Abstract
Background:
Total HIV-DNA load in peripheral blood cell (PBMCs) reflects the global viral reservoir that seems not to be affected by antiretroviral treatment. However, some studies report-ed a different permeability of different drugs in cellular compartments.
Objective:
To investigate the relation between the amount of total HIV-1 DNA and different treatment strategies.
Methods:
Total HIV-1 DNA was quantified by real time PCR in PBMCs collected from 161 patients with long-term undetectable HIV-RNA receiving different therapy schedules (3-drug regimens or 2-drug regimen containing Raltegravir as integrase inhibitor).
Results:
Overall, HIV patients who started therapy with a median pre-ART CD4+ cell count >400 cells/mm3 and HIV viral load of 3 log10 copies/ml, achieved a lower amount of HIV total DNA. No significant correlation was found in DNA size when patients were stratified on the basis of different therapeutic protocols. However, HIV DNA load analysis, when only performed in HIV patients with a median pre-ART CD4+ cell count >200 cells/mm3 and HIV viral load < 3 log10 copies/ml, showed a significative DNA decrease in Raltegravir treated group with respect to the NNRTIs-treated group.
Conclusion:
The data emphasize that HIV-DNA level represents a predictive factor in long-term sup-pressive therapy patients. In addition, the diminished reservoir, only observed in patients treated with the NRTI-sparing regimen RAL plus PI/r before immunological and virological derangement, sug-gests that latest generation drugs, such as integrase inhibitors, might represent an optimal chance in the management of HIV infection.
Keywords: HIV-1 DNA, HIV-RNA, CD4+ cells, antiretroviral therapy, reservoir, NRTI-sparing regimen
1. Introduction
Despite the development of more effective antiretroviral protocols, long-lived viral reservoirs represent the main obstacle to eradicating HIV infection [1-3].
Antiretroviral therapy (ART) quickly reduces the pool of activated CD4 cells in peripheral blood and increases the CD4+ T cell count to levels closer to normalcy, but the persistence of competent viral DNA in long-lived CD4 cells seems to be extremely refractory to antiretroviral treatment and immune intervention [4].
HIV-1 DNA, a marker of viral reservoir size, can be found integrated and unintegrated (linear and circular forms) into the host genome in infected cells [3, 5, 6]. Integrated and unintegrated DNA have been extensively studied as different predictive markers of disease progression [7-10]. Independently from RNA level and CD4 counts, a high amount of total HIV-1 DNA seems to be correlated with a rapid progression to AIDS in naïve patients [11, 12] and with viral rebound after treatment interruption in patients receiving therapy [12, 13].
Reservoir dynamics during a suppressive treatment shows a significant initial decay during the first years [14, 15], probably due to the disappearance of non-integrated linear and circular forms [16], but the integrated DNA (also called “provirus”) is extremely stable and persists indefinitely in the long life span of CD4 memory [3].
A critical factor limiting reservoir replenishment remains the timing of the first therapeutical intervention [17, 18]. Whereas the kinetics of plasma viremia is obvious and well-described during an efficient ART, changes in the long-term integrated and episomal HIV-1 DNA are not so clear [19].
In this scenario, the impact of specific ART composition on reservoir size in long-term suppressed antiretroviral therapy patients has been studied to verify a possible correlation between a specific antiretroviral treatment and DNA amount [20-22].
Some therapeutical options seem to influence HIV burden size. In particular, intensification regimens with integrase inhibitors (INI) [23-26] or regimens containing a non-nucleoside reverse transcriptase inhibitor [27] or unconventional simplification regimens like dual or monotherapies with PI [28, 29] have been investigated. Even if different classes of drugs seem to be associated with lower levels of residual viremia [30], NNRTI and Raltegravir-based regimens appear more effective to contain the size of the cellular reservoir [26, 31, 32].
Since unintegrated viral DNA contributes to the total HIV DNA signal, but adds very little to the viral reservoir (due to its limited transcription potential), most attention focused on total proviral DNA [3, 33] correlated to viral rebound in HIV patients on long-term treatment.
On these basis, the quantitative determination of total HIV-l proviral DNA in peripheral blood mononuclear cells (PBMCs) can yield important information on the reservoir and dynamics of HIV infection [34-36] even in the presence of undetectable levels of HIV-RNA viral load in plasma. However, HIV-1 DNA represents a unique biomarker of viral activity, especially when the HIV RNA level drops below detectable limits [20].
PBMCs from HIV-1 patients on long-lasting stable ART have been collected with the dual aim to investigate i) the relationship between the amount of total HIV-1 DNA and immunological/virological parameters, and ii) the possible correlation between the values of HIV-1 DNA load and different antiretroviral regimens (3-drug regimens or 2-drug regimen containing an integrase inhibitor).
2. Materials and Methods
2.1. Patients
We performed an observational, cross-sectional study evaluating cellular HIV DNA in PBMCs from HIV-1 infected patients on stable treatment in the last 4 years.
