Abstract
Recently, two transcriptome-wide studies identified 40 genes that were differentially expressed in relation to blood pressure. However, to what extent these BP related gene expression signatures as well as their associations with BP are driven by genetic or environmental factors has not been investigated. In this study of 391 twins (193 twin pairs and 5 singletons; age 55–69; 40% male, 57 % monozygous) recruited from the Finnish Twin Cohort, transcriptome-wide data on peripheral leukocytes were obtained using the Illumina HT12 V4 array. Our transcriptome-wide analysis identified one gene (MOK, P=7.16×10−8) with its expression levels associated with systolic BP at the cutoff of false discovery rate <0.05. This association was further replicated in the Framingham Heart Study (P=1.02×10−5). Out of the 40 genes previously reported, 12 genes could be replicated in the twin cohort with FDR<0.05 and consistent direction of effect. Univariate twin modeling showed that genetic factors contributed to the expression variations of 12 genes with heritability estimates ranging from 6–65%. Bivariate twin modeling showed that 53% of the phenotypic association between systolic BP and MOK expression, and 100% of the phenotypic association of systolic and diastolic BP with CD97, TIPARP, and TPPP3 expression could be explained by genetic factors shared in common. In this study of adult twins, we identified one more gene, MOK, with its expression level associated with BP, and replicated several previously identified signals. Our study further provides new insights into the genetic and environmental sources of BP related gene expression signatures.
Keywords: Blood pressure, twin study, gene expression, genome wide study, heritability
INTRODUCTION
High blood pressure (BP) is the leading risk factor for cardiovascular disease worldwide1. Systolic and diastolic blood pressure (SBP and DBP) are complex physiological traits which are affected by both genetic and environmental factors2,3. Recently, two genome-wide gene expression studies on peripheral leukocytes identified 40 genes that were differentially expressed in relation to BP4,5. However, the causal nature of these associations remains unclear. In addition to the possibility of one-way causation, their relationships can also be bidirectional. For example, the cellular processes underlying BP regulation may result in changes in gene expression. In turn, the gene expression or its downstream molecular products may increase the risk of hypertension. It is also possible that common genetic effects account for the observed associations between BP and gene expression. Similar to BP, the expression level of a gene is determined by both genetic and environmental factors 6, although the extent to which the BP related gene expression signatures are driven by genetic factors has not been investigated. Discovery of a common genetic substrate may point towards a common etiological pathway and improve our understanding of the molecular mechanisms underlying BP regulation. In the present study of a transcriptome-wide analysis in leukocytes from 391 adult twins, we first identified new transcriptional signals associated with BP, next replicated previous signals associated with BP, third conducted twin modeling to estimate the heritability of gene expression signatures of BP, and finally assessed whether shared genetic effects play a role in the link between BP and gene expression.
METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Subjects
In 2011, a comprehensive questionnaire was sent to all the living twins of the Finnish Twin Cohort born between 1945 and 1957 (11738 twins). A total of 8501 twins returned the questionnaire with a response rate of 72% 7. Three questions in this questionnaire are related to hypertension: (1) When was your BP last measured? (2) Have you ever been told that you have elevated BP or hypertension? and (3) On how many days in the past year have you used antihypertensive medication? Based on the questionnaire, twins were defined as hypertensive if they had been diagnosed with hypertension or had taken antihypertensive medication for at least 2 months, while twins were defined as normotensive if they had not been diagnosed with hypertension and did not take antihypertensive medication. Based on the replies, 330 same-sex twin pairs free of self-reported previous diagnosis of myocardial infarction, congestive heart failure or stroke were defined as potentially discordant for hypertension. These twin pairs were phone interviewed and asked to participate in clinical assessments at the University of Helsinki during 2012–2015. A total of 222 twin pairs and 3 singletons (n=447) participated in the clinical protocol. Informed consent was obtained from each subject, and the study was approved by the Ethics Committee of the University Central Hospital of Helsinki.
Measurements
During the clinical testing, the twins had a comprehensive physical examination, and their health history was recorded, including questions again about previous diagnoses of hypertension and use of anti-hypertensive medication. Resting BPs were measured four times during the visit, seated by a sphygmomanometer according to the JNC7 guidelines. The average of the last two readings of each measurement occasion was used to represent BP values. Information on medications was further complemented with data from community pharmacies. Based on the fact that only 50 twin pairs met the criteria for current discordance for hypertension (one twin on anti-hypertensive medication or with SBP≥140mmHg or DBP≥90mmHg and his/her co-twin not on anti-hypertensive medication, and with SBP<120mmHg and DBP<80mmHg), we included all the twins for the current analysis by using SBP and DBP as continuous variables. If antihypertensive medication was used, 15mmHg and 10mmHg were added to the measured SBP and DBP levels, respectively 8. Fasting peripheral blood samples were obtained from 402 participants. Zygosity was determined by genotyping using Illumina HumanCoreExome BeadChip.
