potential role of RUNX1 in postnatal pulmonary homeostasis was a recent study linking airway hyper-responsiveness to the pathogenesis of asthma [58]. The study showed that maternal smoke exposure increased the expression of RUNX1 during human fetal lung development. However, the author in this study only indicated a phenomenon without fully illustrating the mechanism because of the limitation of human studies. However, this study still reminds us that RUNX1 may play a role in lung diseases.
The RUNX1 gene has come to prominence because of its role as an essential regulators of cell fates in development and its paradoxical effects in disease. In 2011, Masahiro Nakagawa et al. demonstrated the relationship between RUNX1 and the NF-κB signaling pathway. They found that RUNX1 inhibits NF-κB signaling in the hematopoietic cells by using homozygous RUNX1 floxed mice (RUNX1 f/f) and Mx-Cre-expressing homozygous AML1 floxed mice. This repression is achieved by inhibition of the kinase activity of IKK through physical interaction between RUNX1 and IKK in the cytoplasm [60], demonstrating that RUNX1 can act as a cytoplasmic attenuator of NF-κB signaling in the hematopoietic cells. However, Maocai Luo et al. recently reported that RUNX1 interacted with the NF-κB subunit p50 in macrophages and thus deteriorates septic shock by enhancing TLR4-triggered inflammation. Silencing of RUNX1 attenuated the LPS induced IL-1β and IL-6 production levels and overexpression of RUNX1 promoted IL-1β and IL-6 production in macrophages in response to LPS stimulation [61], indicating that RUNX1 acts as a enhancer of NF-κB signaling in macrophages. In conclusion, these studies indicated to us that RUNX1 may play different roles in different cells. Respiratory epithelium has a remarkable capacity to respond to acute lung injury and plays important roles in repair of the airway after injury. Therefore, it is necessary for us to investigate the function of RUNX1 in respiratory epithelial cells.
In our study, the RUNX1 gene was efficiently deleted from respiratory epithelial cells to produce RUNX1 conditional knockout mice. To determine whether RUNX1 modulates the fetal lung development, lungs from RUNX1 conditionally knockout mice were harvested at E18.5. Although lung maturation was delayed, RUNX1 conditional knockout mice survived postnatal and subsequent growth and maturation of the lung proceeded normally, proving that RUNX1 is not absolutely required for lung morphogenesis during embryonic periods. Acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) are devastating clinical problems with high mortality [62], characterized by diffuse damage to the alveoli resulting in disruption of the endothelial and alveolar epithelial barriers causing microvascular permeability, with protein-rich pulmonary edema [63]. The role of RUNX1 in respiratory epithelial cells during ALI or ARDS has not been reported so far. To better understand the function of RUNX1 in ALI, RUNX1 knockout mice and control mice were exposed to lipopolysaccharide (LPS), an inflammatory stimulus, by intra-tracheal instillation. After LPS exposure, increased respiratory distress, inflammation, and pro-inflammatory cytokines were observed in the RUNX1-deleted mice [59], demonstrating RUNX1 deletion in alveolar epithelial cells enhanced pulmonary inflammation following exposure to LPS. These findings provide insights into the relationship between the expression of RUNX1 in respiratory epithelium and ALI or ARDS, and reveal a protective role of RUNX1 in respiratory epithelial cells.