Among all the patients referred to our laboratory or Infectious diseases Unit, we only selected individuals with undetectable HIV RNA load and available PBMC sample at the end of the study and accessible data concerning age, gender, therapy, transmission route, immunological parameters.
The study was authorized by the local ethics committee (Fellowship Study, Prot 104/2013/U/OSS) and written informed consent was obtained from all patients. Our cohort was characterized by long-lasting HIV-1 infection with a median time of 15.0 years (IQR, 11.8-18.6) and a median cART duration of 8.1 years (IQR, 4.4-10.7). Intensive medical evaluation excluded a history of drug abuse and transmission was established to have occurred by sexual contact. Exclusion criteria were acute infectious diseases, inflammatory, cardiovascular or peripheral vascular diseases.
On the basis of therapeutic schedules, patients were divided into 4 groups: patients receiving abacavir/lamivudine (ABC/3TC) or emtricitabine/tenofovir diproxil (FTC/TDF) plus one non-nucleoside reverse trascriptase inhibitor [NNRTI, efavirenz (EFV) or etravirine (ETV) or Rilpivirine RPV)] (group 1) or nevirapine (NVP) (group 2); patients receiving abacavir/lamivudine (ABC/3TC) or emtricitabine/ tenofovir diproxil (FTC/TDF) plus one boosted protease inhibitor (PI/r, darunavir/ritonavir) (group 3) and patients receiving an integrase inhibitor (INI) such Raltegravir (RAL) plus darunavir/ritonavir (group 4).
2.2. CD4+ Cells Count and RNA Viral Load Determination
CD4+T lymphocytes were determined by flow cytometry (FACScan, Becton & Dickinson, Mountain View, CA, USA) using commercially available monoclonal antibodies and plasma HIV-RNA load was detected by standard commercial viral RNA detection assay (COBAS® AMPLICOR, Roche Molecular Systems, Inc., Branchburg, NJ, USA). All samples were tested for CD4+ cell counts and viral RNA at baseline and at time of observation.
2.3. DNA Proviral Load
Total HIV DNA levels were measured in duplicate in peripheral blood mononuclear cells (PBMCs) by a quantitative real time PCR assay, targeting the long terminal repeat region (Biocentric, Bandol, France). Viral and cellular DNA were extracted from PBMCs, separated by density gradient centrifugation, using a QIAamp DNA mini kit (QIAGEN, Courtaboeuf, France) to obtain 100 μl of eluate, according to the manufacturer ’s instructions. The DNA concentration was quantified using fluorescence readings at 260 nm (Nanodrop, Labtech, Ringmer, UK) and the samples were eventually diluted in distilled water to reach 1μg of DNA /PCR tube (considered to be equivalent to 150,000 cells) [37]. Results were expressed as number of HIV DNA copies/106 PBMCs. For a given sample, an HIV-1 DNA concentration “C” is calculated as reported: C= HIV DNA copies/PCR test (mean value) x 1.000,000/150,000 (copies/106 cells). The quantitative amount of total DNA proviral load was only performed at our last observation.
2.4. Statistical Analysis
The continuous variables were expressed as the median [interquartile range (IQR)], and the categorical variables were expressed as percentages. The univariable linear regression model was used to examine the association between HIV-1 DNA and pre-HAART viroimmunological data (nadir CD4 cell count and plasma HIV-RNA zenith) and the HIV-1 DNA and the duration of suppressive ART.
Differences between nominal data were tested for statistical significance by non-parametric test (Mann- Whitney).
A p-value of <0.05 was considered significant.
3. Results
3.1. Pre-ART HIV-RNA Viral Load and CD4+ Cell Counts are Correlated with HIV DNA Levels at Time of Observation
To evaluate the relation between zenith values of HIV RNA and DNA load levels, HIV-1 patients were stratified on the basis of different pre-ART viremia levels (< 3 log10 copies/ml, 4 log10 copies/ml and > 5log10 copies/ml of HIV-RNA).
In particular, 38 patients showed pre-ART viral load <103 log10 copies/ml, (median 3.90, IQR, 3.45-3.95), 70 patients had 104 log10 copies/ml (median 4.50, IQR, 4.25-4.80) and 53 patients 105 log10 copies/ml (median 5.48, IQR, 5.23-5.65).