RNA extraction and transcriptome-wide gene expression assays
RNA samples were extracted from the peripheral leukocytes stored in the RNA cell protection reagents (QIAGEN Inc. Valencia, CA, USA) using the miRNeasy Mini Kit (QIAGEN Inc. Valencia, CA, USA). RNA concentration and purity were evaluated on a NanoDrop spectrophotometer 2000 (Thermo Scientific Inc. Waltham, MA, USA). RNA integrity (RIN) was evaluated on a Bioanalyzer 2100 (Agilent Inc. Santa Clara, CA, USA), and samples with a RIN score ≥ 8 qualified for gene expression analysis. Out of the 402 peripheral blood samples, high quality RNA was obtained for 391 subjects (116 monozygotic [MZ], 77 dizygotic [DZ] twin pairs, and 5 singletons).
Transcriptome-wide gene expression data were obtained using the Illumina HumanHT-12 v4 Expression BeadChip (Illumina Inc). This chip targets more than 48,000 probes that provide transcriptome-wide coverage of well-characterized genes, gene candidates and splice variants. A block design was used to keep the distributions of sex and zygosity similar across chips (12 samples/chip) with the co-twins assigned to the same chip. The twin pairs were randomly assigned to the 12 positions on each chip. The Genome-Studio Gene Expression Module (Illumina, San Diego, CA, USA) was used for initial quantification and the lumi package 9 for data preprocessing and quality control, which included the following key steps: (1) Probes with detection P-value <0.05 in more than 50% of the samples were defined as “present”; (2) Log2 transformation and quantile normalization were applied to the gene expression data. There were 19530 probes that passed the quality control steps and they were used as indices of gene expression levels in further analyses.
Statistical analysis
The purposes of our analyses were: (1) to identify novel and replicate previously identified differentially expressed genes associated with BP in the Finnish twin cohort 4,5; (2) to disentangle genetic and environmental sources of expression variation of the BP associated genes; and (3) to test whether common genetic or environmental effects account for the observed association between BP and gene expression. We used linear mixed model to answer the first aim and structural equation modeling to answer the other two aims of our study.
Linear mixed model
In the entire sample treating twins as individuals, mixed-effect linear regression models with intercept varying among chips and varying among twin pairs within chips to account for the block design and the relatedness of twins, were performed to examine the relationship between BP (explanatory variables) and gene expressions (outcome variables), adjusting for age, sex, and BMI. The differentially expressed genes identified for BP with a false discovery rate (FDR) <0.05 were carried forward for replication in the Framingham Heart Study 5 (n=3679, 42% males, aged 51±12). RNA was isolated from whole blood samples that were collected in PaxGene tubes (PreAnalytiX, Hombrechtikon, Switzerland) and the Affymetrix Exon Array ST 1.0 was used in the Framingham Heart Study. Replication was defined as consistent direction of the β-coefficient and FDR<0.05. We also checked if we could replicate any of the 40 genes identified by previous studies 5,10 that showed association of their expression with BP. Replication was defined successful if the direction of the BP associated gene expression (β-coefficient) was consistent and FDR<0.05.
Univariate structural equation modeling
To estimate the relative contributions of genetic and environmental factors on the expression variation of the BP associated genes, structural equation modeling was conducted using the R package OpenMx 11,12. Prior to analysis, age, sex, BMI, and chip effects were regressed out and the gene expression residuals were used in model fitting. The univariate model (Figure 1a) allows separation of the observed phenotypical variance into underlying additive genetic variance (A), common environmental variance shared by a twin pair (C), and unique environmental variance specific to individuals (E). Under this model, we assumed that the MZ twins share 100% of their genes whereas DZ twins share 50% of their segregating genes on average. We define shared environmental factors to be those that are the same effects on the co-twins irrespective of zygosity while unique environmental factors are by definition not shared by the co-twins, whether MZ or DZ. Significance tests of the individual path (A, C or E) were conducted by constraining paths to zero and applying χ2 test (p<0.05).
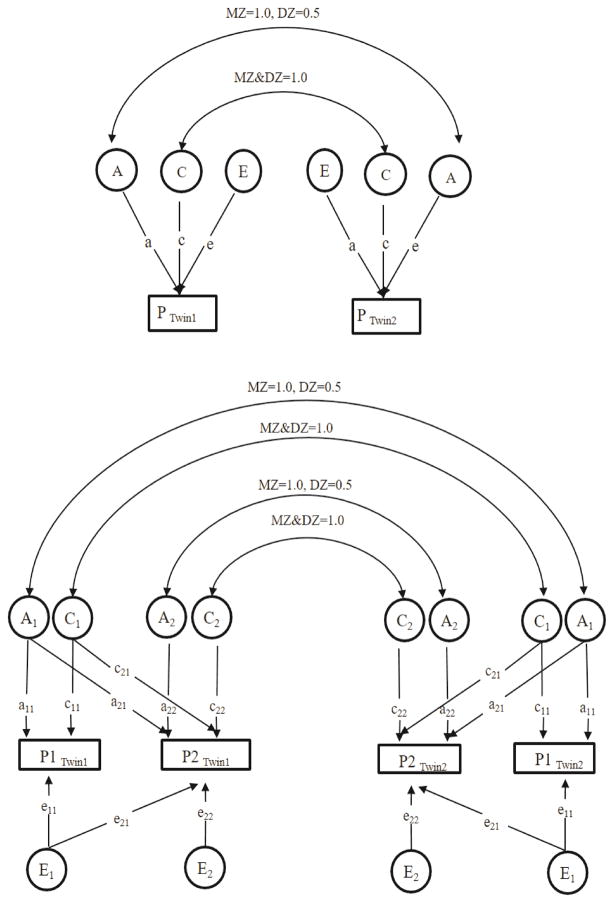
Figure 1.