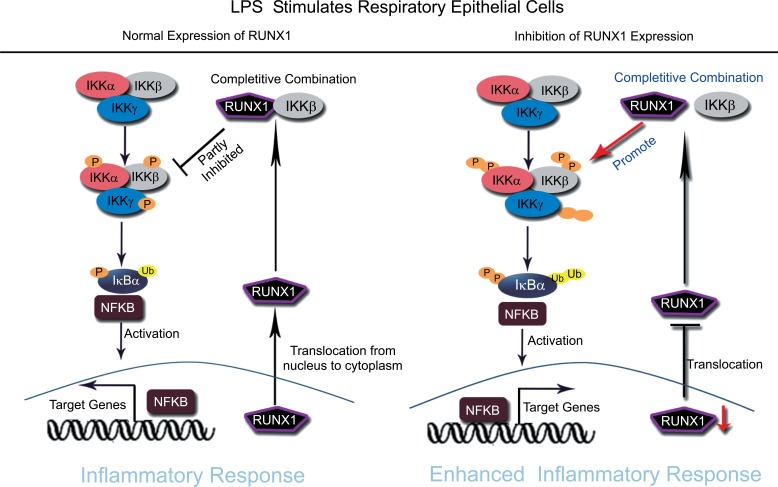
Previous studies have reported that RUNX1 can regulate inflammation response through regulating the NF-κB signaling pathway. NF-κB signaling is also a key regulator of LPS-induced pulmonary inflammation during ALI. Therefore, it is important to understand the relationship between RUNX1 and NF-κB signaling in ALI. We found that the conditional deletion of RUNX1 in alveolar epithelial cells resulted in activation of NF-κB signaling as early as E18.5 and that RUNX1 suppressed NF-κB activation through interaction with IKKβ in cytoplasm of respiratory epithelial cells (Fig. 2) [59]. The results are similar to those observed in hematopoietic cells by Masahiro Nakagawa [60], indicating that RUNX1 might also act as a cytoplasmic attenuator of NF-κB signaling in respiratory epithelial cells. However, the study only investigated the regulation of RUNX1 on one side. RUNX1 usually plays a dual role in a disease. RUNX1 can act as activator or repressor of target genes expression depending upon the large number of transcription factors, activators and repressors that interact with it. This study demonstrated the function of RUNX1 in the cytoplasm, but did not investigate the transcriptional regulation of RUNX1 in the nucleus of respiratory epithelial cells.
Fig. (2).
RUNX1 acts as a cytoplasmic attenuator of NF-κB signaling in respiratory epithelial cells. After respiratory epithelial cells exposure to LPS, RUNX1, the nuclear transcription factor, translocates from the nucleus to the cytoplasm, and competitively binds to IKKβ to form complexes, which resulted in partly inhibiting of the activation of the NF-κB signaling pathway (left). The loss or inhibition of RUNX1 expression in respiratory epithelial cells can enhance LPS induced inflammatory response by promoting the activation of NF-κB signaling (right).
Conclusion
The RUNX1 gene has two promoters coding four different isoforms with distinct amino-terminal sequences. Tissue-specific expression of RUNX1 genes is tightly regulated at the transcriptional and post-transcriptional levels, but the molecular mechanisms that control the spatial and temporal patterns of RUNX1 expression are still not clear. RUNX1 is expressed in both mesenchymal and epithelial cell compartments of the developing and postnatal lung. Recent studies provide insights into a relationship between RUNX1 and ALI/ARDS. RUNX1 binds IKK in the cytoplasm to inhibit NF-κB activity that serves to modulate the innate immune response in the airway. Maintaining precise intracellular levels of RUNX1 in respiratory epithelial cells may be important to the pathogenesis of ALI/ARDS. However, the roles of RUNX1 in regulating other pulmonary diseases are still largely unknown at present.
Consent for Publication
Not applicable.
Acknowledgements
This work was supported by National Nature Science Foundation of China grant nos. 81470268 and 81270129 (F.M.L.), Technology Service Demonstration Project of Sichuan nos.2016GFW0175, Science and Technology Support Program of Sichuan province nos.2016SZ0002.
conflict of interest
The authors declare no conflict of interest, financial or otherwise.