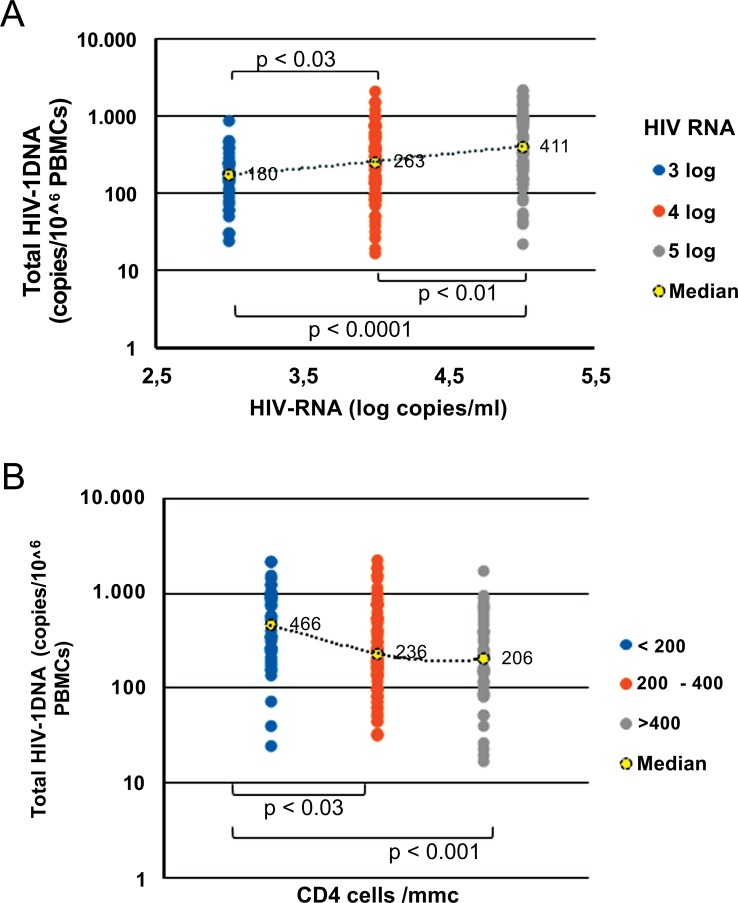
Patients who started therapy with lower RNA viral replication (<3 log10 copies/ml) showed lower DNA amounts [180 copies/106 PBMCs (IQR, 95-255)] in comparison with HIV patients who entered therapy with a higher level of viral replication (4 log10 copies/ml and > 5log10 copies/ml) who reached median DNA levels of 263 copies/106 PBMCs (IQR, 114-595) and 411 copies/106 PBMCs (IQR, 156-975) respectively [p<0.03 and p<0.0001] (Fig. 1A).
Fig. (1).
Correlation between total HIV-DNA and zenith HIV-RNA (A) and nadir CD4+ cell count (B). Fig. (1A). 180, 263 and 411 represent the median copy numbers of HIV DNA load detected in HIV patients regardless of therapy protocols. Fig. (1B): 466, 236 and 206 represent the median CD4 cell count/mmc detected in HIV patients regardless of therapy protocols.
Subjects were also stratified on the basis of nadir CD4+ cell counts. In particular, 40 patients showed pre-ART CD4+ cell counts >400 cells/mm3 (median 476, IQR, 436-520 cells/mm3), 79 patients had CD4+ cell counts from 200 to 400 cells/mm3 (median 302, IQR, 263-344 cells/mm3) and 42 patients CD4+ cell counts <200 cells/mm3 (median 93, IQR, 61-134 cells/mm3). Results showed that nadir CD4+ cell values are inversely related to median HIV DNA levels, showing that patients with pre-therapy lower nadir CD4+ counts achieved higher total HIV DNA amount [(466 copies/106 PBMCs, (IQR 243-968)] than patients with intermediate or higher CD4+ counts [236 copies/106 PBMCs (IQR, 116-544, p< 0.04) and 206 copies/106 PBMCs (IQR, 96-407, p<0.001), respectively] (Fig. 1B).
3.2. Total HIV-1 DNA Level is Associated with Different Therapeutic Protocols
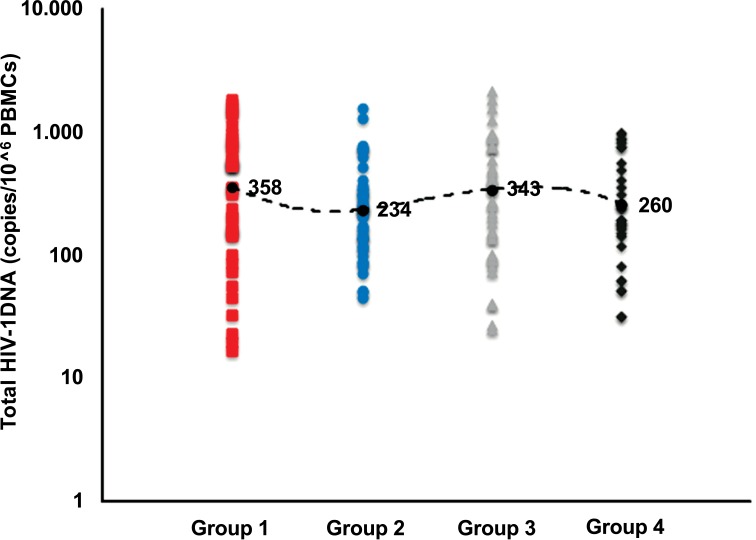
HIV-1 patients were further divided on the basis of specific regimens: group 1) 39 patients receiving an NRTI backbone (ABC/3TC or FTC/TDF) plus an NNRTI (EFV or ETV or RPV); group 2) 42 patients receiving an NRTI backbone (ABC/3TC or FTC/TDF) plus nevirapine (NNRTIs); group 3) 41 patients receiving an NRTI backbone (ABC/ 3TC or FTC/TDF) plus darunavir/ritonavir (PI/r), and group 4) 39 patients receiving a dual therapy of Raltegravir (INI) plus darunavir/ritonavir (Table 1). No significant differences in median levels of total HIV DNA were observed in these groups (Fig. 2), even if lower DNA values were reached in patients belonging to groups 2 and 4 (NRTI backbone plus nevirapine and Raltegravir plus darunavir/ritonavir, respectively).