Figure 1a: Path diagram for univariate structural equation model
The observed phenotypes (P) for co-twins are shown in squares, and latent factors are shown in circles. Correlations between genetic factors (A) are 1 in monozygotic (MZ) twin pairs and 0.5 in dizygotic (DZ) twin pairs. Correlations between common (or shared) environmental factors (C) are 1 for MZ/DZ twin pairs. Unique environmental factors (E) are always uncorrelated.
Figure 1b: Path diagram for bivariate structural equation model (Cholesky model)
The two phenotypes observed (P1, P2) for co-twins are shown in squares, and latent factors are shown in circles. Correlations of the same phenotype between genetic factors (A1) are 1 in monozygotic (MZ) twin pairs and 0.5 in dizygotic (DZ) twin pairs. The path of genetic factors between two phenotypes is a21. Correlations between common (or shared) environmental factors (C) are 1 for MZ/DZ twin pairs. The path of common environmental factors between two phenotypes is c21. Unique environmental factors (E) are always uncorrelated. The path of unique environmental factors between two phenotypes is e21.
Bivariate structural equation modeling
A bivariate Cholesky decomposition was used to model the covariance between gene expression and BP. This model can determine to what extent the covariance can be explained by common genetic or environmental factors. Details of this model have been described previously13. As shown in Figure 1b, similar to the univariate model, the variation of gene expression and the variation of BP were decomposed into A, C and E variance components. The bivariate model allows determination of the sources of the observed covariance between gene expression and BP by using a sequence of models that testing which paths (a21 for genetic covariance, c21 for shared environmental covariance or e21 for covariance of unshared environmental effects) can be set to 0. For example, if a21 cannot be set to 0, it allows further determination of the amount of overlap between the genetic factors influencing gene expression and BP by calculating the genetic correlation between the traits: rg = COVA(gene expression and BP)/√(VA gene expression * VA BP). Shared and unique environmental correlations can be calculated in a similar fashion.
Functional analysis
Gene set enrichment analysis (GSEA) 14,15 was used to identify sets of genes representing biological processes and pathways associated with gene expression changes associated with BP. GSEA was performed on an unfiltered ranked lists of genes (ranked by the p values), and a running-sum statistic was used to determine the enrichment of a prior defined gene sets (pathways) based on the gene ranks. Statistical significance of pathway enrichment score was ascertained by permutation testing over size-matched random gene sets, and multiple testing was controlled by FDR. A FDR <0.05 was considered as significant. We tested for enrichment of any gene ontology (GO) biological processes or KEGG pathways.
RESULTS
The general characteristics of participants are presented for all and by zygosity are presented in Table 1. We identified one gene (MOK) that was differentially expressed in relation to SBP (Table 2) at FDR<0.05. For this gene, higher expression was associated with higher SBP levels. Similar association of this gene with DBP was also observed with a nominal p value of 1.34×10−5. Limiting the analysis to participants with no anti-hypertensive medication revealed the same results with higher MOK expression associated with higher BP levels (p =0.02 and 0.04 for SBP and DBP, respectively). The associations of MOK expression with SBP and DBP were further replicated in the Framingham Heart Study (P=1.02×10−5 and 6.11×10−4, respectively) (Table 2). We did not identify any signals surviving multiple testing for DBP in the Finnish Twin Cohort.
Table 1.
General characteristics of the participants
| Variables | Overall | MZ | DZ |
|---|---|---|---|
| N | 391 | 236 | 155 |
| Age, years (mean, sd) | 62.3 (3.8) | 61.7 (3.6) | 63.2(3.9) |
| Females, % | 59.3 | 61.9 | 55.5 |
| Antihypertensive medications, % | 43.5 | 39.8 | 49.0 |
| SBP, mmHg (mean,sd) * | 150.2 (18.6) | 150.8(17.9) | 149.2(19.7) |
| DBP, mmHg (mean,sd) * | 87.8 (11.0) | 89.0(11.1) | 86.0(10.7) |
SBP and DBP are adjusted (+15 mmHg for SBP and +10 mmHg for DBP) for taking antihypertensive medication.
Table 2.
Gene with expression level associated with SBP and DBP
| Probe ID | Gene | Chr. | Trait | Finland Twin cohort
|
Framingham Heart Study
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | FDR | Beta | SE | P value | ||||
|
|
|
|||||||||
| ILMN_1745282 | MOK* | 14 | SBP | 2.67E-03 | 4.75E-04 | 7.16E-08 | 1.40E-03 | 8.16E-04 | 1.85E-04 | 1.02E-05 |
| DBP | 3.58E-03 | 8.00E-04 | 1.34E-05 | 1.74E-01 | 9.56E-04 | 2.79E-04 | 6.11E-04 | |||
Trans ID 3580234 in Framingham heart study, annotated using the Affymetrix annotation file from Netaffx (www.netaffx.com, HuEx-1_0-st-v2.na29.hg18.probeset.csv)5.