ReferenceS
- 1.Blyth K., Cameron E.R., Neil J.C. The RUNX genes: Gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5(5):376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 2.Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA. 1991;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Braekeleer E., Douet-Guilbert N., Morel F., Le Bris M.J., Ferec C., De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7(1):77–91. doi: 10.2217/fon.10.158. [DOI] [PubMed] [Google Scholar]
- 4.Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 1996;93(8):3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleford P.J., Woollard A. RUNX genes find a niche in stem cell biology. J. Cell. Biochem. 2009;108(1):14–21. doi: 10.1002/jcb.22249. [DOI] [PubMed] [Google Scholar]
- 7.Liu H.P., Cao A.T., Feng T., Li Q., Zhang W., Yao S., Dann S.M., Elson C.O., Cong Y. TGF-beta converts Th1 cells into Th17 cells through stimulation of Runx1 expression. Eur. J. Immunol. 2015;45(4):1010–1018. doi: 10.1002/eji.201444726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong W.F., Kohu K., Nakamura A., Ebina M., Kikuchi T., Tazawa R., Tanaka K., Kon S., Funaki T., Sugahara-Tobinai A., Looi C.Y., Endo S., Funayama R., Kurokawa M., Habu S., Ishii N., Fukumoto M., Nakata K., Takai T., Satake M. Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J. Immunol. 2012;188(11):5408–5420. doi: 10.4049/jimmunol.1102991. [DOI] [PubMed] [Google Scholar]
- 9.Scheitz C.J., Tumbar T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J. Cell. Biochem. 2013;114(5):985–993. doi: 10.1002/jcb.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levanon D., Brenner O., Negreanu V., Bettoun D., Woolf E., Eilam R., Lotem J., Gat U., Otto F., Speck N., Groner Y. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 2001;109(2):413–417. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 11.Kagoshima H., Shigesada K., Satake M., Ito Y., Miyoshi H., Ohki M., Pepling M., Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9(10):338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 1996;16(7):3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanno T., Kanno Y., Chen L.F., Ogawa E., Kim W.Y., Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol. Cell. Biol. 1998;18(5):2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levanon D., Glusman G., Bangsow T., Ben-Asher E., Male D.A., Avidan N., Bangsow C., Hattori M., Taylor T.D., Taudien S., Blechschmidt K., Shimizu N., Rosenthal A., Sakaki Y., Lancet D., Groner Y. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262(1-2):23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- 15.Bernardin F., Friedman A.D. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene. 2002;21(20):3247–3252. doi: 10.1038/sj.onc.1205447. [DOI] [PubMed] [Google Scholar]
- 16.Otto F., Lubbert M., Stock M. Upstream and downstream targets of RUNX proteins. J. Cell. Biochem. 2003;89(1):9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- 17.Bae S.C., Lee J. cDNA cloning of run, a Caenorhabditis elegans Runt domain encoding gene. Gene. 2000;241(2):255–258. doi: 10.1016/s0378-1119(99)00488-6. [DOI] [PubMed] [Google Scholar]
- 18.Westendorf J.J., Hiebert S.W. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J. Cell. Biochem. 1999;32-33:51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G., Liu P.P., Wynshaw-Boris A., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 20.L’Abbate A., Tolomeo D., De Astis F., Lonoce A., Lo Cunsolo C., Muhlematter D., Schoumans J., Vandenberghe P., Van Hoof A., Palumbo O., Carella M., Mazza T., Storlazzi C.T. t(15;21) translocations leading to the concurrent downregulation of RUNX1 and its transcription factor partner genes SIN3A and TCF12 in myeloid disorders. Mol. Cancer. 2015;14:211. doi: 10.1186/s12943-015-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn M.Y., Huang G., Bae S.C., Wee H.J., Kim W.Y., Ito Y. Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO. Proc. Natl. Acad. Sci. USA. 1998;95(4):1812–1817. doi: 10.1073/pnas.95.4.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed-Inderbitzin E., Moreno-Miralles I., Vanden-Eynden S.K., Xie J., Lutterbach B., Durst-Goodwin K.L., Luce K.S., Irvin B.J., Cleary M.L., Brandt S.J., Hiebert S.W. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25(42):5777–5786. doi: 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- 23.Telfer J.C., Hedblom E.E., Anderson M.K., Laurent M.N., Rothenberg E.V. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 2004;172(7):4359–4370. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- 24.Stein G.S., Lian J.B., Stein J.L., van Wijnen A.J., Montecino M., Pratap J., Choi J., Zaidi S.K., Javed A., Gutierrez S., Harrington K., Shen J., Young D. Intranuclear organization of RUNX transcriptional regulatory machinery in biological control of skeletogenesis and cancer. Blood Cells Mol. Dis. 2003;30(2):170–176. doi: 10.1016/s1079-9796(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23(14):2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thandla S.P., Ploski J.E., Raza-Egilmez S.Z., Chhalliyil P.P., Block A.W., de Jong P.J., Aplan P.D. ETV6-AML1 translocation breakpoints cluster near a purine/pyrimidine repeat region in the ETV6 gene. Blood. 1999;93(1):293–299. [PubMed] [Google Scholar]