Table 1.
Patients’ characteristics based on antiretroviral therapy regimens administered in the last four years.
| Therapy | ||||
|---|---|---|---|---|
|
Group 1
Backbone plus NNRTI (EFV or ETV or RPV) |
Group 2
Backbone plus NNRTI (NVP) |
Group 3
Backbone plus PI/r (DRV/r) |
Group 4
INI (RAL) plus PI/r (DRV/r) |
|
| N° of patients | 39 | 42 | 41 | 39 |
| Age | 41 (IQR, 38-48) | 49 (IQR, 42-55) | 45 (IQR, 42-51) | 48 (IQR, 42-55) |
| Male, no (%) | 87 | 76 | 92 | 79 |
| Risk group, no (%): Homo/bisexual | 62 | 60 | 56 | 52 |
| Risk group, no (%): Heterosexual | 34 | 38 | 37 | 43 |
| Risk group, no (%): Drug user | 4 | 2 | 7 | 5 |
| Duration of current cART (years) | 4.4 (IQR, 3.9-4.8) | 4.5 (IQR, 4.1-5.2) | 4.2 (IQR, 3.6-5.5) | 4.3 (IQR, 3.6-5.1) |
| Zenith HIV RNA (log copies/ml) | 4.96 (IQR, 4.58-5.28) | 4.80 (IQR, 4.30-5.38) | 4.45 (IQR, 4.00-4.95) | 4.00 (IQR, 4.00-5.08) |
| Current CD4 cell count (cells/mm3) | 706 (IQR, 589-925) | 660 (IQR, 521-849) | 776 (IQR, 577-943) | 789 (IQR, 589-1045) |
| Nadir CD4 cell count (cells/mm3) | 319 (IQR, 257-400) | 296 (IQR, 160-240) | 290 (IQR, 176-396) | 284 (IQR, 71-390) |
Data are median (IQR, interquartile range) and values are expressed as n (%); cART, combination antiretroviral therapy; backbone: abacavir/lamivudine (ABC/3TC) or emtricitabine/tenofovir diproxil (FTC/TDF); NNRTI: non-nucleoside reverse transcriptase inhibitors; EFV: Efavirenz; ETV: Etravirine; RPV: rilpivirine; NVP: Nevirapine; DRV/r: darunavir/ritonavir; INI: integrase inhibitor; RAL: Raltegravir.
Fig. (2).
Median levels of total HIV-DNA in 161 patients on stable ART stratified on the basis of therapy protocols. 358, 234, 343 and 260 represent the median copy numbers of HIV DNA load detected in HIV patients undergoing different therapy protocols. From left to right: patients receiving abacavir/lamivudine (ABC/3TC) or emtricitabine/tenofovir diproxil (FTC/TDF) plus one non-nucleoside reverse trascriptase inhibitor (NNRTI, efavirenz or etravirine or rilpivirine) (group I) or nevirapine (group II); patients receiving abacavir/lamivudine (ABC/3TC) or emtricitabine/tenofovir diproxil (FTC/TDF) plus one boosted protease inhibitor (PI/r, darunavir/ritonavir) (group III), and patients receiving an integrase inhibitor (INI) such Raltegravir (RAL) plus darunavir/ritonavir (group IV).
In particular, the median values of DNA load at the time of observation were 358 (IQR, 148-800), 234 (IQR, 123-344), 343 (IQR, 135-767) and 260 (IQR, 159-787) copies/106 PBMCs in groups 1, 2, 3 and 4 respectively, suggesting that only two antiretroviral regimens analyzed seem to have a moderate impact on achieving low level proviral cellular DNA.