Out of the 40 genes of which their expression levels were previously reported to be associated with BP, 12 genes could be replicated with consistent direction of effects and FDR<0.05 in our twin cohort. Table 3 lists the association of these 12 genes with both SBP and DBP in our cohort although the original transcriptome-wide signals for certain genes were reported only for SBP (TAGLN2 and TAGAP) or DBP (S100A10). The higher expression levels of these 12 genes were associated with higher BP levels.
Table 3.
Replication of previously reported genes with their expression levels associated with BP
| Probe ID | Gene | Previously associated traits | Association with SBP
|
Association with DBP
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | FDR | Beta | SE | P value | FDR | |||
| ILMN_2413508 | CD97 | SBP/DBP | 2.97E-03 | 8.58E-04 | 6.75E-04 | 1.35E-03 | 3.93E-03 | 1.43E-03 | 6.76E-03 | 7.38E-03 |
| ILMN_1656920 | CRIP1 | SBP/DBP | 2.71E-03 | 1.01E-03 | 8.22E-03 | 8.22E-03 | 4.63E-03 | 1.69E-03 | 6.58E-03 | 7.38E-03 |
| ILMN_1671766 | F12 | SBP | 1.52E-03 | 5.60E-04 | 7.07E-03 | 7.72E-03 | 1.59E-03 | 9.37E-04 | 9.12E-02 | 9.12E-02 |
| ILMN_1696749 | LMNA | SBP/DBP | 2.25E-03 | 6.04E-04 | 2.57E-04 | 6.17E-04 | 4.36E-03 | 9.89E-04 | 1.78E-05 | 2.14E-04 |
| ILMN_2308849 | MYADM | SBP/DBP | 3.80E-03 | 8.95E-04 | 3.37E-05 | 2.02E-04 | 4.70E-03 | 1.50E-03 | 1.96E-03 | 3.69E-03 |
| ILMN_2046730 | S100A10 | DBP | 1.95E-03 | 6.62E-04 | 3.59E-03 | 4.78E-03 | 3.11E-03 | 1.11E-03 | 5.61E-03 | 7.38E-03 |
| ILMN_1758938 | SLC31A2 | SBP/DBP | 4.65E-03 | 1.00E-03 | 6.75E-06 | 8.10E-05 | 5.58E-03 | 1.70E-03 | 1.22E-03 | 3.65E-03 |
| ILMN_1676408 | TAGAP | SBP | 3.91E-03 | 1.39E-03 | 5.34E-03 | 6.41E-03 | 7.82E-03 | 2.28E-03 | 7.34E-04 | 2.93E-03 |
| ILMN_2090105 | TAGLN2 | SBP | 3.27E-03 | 8.56E-04 | 1.82E-04 | 5.45E-04 | 4.20E-03 | 1.43E-03 | 3.82E-03 | 5.72E-03 |
| ILMN_1765578 | TIPARP | SBP/DBP | 2.09E-03 | 6.73E-04 | 2.17E-03 | 3.26E-03 | 3.48E-03 | 1.12E-03 | 2.15E-03 | 3.69E-03 |
| ILMN_1797744 | TPPP3 | SBP/DBP | 2.03E-03 | 5.15E-04 | 1.17E-04 | 4.67E-04 | 3.27E-03 | 8.63E-04 | 2.03E-04 | 1.22E-03 |
| ILMN_2276952 | TSC22D3 | SBP/DBP | 3.92E-03 | 1.17E-03 | 9.33E-04 | 1.60E-03 | 6.16E-03 | 1.96E-03 | 1.94E-03 | 3.69E-03 |
Twin correlations for the gene expression levels of the newly identified gene (MOK) and the 12 previously reported genes are shown in Table 4. With the exception of MYADM and TIPARP, twin correlations in MZ twin pairs were larger than those in DZ twin pairs, indicating genetic influences. For the expression of 7 genes including CRIP1, F12, LMNA, MOK, S100A10, TAGAP and TSC22D3, the best fitting models are AE models, with heritability estimates ranging from 0.27 to 0.65. The remaining part of the variation for the expression of these 7 genes is explained by environmental influences that are unique to the individual. For the expression of MYADM, the best fitting model is the CE model, with the familiar aggregation best explained by the shared environment factor that explains 47% of the expression variation of the MYADM gene. For the expression of the other 5 genes including CD97, SLC31A2, TAGLN2, TIPARP and TPPP3 the best fitting models are ACE models. For these genes, the model assuming either the absence of A component or absence of C component fitted the data well, but the model assuming the absence of both components fitted the data significantly worse. This indicates that the expression of these genes did show familial aggregation, either due to genetic and/or shared environment influences and their variations in the population could not be explained by unique environment effects alone.
Table 4.