- 27.Ghozi M.C., Bernstein Y., Negreanu V., Levanon D., Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. USA. 1996;93(5):1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara M., Tagashira S., Harada H., Ogawa S., Katsumata T., Nakatsuka M., Komori T., Takada H. Isolation and characterization of the distal promoter region of mouse Cbfa1. Biochim. Biophys. Acta. 1999;1446(3):265–272. doi: 10.1016/s0167-4781(99)00113-x. [DOI] [PubMed] [Google Scholar]
- 29.Telfer J.C., Rothenberg E.V. Expression and function of a stem cell promoter for the murine CBFalpha2 gene: Distinct roles and regulation in natural killer and T cell development. Dev. Biol. 2001;229(2):363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 30.Bangsow C., Rubins N., Glusman G., Bernstein Y., Negreanu V., Goldenberg D., Lotem J., Ben-Asher E., Lancet D., Levanon D., Groner Y. The RUNX3 gene--sequence, structure and regulated expression. Gene. 2001;279(2):221–232. doi: 10.1016/s0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- 31.Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G.A. Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13(8):1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20(9):2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stricker S., Fundele R., Vortkamp A., Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 2002;245(1):95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 34.Yamashiro T., Aberg T., Levanon D., Groner Y., Thesleff I. Expression of Runx1, -2 and -3 during tooth, palate and craniofacial bone development. Mech. Dev. 2002;119(Suppl. 1):S107–S110. doi: 10.1016/s0925-4773(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 35.Rini D., Calabi F. Identification and comparative analysis of a second runx3 promoter. Gene. 2001;273(1):13–22. doi: 10.1016/s0378-1119(01)00579-0. [DOI] [PubMed] [Google Scholar]
- 36.Harrington K.S., Javed A., Drissi H., McNeil S., Lian J.B., Stein J.L., Van Wijnen A.J., Wang Y.L., Stein G.S. Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J. Cell Sci. 2002;115(Pt 21):4167–4176. doi: 10.1242/jcs.00095. [DOI] [PubMed] [Google Scholar]
- 37.Levanon D., Glusman G., Bettoun D., Ben-Asher E., Negreanu V., Bernstein Y., Harris-Cerruti C., Brenner O., Eilam R., Lotem J., Fainaru O., Goldenberg D., Pozner A., Woolf E., Xiao C., Yarmus M., Groner Y. Phylogenesis and regulated expression of the RUNT domain transcription factors RUNX1 and RUNX3. Blood Cells Mol. Dis. 2003;30(2):161–163. doi: 10.1016/s1079-9796(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 38.Pozner A., Goldenberg D., Negreanu V., Le S.Y., Elroy-Stein O., Levanon D., Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 2000;20(7):2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozner A., Lotem J., Xiao C., Goldenberg D., Brenner O., Negreanu V., Levanon D., Groner Y. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev. Biol. 2007;7:84. doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sroczynska P., Lancrin C., Kouskoff V., Lacaud G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009;114(26):5279–5289. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- 41.Kim J.H., Lee S., Rho J.K., Choe S.Y. AML1, the target of chromosomal rearrangements in human leukemia, regulates the expression of human complement receptor type 1 (CR1) gene. Int. J. Biochem. Cell Biol. 1999;31(9):933–940. doi: 10.1016/s1357-2725(99)00048-5. [DOI] [PubMed] [Google Scholar]
- 42.Markova E.N., Kantidze O.L., Razin S.V. Transcriptional regulation and spatial organisation of the human AML1/RUNX1 gene. J. Cell. Biochem. 2011;112(8):1997–2005. doi: 10.1002/jcb.23117. [DOI] [PubMed] [Google Scholar]
- 43.Nottingham W.T., Jarratt A., Burgess M., Speck C.L., Cheng J.F., Prabhakar S., Rubin E.M., Li P.S., Sloane-Stanley J., Kong A.S.J., de Bruijn M.F. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110(13):4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markova E.N., Razin S.V., Kantidze O.L. Fragment of intron 5.2 of the human RUNX1 gene important for transcription activation is neither enhancer nor MAR-element. Dokl. Biochem. Biophys. 2012;442:26–29. doi: 10.1134/S1607672912010085. [DOI] [PubMed] [Google Scholar]