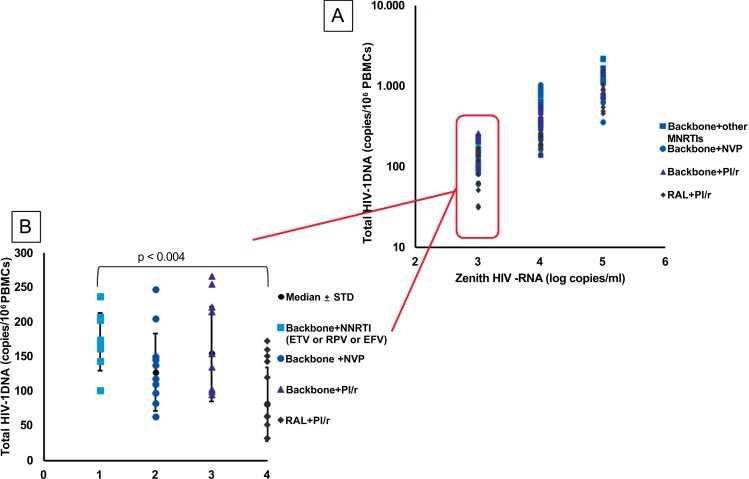
Finally, to investigate whether different levels of viral replication and/or CD4+ cell counts could predict therapy success (in terms of smaller reservoir size), we restricted the analysis to HIV-1 patients with CD4+ values > 200 cells/mm3 divided on the basis of basal viremia levels (3 log10, 4 log10 and 5 log10 HIV-RNA). Results (Fig. 3A) showed that the lower levels of proviral load were only
Fig. (3).
(A). Total HIV-DNA amount in HIV patients treated with different antiretroviral therapy and stratified on zenith HIV-RNA (3 log10, 4 log10 and 5 log10 HIV-RNA) and CD4+ >200 cells/mm3. (B). Total HIV-DNA amount in HIV patients treated with different antiretroviral therapy protocols, selected on restricted parameters (T CD4 cells > 200 cells/mm3 and HIV-RNA level <3 log10). Backbone: abacavir/lamivudine (ABC/3TC) or emtricitabine/tenofovir diproxil (FTC/TDF); NNRTI:non-nucleoside reverse transcriptase inhibitors (EFV: Efavirenz; ETV: Etravirine; RPV: Rilpivirine; NVP: nevirapine); PI/r: protease inhibitors (darunavir/ritonavir); RAL: Raltegravir
obtained in subjects who started therapy with CD4+> 200 cells /mm3 and HIV-RNA viral load <3 Log.
Taking this concomitant condition (T CD4+ > 200 cells/mm3 and HIV-RNA level <3 log10) as the reference baseline and focusing attention on the amount of HIV DNA in patients treated with specific therapeutical protocols, we did not observe a strong statistical association among the 4 regimens and proviral amount. Neverthelss, HIV DNA load analysis, when only performed in HIV patients with a median pre-ART CD4+ cell count >200 cells/mm3 and HIV viral load < 3 log10 copies/ml, showed a significative DNA decrease in Raltegravir treated group respect to the NNRTIs-treated group (95 vs. 174 copies/106PBMCs, p<0.004) (Fig. 3B).
4. Discussion
HIV persistence in cellular and anatomic reservoirs is a major obstacle to HIV-1 infection eradication. Current antiretroviral therapy successfully suppresses replication in plasma and reduces viral transmission, but is not able to definitively flush out the “hidden virus” established in the first phase of infection [38].
Since the measurement of HIV DNA level, a surrogate marker able to predict viral rebound and disease progression [12, 13], might shed more light on the meaning and dynamics of cellular reservoir [39, 40] our first analysis focused on all patients, regardless of specific therapy. Results demonstrated the close association between HIV-1 DNA and the pre-therapy immune-virological markers, confirming several data on this topic [12, 22]. The HIV-DNA amount is negatively associated with the number of nadir CD4+ cells and directly correlated with the pre-therapy viral load, further confirming that patients who underwent therapy before immune system derangement were more likely to reach more contained proviral DNA levels.
Although available data on the correlation between proviral HIV DNA and different antiretroviral regimens warrant further investigation, much recent research interest has focused on understanding the best therapeutic combinations with better penetration of sanctuaries and lower long-term side-effects in patients on stable virological control [21, 23-29, 39].
HIV DNA is already present after the onset of infection and persists over time, representing a virtually endless reservoir, ready to produce new virus. Hence our study also undertook a quantitative analysis of cellular proviral HIV DNA in patients on stable combination antiretroviral therapy for over 4 years, including four different antiretroviral regimens containing a classic triplice cocktail [two NRTI plus one NNRTI [Efavirenz or rilpivirine or etravirine (group1) or nevirapine (group 2)], two NRTI plus a protease inhibitor with booster of ritonavir (darunavir/r) (group 3)] or a regimen containing two drugs [INI (Raltegravir) plus PI (darunavir/ritonavir) (group 4).
To this aim, we only selected HIV patients on stable treatment with undetectable levels of circulating virus in the last four years with available data concerning age, gender, therapy, transmission route, immunological parameters and PBMC sample available for HIV proviral quantification.