Best fitting models of the univariate SEM analysis
| Probe ID | Gene | Correlation | Best Model* | Standardized Variance Components | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MZ | DZ | a2 | c2 | e2 | |||
| ILMN_2413508 | CD97 | 0.48 | 0.34 | ACE | 0.34(0.00,0.612) | 0.16(0.00,0.49) | 0.50(0.39,0.65) |
| ILMN_1656920 | CRIP1 | 0.46 | 0.24 | AE | 0.50(0.35,0.62) | -- | 0.50(0.38,0.65) |
| ILMN_1671766 | F12 | 0.53 | 0.36 | AE | 0.55(0.41,0.65) | -- | 0.45(0.35,0.58) |
| ILMN_1696749 | LMNA | 0.58 | 0.37 | AE | 0.61(0.49,0.70) | -- | 0.39(0.30,0.51) |
| ILMN_1745282 | MOK | 0.52 | 0.26 | AE | 0.53(0.40,0.64) | -- | 0.47(0.36,0.60) |
| ILMN_2308849 | MYADM | 0.47 | 0.48 | CE | -- | 0.47(0.35,0.57) | 0.53(0.42,0.64) |
| ILMN_2046730 | S100A10 | 0.39 | 0.13 | AE | 0.39(0.22,0.54) | -- | 0.61(0.46,0.78) |
| ILMN_1758938 | SLC31A2 | 0.36 | 0.33 | ACE | 0.08(0.00,0.48) | 0.28(0.00,0.45) | 0.64(0.50,0.79) |
| ILMN_1676408 | TAGAP | 0.67 | 0.42 | AE | 0.65(0.55,0.72) | -- | 0.35(0.27,0.45) |
| ILMN_2090105 | TAGLN2 | 0.42 | 0.26 | ACE | 0.36(0.00,0.56) | 0.07(0.00,0.43) | 0.57(0.44,0.73) |
| ILMN_1765578 | TIPARP | 0.40 | 0.41 | ACE | 0.06(0.00,0.52) | 0.67(0.00,0.52) | 0.57(0.44,0.71) |
| ILMN_1797744 | TPPP3 | 0.30 | 0.19 | ACE | 0.26(0.00,0.47) | 0.06(0.00,0.38) | 0.67(0.52,0.85) |
| ILMN_2276952 | TSC22D3 | 0.28 | 0.11 | AE | 0.27(0.10,0.41) | -- | 0.73(0.58,0.90) |
“A” means additive genetic factor. “C” means common environmental factor. “E” means unique environmental factor.
The best fitting model for both SBP and DBP is the AE model, with heritabilities of 0.45(0.29,0.58) for SBP and 0.40(0.24,0.53) for DBP respectively.
Next, we estimated the relative contributions of genetic and environmental factors to the association between gene expressions and BP. Due to the fact that the AE model has generally been the best fitting model in previous twin studies of BP 2,16 which was again confirmed in the current study (Heritability was 45% for SBP and 40% for DBP respectively), the bivariate modeling was only conducted on those genes with expression levels determined by ACE or AE models. The results are listed in Table 5. For the association of MOK expression with SBP, both a21 and e21 could not be set to 0, indicating the phenotypic correlation is determined by both shared genetic factors and shared unique environment with common genetic factors accounting for 53% of the phenotypic correlation. For the association of LMNA, SLC31A2 and TSC22D3 expression with BP and the association of TAGLN2 expression with SBP, a21 could but e21 could not be set to 0, indicating the phenotypic correlation is completely determined by shared unique environmental factors. On the other hand, the phenotypic correlations of CD97, TIPARP and TPPP3 expression with both SBP and DBP were completely determined by shared genetic factors. For the associations of the other gene expressions with BP, the model assuming either the absence of a21 component or e21 component fitted the data well, but the model assuming the absence of both components fitted the data significantly worse, indicating a larger sample size is needed to increase the power to distinguish the relative contributions of genetic and environmental factors to the observed phenotypic correlations.
Table 5.
Best fitting models of the bivariate SEM analysis
| Probe ID | Trait 1 | Trait 2 | Best Model | rg † | re † | Standardized rg ‡ | Standardized re ‡ |
|---|---|---|---|---|---|---|---|
| ILMN_2413508 | CD97 | SBP | a21≠0 | 0.39(0.18,0.59) | -- | 1.00 | -- |
| ILMN_2413508 | CD97 | DBP | a21≠0 | 0.34(0.12,0.56) | -- | 1.00 | -- |
| ILMN_1656920 | CRIP1 | SBP | a21≠0 | e21≠0 * | 0.22(−0.26,0.98) | 0.06(−0.11,0.23) | 0.77 | 0.23 |
| ILMN_1656920 | CRIP1 | DBP | a21≠0 | e21≠0 * | 0.28(0.00,0.54) | 0.05(−0.11,0.22) | 0.80 | 0.20 |
| ILMN_1671766 | F12 | SBP | a21≠0 | e21≠0 * | 0.015(−0.09,0.40) | 0.07(−0.10,0.24) | 0.69 | 0.31 |
| ILMN_1696749 | LMNA | SBP | e21≠0 | -- | 0.24(0.09, 0.37) | -- | 1.00 |
| ILMN_1696749 | LMNA | DBP | e21≠0 | -- | 0.28(0.14,0.41) | -- | 1.00 |
| ILMN_1745282 | MOK | SBP | a21≠0 & e21≠0 | 0.27(0.01,0.49) | 0.26(0.09,0.41) | 0.52 | 0.48 |
| ILMN_2046730 | S100A10 | DBP | a21≠0 | e21≠0 * | 0.14(−0.20,0.45) | 0.07(−0.10,0.24) | 0.56 | 0.44 |
| ILMN_1758938 | SLC31A2 | SBP | e21≠0 | -- | 0.27(0.14,0.39) | -- | 1.00 |
| ILMN_1758938 | SLC31A2 | DBP | e21≠0 | -- | 0.17(0.03,0.30) | -- | 1.00 |
| ILMN_1676408 | TAGAP | SBP | a21≠0 | e21≠0 * | 0.08(−1.00,1.00) | 0.17(−0.01,0.33) | 0.36 | 0.64 |
| ILMN_2090105 | TAGLN2 | SBP | e21≠0 | -- | 0.29(0.15,0.41) | -- | 1.00 |
| ILMN_1765578 | TIPARP | SBP | a21≠0 | 0.46(0.20,0.67) | -- | 1.00 | -- |
| ILMN_1765578 | TIPARP | DBP | a21≠0 | 0.40(0.18,0.66) | -- | 1.00 | -- |
| ILMN_1797744 | TPPP3 | SBP | a21≠0 | 0.41(0.18,0.65) | -- | 1.00 | -- |
| ILMN_1797744 | TPPP3 | DBP | a21≠0 | 0.45(0.21,0.69) | -- | 1.00 | -- |
| ILMN_2276952 | TSC22D3 | SBP | e21≠0 | -- | 0.22(0.09,0.34) | -- | 1.00 |
| ILMN_2276952 | TSC22D3 | DBP | e21≠0 | -- | 0.21(0.08,0.33) | -- | 1.00 |
a21 and e21 cannot be set to 0 at the same time
rg is genetic correlation.( ) -- indicates that it can be set to 0 in the best fitting model.