- 45.Levanon D., Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23(24):4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 46.Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J. Cell. Biochem. 2003;88(5):899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa E., Maruyama M., Kagoshima H., Inuzuka M., Lu J., Satake M., Shigesada K., Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA. 1993;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae S.C., Ogawa E., Maruyama M., Oka H., Satake M., Shigesada K., Jenkins N.A., Gilbert D.J., Copeland N.G., Ito Y. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell. Biol. 1994;14(5):3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae S.C., Takahashi E., Zhang Y.W., Ogawa E., Shigesada K., Namba Y., Satake M., Ito Y. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene. 1995;159(2):245–248. doi: 10.1016/0378-1119(95)00060-j. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T., Tanaka K., Ogawa S., Kurokawa M., Mitani K., Nishida J., Shibata Y., Yazaki Y., Hirai H. An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 1995;14(2):341–350. doi: 10.1002/j.1460-2075.1995.tb07008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levanon D., Negreanu V., Bernstein Y., Bar-Am I., Avivi L., Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23(2):425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 52.Levanon D., Bernstein Y., Negreanu V., Ghozi M.C., Bar-Am I., Aloya R., Goldenberg D., Lotem J., Groner Y. A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol. 1996;15(3):175–185. doi: 10.1089/dna.1996.15.175. [DOI] [PubMed] [Google Scholar]
- 53.Markova E.N., Petrova N.V., Razin S.V., Kantidze O.L. 2012. [PubMed]
- 54.Zhang Y.W., Bae S.C., Huang G., Fu Y.X., Lu J., Ahn M.Y., Kanno Y., Kanno T., Ito Y. A novel transcript encoding an N-terminally truncated AML1/PEBP2 alphaB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol. Cell. Biol. 1997;17(7):4133–4145. doi: 10.1128/mcb.17.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23(24):4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 56.Huang G., Zhang P., Hirai H., Elf S., Yan X., Chen Z., Koschmieder S., Okuno Y., Dayaram T., Growney J.D., Shivdasani R.A., Gilliland D.G., Speck N.A., Nimer S.D., Tenen D.G.P.U. 1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 2008;40(1):51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 57.Growney J.D., Shigematsu H., Li Z., Lee B.H., Adelsperger J., Rowan R., Curley D.P., Kutok J.L., Akashi K., Williams I.R., Speck N.A., Gilliland D.G. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haley K.J., Lasky-Su J., Manoli S.E., Smith L.A., Shahsafaei A., Weiss S.T., Tantisira K. RUNX transcription factors: association with pediatric asthma and modulated by maternal smoking. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011;301(5):L693–L701. doi: 10.1152/ajplung.00348.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang X., Sun L., Jin X., Chen Y., Zhu H., Liang Y., Wu Q., Han X., Liang J., Liu X., Liang Z., Wang G., Luo F. Runt-related transcription factor 1 regulates LPS-induced acute lung injury via NF-kappaB signaling. Am. J. Respir. Cell Mol. Biol. 2017;57(2):174–183. doi: 10.1165/rcmb.2016-0319OC. [DOI] [PubMed] [Google Scholar]
- 60.Nakagawa M., Shimabe M., Watanabe-Okochi N., Arai S., Yoshimi A., Shinohara A., Nishimoto N., Kataoka K., Sato T., Kumano K., Nannya Y., Ichikawa M., Imai Y., Kurokawa M. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-kappaB signaling in the repression of myeloid tumors. Blood. 2011;118(25):6626–6637. doi: 10.1182/blood-2010-12-326710. [DOI] [PubMed] [Google Scholar]
- 61.Luo M.C., Zhou S.Y., Feng D.Y., Xiao J., Li W.Y., Xu C.D., Wang H.Y., Zhou T. Runt-related transcription factor 1 (RUNX1) binds to p50 in macrophages and enhances TLR4-triggered inflammation and septic shock. J. Biol. Chem. 2016;291(42):22011–22020. doi: 10.1074/jbc.M116.715953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu G., Sun Y., Wang K., Chen Z., Wang X., Chang F., Li T., Feng P., Xia Z. Relationship between elevated soluble CD74 and severity of experimental and clinical ALI/ARDS. Sci. Rep. 2016;6:30067. doi: 10.1038/srep30067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heijink I.H., Nawijn M.C., Hackett T.L. Airway epithelial barrier function regulates the pathogenesis of allergic asthma. Clin. Exp. Allergy. 2014;44(5):620–630. doi: 10.1111/cea.12296. [DOI] [PubMed] [Google Scholar]