In our experimental conditions, HIV-DNA amount was not significantly altered in PBMCs from patients stratified on the basis of different therapeutic protocols, albeit a slightly smaller reservoir has been documented in nevirapine-treated patients (NNRTI), showing a better penetration score in different anatomical districts [30, 32] and in Raltegravir-treated patients, affecting the viral integration step [41] indicating a partial but not definitive role of nevirapine or Raltegravir regimens in reducing the viral reservoir [17, 26, 31, 42].
In any case, the slight, but not drastic, decrease in PBMC DNA in patients on therapies containing an anti-integrase drug might be due to several reasons, such as the persistence of ongoing replication leading to an transient increase in unintegrated forms (2-LTR) of HIV-1 DNA or the migration of cells with unintegrated DNA from other tissue reservoirs to the peripheral blood [26, 31].
Recently, a decrease in DNA load was demonstrated in HIV patients only after the initiation of the RAL-containing regimen, but not maintained over time, suggesting that most HIV-1 DNA is integrated in the majority of well virologically controlled patients [26, 43-46]. Moreover, we found that the concomitant presence of an HIV RNA value < 3 log and CD4+ cell count nadir > 200 cells/mm3 at baseline could offer optimistic information on the effectiveness of specific treatment to contain reservoir size. In this connection, the lower HIV-DNA load observed in the RAL-treated group, underlying that a dual therapy containing Raltegravir could have a good efficacy to control the DNA amount in PBMCs and could represent an interesting alternative simplification strategy, especially in patients with comorbidity, due to the good long-term tolerability of Raltegravir and high genetic barrier of protease inhibitors [47].
Our study has some limitations such as: i) the evaluation of HIV-DNA using a PCR-based assay is not able to distinguish replication-competent viruses from viral defective forms, ii) the detection of DNA in PBMCs, representing only a small part of viral reservoir, iii) the lack of analysis of chronic inflammation markers (PCR, D-dimer, Il-6, TNF), iiii) the lack of randomization in the enrollment of patients.
Conclusion
In this cohort of virologically suppressed HIV-infected subjects, the amount of HIV DNA was comparable in patients receiving different type of NRTI-based cART including non-nucleoside analogues and PI/r; however, in patients who started therapy with T CD4+ > 200 cells/mm3 and HIV-RNA level <3 log10, total HIV-DNA was significantly lower Raltegravir plus PI/r treated group respect to NNRTI-group. These data open the way to exploring optimization models of therapy, indicating a potential role of Raltegravir, or other integrase inhibitors, in reducing the size of viral reservoir when switching from a suppressive successful therapy. So, much larger studies will be needed to carefully select patients for NRTI-sparing regimens, to avoid a potentially “re-seeding” of reservoir in well-controlled patients.
Ethics Approval and Consent to Participate
The study was approved by the local ethics committee [Comitato Etico indipendente dell’Azienda Ospedaliera Universitaria di Bologna, Italia, (Fellowship Study, Prot 104/2013/U/OSS)].
Human and Animal Rights
No animal were used in this research. All humans research procedures followed were in accordance with the standards set forth in the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/) principles of 1975, as revised in 2008 (http://www.wma.net/en/20activities/10ethics/10helsinki/).
Consent for Publication
Not applicable.
Acknowledgements
We are grateful to Dr. Paolo Bonini for statistical assistance and Drs. Cristina Coppola and Maria Luce Nemi for technical assistance throughout the experimental procedures.
The work was performed at the Microbiology Section of the Department of Experimental, Diagnostic and Specialty Medicine, School of Medicine, University of Bologna.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
Funding
This work was supported by Fondazione del Monte di Bologna e Ravenna (CON20RE14), Fondazione Fanti Melloni 2015 and RFO grants 2015 and 2016, University of Bologna, Italy.
Author contribution
Isabella Bon acquired, analyzed the data and drafted the manuscript.
Serena Longo, Giuseppina Musumeci and Davide Gibellini contributed to the analysis of results obtained.
Alessia Bertoldi and Vanessa D’Urbano collected and processed plasma samples.
Leonardo Calza and Eleonora Magistrelli attended the patients and helped in data interpretation.
Pier Luigi Viale contributed to the study design.
Maria Carla Re contributed to the study design, supervised the project and wrote the manuscript.
All authors contributed to the revision of the manuscript and approved the final version.