re is nonshared-environmental correlation.( )--indicates that it can be set to 0 in the best fitting model.
Standardized rg = a11*a21/rp, standardized re = e11*e21/rp, where rp= a11*a21 + c11*c21 + e11*e21
The pathway analyses yielded significant (FDR<0.05) enrichment of 16 KEGG and GO biology process pathways for SBP related gene expression changes and 18 for DBP related gene expression changes (with 10 pathways overlapped between SBP and DBP) in peripheral leukocytes (Table S1 in supplement, please see http://hyper.ahajournals.org). These pathways represent processes involved in inflammatory pathways (e.g. antigen processing and presentation, innate immune response, defense response to virus, defense response to other organism, regulation of type I interferon, and mediated signaling pathway) and autoimmune response (e.g. systemic lupus erythematosus, Type 1 diabetes, allograft rejection, graft versus host disease, auto-immune thyroid disease).
DISCUSSION
In this study of adult twins, we identified one more gene with its expression level associated with BP and replicated 12 previously identified signals. The newly identified signal is MOK (MAPK/MAK/MRK overlapping kinase gene), which is also known as RAGE-1 (renal tumor antigen-1). It was first identified in a renal carcinoma cell line 17 and has been considered as a tumor-associated antigen for its wide expression in various tumors including renal carcinoma, melanoma, head and neck cancer, mesothelioma, hepatocellular carcinoma and leukemia. Later studies found that it was also widely expressed in the cytoplasm of normal tissues including blood leukocytes18. Although it is suspected that the signaling kinase that belongs to the MAPK superfamily may have important biological functions in different physiologic processes as well as in disease, its biological roles are still largely unknown. We did not find literature support for a direct role of MOK in BP regulation. Based on the findings by Oehlrich et al. 19 that MOK could be recognized by cytotoxic T lymphocytes to induce immune response and its participation in the inflammation response in microglia by extracellular stimulation 20, we speculate that MOK may participate in BP regulation through inflammation pathway, a mechanism that has been implicated in the development of hypertension. Further experimental validation is needed.
As opposed to the instructions encoded in the genome sequence, actual gene expression (i.e., the transcriptome) is tissue specific. The study of the transcriptome in disease requires focusing on the specific tissues involved in the pathogenesis of this disease. In addition to vasculature, kidney, adrenals and central nervous system which have long been implicated in the regulation of BP, recent evidence has also pointed to the involvement of oxidative stress and low-grade inflammation in the pathophysiology of hypertension and suggests that the immune system might be the missing mechanism that links these various organs 21. Both our study and the study from Huan et al.5 obtained transcriptome-wide gene expression data from peripheral leukocytes and the functional analysis of the differentially expressed BP genes from both studies yielded significant enrichment of pathways representing processes involved in inflammation and immune response, providing another strong piece of evidence supporting the involvement of inflammation pathways in the pathogenesis of hypertension. One previous study 10 conducted a transcriptome profiling on kidney and identified 14 differentially expressed genes in renal medulla and 46 differentially expressed genes in renal cortex between 15 hypertensive patients and 7 normotensive controls. Out of these 60 genes, the expression data of 48 are available from the Illumina HumanHT-12 v4 Expression BeadChip. However, none of the associations observed in renal tissue could be replicated in the current study using peripheral leukocytes. In addition to the difference in tissues, this previous study was based on a small sample size and lacked a replication cohort. Nevertheless, we believe that using similar approaches (large discovery panel plus replication) in tissues such as the kidney, which are potentially more relevant for BP other than peripheral leukocytes (i.e. the most accessible cells) is a promising approach and may yield additional differentially expressed BP signature genes.