References
- 1.Chun T.W. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archin N.M., Vaidya N.K., Kuruc J.D., et al. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. USA. 2012;109:9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharaf R.R., Li J.Z. The Alphabet Soup of HIV Reservoir Markers. Curr. HIV/AIDS Rep. 2017;14:72–81. doi: 10.1007/s11904-017-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buzon M.J., Martin-Gayo E., Pereyra F., et al. Long-term antiretroviral treatment at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1 infected CD4 T cells. J. Virol. 2014;88:10056–10065. doi: 10.1128/JVI.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler S.L., Hansen M.S., Bushman F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 6.Vandegraaff N., Kumar R., Burrell C.J. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 2001;1:11253–11260. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell P., Montaner L.J., Maul G.G. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 2001;75:7683–7691. doi: 10.1128/JVI.75.16.7683-7691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koelsch K.K., Liu L., Haubrich R., et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J. Infect. Dis. 2008;197:411–417. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 9.Re M.C., Vitone F., Biagetti C., et al. HIV-1 DNA proviral load in treated and untreates HIV 1 seropositive patients. Clin. Microbiol. Infect. 2010;16:640–646. doi: 10.1111/j.1469-0691.2009.02826.x. [DOI] [PubMed] [Google Scholar]
- 10.Suspène R., Meyerhans A. Quantification of unintegrated HIV-1 DNA at the single cell level in vivo. PLoS One. 2012;7:e36246. doi: 10.1371/journal.pone.0036246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goujard C., Bonarek M., Meyer L., et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin. Infect. Dis. 2006;42:709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- 12.Williams J.P., Hurst J., Stohr W. HIV-1 DNA predicts disease progression and post-treatment virological control. eLife. 2014;3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yerly S., Gunthard H.F., Fagard C., et al. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS. 2004;18:1951–1953. doi: 10.1097/00002030-200409240-00011. [DOI] [PubMed] [Google Scholar]
- 14.Chun T.W., Justement J.S., Moir S., et al. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 2007;195:1762–1764. doi: 10.1086/518250. [DOI] [PubMed] [Google Scholar]
- 15.Besson G.J., Lalama C.M., Bosch R.J., et al. HIV-1 decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 2014;59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agosto L.M., Liszewski M.K., Mexas A., et al. Patients on HAART often have an excess of unintegrated HIV DNA: implications for monitoring reservoirs. Virology. 2011;409:46–53. doi: 10.1016/j.virol.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ananworanich J., Dubè K., Chomont N. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr. Opin. HIV AIDS. 2015;10:18–28. doi: 10.1097/COH.0000000000000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ananworanich J., Chomont N., Eller L.A., et al. HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine. 2016;11:68–72. doi: 10.1016/j.ebiom.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgard M., Boufassa F., Viard J.P., et al. Factors influencing peripheral blood mononuclear cell-associated HIV-1 DNA level after long-term suppressive antiretroviral therapy in 236 patients. AIDS. 2009;23:2165–2171. doi: 10.1097/QAD.0b013e32833032d4. [DOI] [PubMed] [Google Scholar]
- 20.Debiaggi M., Zara F., Pistorio A., et al. Quantification of HIV-l proviral DNA in patients with undetectable plasma viremia over long-term highly active antiretroviral therapy. Int. J. Infect. Dis. 2000;4:187–193. doi: 10.1016/s1201-9712(00)90107-3. [DOI] [PubMed] [Google Scholar]
- 21.Nicastri E., Palmisano L., Sarmati L., et al. HIV-1 residual viremia and proviral DNA in patients with suppressed plasma viral load (< 400 HIV-RNA cp/ml) during different antiretroviral regimens. Curr. HIV Res. 2008;6:261–266. doi: 10.2174/157016208784325010. [DOI] [PubMed] [Google Scholar]
- 22.Parisi S.G., Andreis S., Mengoli C., et al. Baseline cellular HIV DNA load predicts HIV DNA decline and residual HIV plasma levels during effective antiretroviral therapy. J. Clin. Microbiol. 2012;50:258–263. doi: 10.1128/JCM.06022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzón M.J., Massanella M., Llibre J.M., et al. HIV-1 replication and immune dynamics are affected by Raltegravir intensification of HAART-suppressed subjects. Nat. Med. 2010;16:460–466. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 24.Charpentier C., Piketty C., Laureillard D., et al. Dynamics of HIV-1 DNA level in highly antiretroviral-experienced patients receiving Raltegravir-based therapy. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:129–133. doi: 10.1007/s10096-011-1284-0. [DOI] [PubMed] [Google Scholar]
- 25.Hatano H., Scherzer R., Wu Y., et al. A randomized controlled trial assessing the effects of Raltegravir intensification on endothelial function in treated HIV infection. J. Acquir. Immune Defic. Syndr. 2012;61:317–325. doi: 10.1097/QAI.0b013e31826e7d0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossetti B., Meini G., Bianco C., et al. Total cellular HIV-1 DNA decreases after switching to Raltegravir-based regimens in patients with suppressed HIV-1 RNA. J. Clin. Virol. 2017;91:18–24. doi: 10.1016/j.jcv.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Haïm-Boukobza S., Morand-Joubert L., Flandre P., et al. Higher efficacy of nevirapine than efavirenz to achieve HIV-1 plasma viral load below 1 copy/ml. AIDS. 2011;25:341–344. doi: 10.1097/QAD.0b013e3283427de3. [DOI] [PubMed] [Google Scholar]