Another difference between the transcriptome and the genome is the plasticity of the transcriptome, i.e. the findings may reflect the consequence rather than the cause of the disease. Theoretically, the cross-sectional association between BP and gene expression observed in the current study and the previous study can be explained in 4 ways. First, higher BP induces the changes in gene expression. Second, changes in gene expression induce higher BP. Third, the effect can be bidirectional. The cellular processes underlying BP regulation may change gene expression, whereas the latter or its downstream molecular products, in turn, may increase the risk of hypertension. Fourth, there could be a underlying factor that influences both BP levels and gene expressions profiles. The twin design of the current study allowed us to explore the last explanation. We first quantified the genetic and environmental sources of the variations of the 13 (one novel and 12 known) BP related gene expression signatures and observed that genetic factors contributed to the variance in expression of 12 out of the 13 genes, with the exception of MYADM. The heritability estimates of the 12 genes ranged from 6–65%. A genetic contribution was not identified for MYADM. The familial aggregation of its expression level was mainly determined by shared environmental factors. This is not uncommon and a previous study 6 has shown that as much as 32% of transcripts from human lymphoblastoid cell lines have a shared environmental component that explain over 30% of the total variance. Since the previous twin studies 2,16 and our current study did not find evidence for influence of shared environmental factors on BP variation, we examined the common etiology hypothesis for BP and the 12 genes (CD97, CRIP1, F12, LMNA, MOK, S100A10, SLC31A2, TAGAP, TAGLN2, TIPARP, TPPP3 and TSC22D3) with their expression variation determined at least partially by genetic factors. Significant genetic correlations were observed between SBP and MOK expression, as well as SBP/DBP and CD97, TIPARP and TPPP3 gene expression. Up to 53% of the phenotypic association between SBP and MOK gene expression can be explained by common genetic factors, while this was 100% for CD97, TIPARP and TPPP3 with both SBP and DBP. These results indicate that shared genes did contribute substantially to the association of BP with expression of certain genes (CD97, MOK, TIPARP, and TPP3).
The identification of common genetic effects accounting for the association between BP and gene expression has important implications for gene-finding studies. In the current gene-finding efforts for BP, univariate analysis has been conducted with BP as the phenotype of interest. Even with the most recent findings from three large-scale genome-wide associate studies (GWAS) 22–24 in over 300,000 subjects which have expanded the list of genetic loci for BP to almost 400, adding all the loci together only explains 3–4% of BP variance although the heritability calculated from all genotyped and imputed SNPs is around 17% 25. With the identification of common genetic effects, bi-variate analysis with both BP and gene expression as the phenotypes of interests can be applied to identify the shared genetic variants. Such an analysis strategy might not only help to identify more BP related loci but also has the potential to shed light on the underlying mechanisms.
Our study has several limitations that need to be recognized. First, the study was designed to have an oversampling of twins from pairs discordant for essential hypertension. However, the univariate twin modeling on BP observed results similar to previous twin studies with twins recruited from the general population, indicating this specific design did not lead to large biases in heritability estimates. Second, our study is cross-sectional, thus limited in the ability to discern the temporal order between BP and gene expression. However, the identification of common genetic precursors for BP and expression of CD97, MOK, TIPARP and TPPP3 genes renders a cause-effect relationship for these genes unlikely. Third, the study only included Caucasian adults, thus one should be cautious extending our results to other ethnic or age groups. Fourth, although this study included almost 400 twins, an even larger sample size is required to tease out the relative contribution of genetic or environmental factor to the associations of BP with gene expression. Fifth, transcriptome data were obtained using traditional chip technology in this study. In comparison with the current state-of-the-art RNA-seq approach, some low abundance transcripts might have been missed.
In conclusion, we identified one more gene with its expression level associated with BP and replicated several previously identified signals. Our study further provides new insights into the genetic and environmental sources of BP related gene expression signatures as well as their associations with BP. The identification of common genetic effects between BP and gene expression of CD97, MOK, TIPARP and TPPP3 will aid in future BP gene finding efforts, and may ultimately point towards more effective prevention and treatment of hypertension.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is New?
Identify one more gene with its expression associated with blood pressure.
Discover that genetic factors contribute to the expression variations of 12 BP associated genes.
Discover that common genetic effects account for the observed associations between BP and the expression of 4 genes.
What Is Relevant?
Discovery of common genetic factors may point towards a common etiological pathway and has the potential to improve our understanding of the molecular mechanisms underlying BP regulation.
Summary.
In this study of adult twins, 391 twins were recruited from the Finnish Twin Cohort. Their gene expression data in peripheral leukocytes were obtained using Illumina HT12 V4 array. Their comprehensive physical examination and health history as well as blood pressure were measured during clinical visit. Linear mixed model was used to identify the differentially expressed genes associated with blood pressure. Univariate and bivariate structural equation modelings were used to further investigate to what extent the genetic factor and environmental factor influence gene expression and blood pressure. Our study identified one more gene (MOK) with its expression level associated with BP and replicated 12 previously identified signals. Univariate twin modeling showed that genetic factors contributed to the expression variations of 12 genes with heritability estimates ranging from 6–65%. Bivariate twin modeling showed that 53% of the phenotypic association between systolic BP and MOK expression, and 100% of the phenotypic association of systolic and diastolic BP with CD97, TIPARP and TPPP3 expression could be explained by genetic factors shared in common. Our study provides new insights into the genetic and environmental sources of BP related gene expression signatures.
Acknowledgments
SOURCES OF FUNDING
This study was supported by NIH/NHLBI (grant HL104125). JK has been supported by the Academy of Finland (grant #s 265240 and 263278).
Footnotes
DISCLOSURES
None
References
- 1.Hao G, Wang X, Treiber FA, Harshfield G, Kapuku G, Su S. Blood Pressure Trajectories From Childhood to Young Adulthood Associated With Cardiovascular Risk: Results From the 23-Year Longitudinal Georgia Stress and Heart Study. Hypertension. 2017 Mar;69(3):435–442. doi: 10.1161/HYPERTENSIONAHA.116.08312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Ding X, Su S, et al. Genetic influences on daytime and night-time blood pressure: similarities and differences. Journal of hypertension. 2009 Dec;27(12):2358–2364. doi: 10.1097/HJH.0b013e328330e84d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Ding X, Su S, Harshfield G, Treiber F, Snieder H. Genetic influence on blood pressure measured in the office, under laboratory stress and during real life. Hypertension research : official journal of the Japanese Society of Hypertension. 2011 Feb;34(2):239–244. doi: 10.1038/hr.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeller T, Schurmann C, Schramm K, et al. Transcriptome-Wide Analysis Identifies Novel Associations With Blood Pressure. Hypertension. 2017 Oct;70(4):743–750. doi: 10.1161/HYPERTENSIONAHA.117.09458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huan T, Esko T, Peters MJ, et al. A meta-analysis of gene expression signatures of blood pressure and hypertension. PLoS genetics. 2015 Mar;11(3):e1005035. doi: 10.1371/journal.pgen.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundberg E, Small KS, Hedman AK, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature genetics. 2012 Oct;44(10):1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaprio J. The Finnish Twin Cohort Study: an update. Twin research and human genetics : the official journal of the International Society for Twin Studies. 2013 Feb;16(1):157–162. doi: 10.1017/thg.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Statistics in medicine. 2005 Oct 15;24(19):2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 9.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008 Jul 01;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 10.Marques FZ, Campain AE, Tomaszewski M, et al. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011 Dec;58(6):1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 11.Neale MC, Hunter MD, Pritikin JN, et al. OpenMx 2.0: Extended Structural Equation and Statistical Modeling. Psychometrika. 2016 Jun;81(2):535–549. doi: 10.1007/s11336-014-9435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritikin JN, Hunter MD, Boker S. Modular Open-Source Software for Item Factor Analysis. Educational and psychological measurement. 2015 Jun 01;75(3):458–474. doi: 10.1177/0013164414554615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Trivedi R, Treiber F, Snieder H. Genetic and environmental influences on anger expression, John Henryism, and stressful life events: the Georgia Cardiovascular Twin Study. Psychosomatic medicine. 2005 Jan-Feb;67(1):16–23. doi: 10.1097/01.psy.0000146331.10104.d4. [DOI] [PubMed] [Google Scholar]
- 14.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003 Jul;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Ding X, Zhang X, et al. Genetic and environmental influences on blood pressure variability: a study in twins. Journal of hypertension. 2013 Apr;31(4):690–697. doi: 10.1097/HJH.0b013e32835e2a4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaugler B, Brouwenstijn N, Vantomme V, et al. A new gene coding for an antigen recognized by autologous cytolytic T lymphocytes on a human renal carcinoma. Immunogenetics. 1996;44(5):323–330. doi: 10.1007/BF02602776. [DOI] [PubMed] [Google Scholar]
- 18.Miyata Y, Akashi M, Nishida E. Molecular cloning and characterization of a novel member of the MAP kinase superfamily. Genes to cells : devoted to molecular & cellular mechanisms. 1999 May;4(5):299–309. doi: 10.1046/j.1365-2443.1999.00261.x. [DOI] [PubMed] [Google Scholar]
- 19.Oehlrich N, Devitt G, Linnebacher M, et al. Generation of RAGE-1 and MAGE-9 peptide-specific cytotoxic T-lymphocyte lines for transfer in patients with renal cell carcinoma. International journal of cancer. 2005 Nov 01;117(2):256–264. doi: 10.1002/ijc.21200. [DOI] [PubMed] [Google Scholar]
- 20.Leal-Lasarte MM, Franco JM, Labrador-Garrido A, Pozo D, Roodveldt C. Extracellular TDP-43 aggregates target MAPK/MAK/MRK overlapping kinase (MOK) and trigger caspase-3/IL-18 signaling in microglia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2017 Mar 23; doi: 10.1096/fj.201601163R. [DOI] [PubMed] [Google Scholar]
- 21.Nosalski R, McGinnigle E, Siedlinski M, Guzik TJ. Novel Immune Mechanisms in Hypertension and Cardiovascular Risk. Current cardiovascular risk reports. 2017;11(4):12. doi: 10.1007/s12170-017-0537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nature genetics. 2017 Mar;49(3):403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann TJ, Ehret GB, Nandakumar P, et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nature genetics. 2017 Jan;49(1):54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wain LV, Vaez A, Jansen R, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension. 2017 Jul 24; doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte IM, van der Most PJ, Alizadeh BZ, et al. Missing heritability: is the gap closing? An analysis of 32 complex traits in the Lifelines Cohort Study. European journal of human genetics : EJHG. 2017 Jun;25(7):877–885. doi: 10.1038/ejhg.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.