- 28.Lambert-Niclot S., Flandre P., Valantin M.A., et al. Similar evolution of cellular HIV-1 DNA level in darunavir/ritonavir monotherapy versus triple therapy in MONOI-ANRS136 trial over 96 weeks. PLoS One. 2012;7:e41390. doi: 10.1371/journal.pone.0041390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reliquet V., Chirouze C., Allavena C., et al. Nevirapine–Raltegravir combination, an NRTI and PI/r sparing regimen, as maintenance antiretroviral therapy in virologically suppressed HIV-1-infected patients. Antivir. Ther. 2014;19:117–123. doi: 10.3851/IMP2691. [DOI] [PubMed] [Google Scholar]
- 30.Sarmati L., Parisi S.G., Montano M., et al. Nevirapine use, prolonged antiretroviral therapy and high CD4 nadir values are strongly correlated with undetectable HIV-DNA and -RNA levels and CD4 cell gain. J. Antimicrob. Chemother. 2012;67:2932–2938. doi: 10.1093/jac/dks331. [DOI] [PubMed] [Google Scholar]
- 31.Michelini Z., Galluzzo C.M., Pirillo M.F., et al. HIV-1 DNA dynamics and variations in HIV-1 DNA protease and reverse transcriptase sequences in multidrug-resistant patients during successful Raltegravir-based therapy. J. Med. Virol. 2016;88:2115–2124. doi: 10.1002/jmv.24581. [DOI] [PubMed] [Google Scholar]
- 32.Kiselinova M., Anna M., Malatinkova E., et al. The impact of nevirapine- versus protease inhibitor-based regimens on virological markers of HIV-1 persistence during seemingly suppressive ART. J. Int. AIDS Soc. 2014;17:19823. doi: 10.7448/IAS.17.4.19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brussel A., Sonigo P. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J. Virol. 2004;78:11263–11271. doi: 10.1128/JVI.78.20.11263-11271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svicher V., Ceccherini-Silberstein F., Antinori A., et al. Understanding HIV compartments and reservoirs. Curr. HIV/AIDS Rep. 2014;11:186–194. doi: 10.1007/s11904-014-0207-y. [DOI] [PubMed] [Google Scholar]
- 35.Alidjinou E.K., Bocket L., Hober D. Quantification of viral DNA during HIV-1 infection: A review of relevant clinical use and laboratori methods. Pathol. Biol. (Paris) 2015;63:53–59. doi: 10.1016/j.patbio.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Gantner P., Mélard A., Damond F., et al. Quantification working group interlaboratory quality control of total HIV-1 DNA load measurement for multicenter reservoir studies. J. Med. Virol. 2017;89:2047–2050. doi: 10.1002/jmv.24874. [DOI] [PubMed] [Google Scholar]
- 37.Dib C., Fauré S., Fizames C., et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 38.Siliciano J.D., Kajdas J., Finzi D., et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 39.Collier A.C., Chun T.W., Maenza J., et al. A pilot study of Raltegravir plus combination antiretroviral therapy in early human immunodeficiency virus infection: challenge and lessons learned. Biores. 2016;5:15–21. doi: 10.1089/biores.2015.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang N.D., Li T.S. Factors associated with the size of HIV DNA reservoir. Chin. Med. J. (Engl.) 2017;130:224–230. doi: 10.4103/0366-6999.198009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Santo R. Inhibiting the HIV integration process: past, present, and the future. J. Med. Chem. 2014;57:539–566. doi: 10.1021/jm400674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandhi R.T., Coombs R.W., Chan E.S., et al. No Effect of raltegravir intensification on viral replication markers in the blood of HIV-1–infected patients receiving antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2012;59:229–235. doi: 10.1097/QAI.0b013e31823fd1f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrowski M., Benko E., Yue F.Y., et al. Intensifying antiretroviral therapy with raltegravir and maraviroc during early human immunodeficiency virus (HIV) infection does not accelerate hiv reservoir reduction. Open Forum Infect. Dis. 2015;2:ofv138. doi: 10.1093/ofid/ofv138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koelsch K.K., Boesecke C., McBride K.L., et al. Impact of treatment with Raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS. 2011;25:2069–2078. doi: 10.1097/QAD.0b013e32834b9658. [DOI] [PubMed] [Google Scholar]
- 45.Lam Y.M., McBride K.L., Amin J., et al. Swithching virally suppressed treatment-experienced patients to a Raltegravir containing regimen does not alter levels of HIV-1 DNA. PLoS One. 2012;7:e31990. doi: 10.1371/journal.pone.0031990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mexas A.M., Graf E.H., Pace M.J., et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26:2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calza L., Danese I., Magistrelli E., et al. Dual raltegravir-darunavir/ritonavir combination in virologically suppressed hiv-1-infected patients on antiretroviral therapy including a ritonavir-boosted protease inhibitor plus two nucleoside/nucleotide reverse transcriptase inhibitors. HIV Clin. Trials. 2016;17:38–47. doi: 10.1080/15284336.2015.1122874. [DOI] [PubMed] [Google Scholar]