Abstract
Human apolipoprotein E4 (apoE4) binds preferentially to lower density lipoproteins, including very low density lipoproteins, and is associated with increased risk of atherosclerosis and neurodegenerative disorders, including Alzheimer's disease. This binding preference is the result of the presence of Arg-112, which causes Arg-61 in the amino-terminal domain to interact with Glu-255 in the carboxyl-terminal domain. ApoE2 and apoE3, which have Cys-112, bind preferentially to high density lipoproteins (HDL) and do not display apoE4 domain interaction. Mouse apoE, like apoE4, contains the equivalent of Arg-112 and Glu-255, but lacks the critical Arg-61 equivalent (it contains Thr-61). Thus, mouse apoE does not display apoE4 domain interaction and, as a result, behaves like human apoE3, including preferential binding to HDL. To assess the potential role of apoE4 domain interaction in atherosclerosis and neurodegeneration, we sought to introduce apoE4 domain interaction into mouse apoE. Replacing Thr-61 in mouse apoE with arginine converted the binding preference from HDL to very low density lipoproteins in vitro, suggesting that apoE4 domain interaction could be introduced into mouse apoE in vivo. Using gene targeting in embryonic stem cells, we created mice expressing Arg-61 apoE. Heterozygous Arg-61/wild-type apoE mice displayed two phenotypes found in human apoE4/E3 heterozygotes: preferential binding to lower density lipoproteins and reduced abundance of Arg-61 apoE in the plasma, reflecting its more rapid catabolism. These findings demonstrate the successful introduction of apoE4 domain interaction into mouse apoE in vivo. The Arg-61 apoE mouse model will allow the effects of apoE4 domain interaction in lipoprotein metabolism, atherosclerosis, and neurodegeneration to be determined.
Apolipoprotein E (apoE) is a 34-kDa plasma protein that participates in diverse biological processes, including plasma lipoprotein metabolism (1), intracellular cholesterol utilization (2), cell growth (3), immunoregulation (4–6), and neuronal growth and repair (7). The three common isoforms of human apoE—apoE2, apoE3, and apoE4—differ at two positions (8). ApoE3 has cysteine at position 112 and arginine at position 158, whereas apoE2 has cysteines at both positions and apoE4 has arginines. Metabolically and physiologically, the most prevalent isoform, apoE3, is considered to be normal. ApoE2 has reduced affinity for the low density lipoprotein (LDL) receptor and is associated with type III hyperlipoproteinemia (9). ApoE4 is associated with elevated plasma cholesterol and LDL levels and predisposes to cardiovascular disease and neurodegenerative disorders, including Alzheimer's disease (9–12). The mechanisms of apoE4's effects on lipoprotein metabolism and neurodegeneration remain elusive. Unlike apoE2 and apoE3, apoE4 associates preferentially with very low density lipoproteins (VLDL) (13–15). This preference contributes to the accelerated catabolism of apoE4 in plasma, which results in lower levels of apoE4 than of apoE3 and apoE2, and to its effects on plasma cholesterol and LDL levels (13, 15).
ApoE possesses two structural domains (16, 17). The amino-terminal domain (22 kDa) contains the binding site for the LDL receptor, and the carboxyl-terminal domain (12 kDa) contains the major lipoprotein-binding region (8). Although the two domains fulfill different biological roles and can function independently, they interact in apoE4 (15, 18, 19). Site-directed mutagenesis and x-ray crystallography studies have shown that Arg-61 in the amino-terminal domain of apoE4 is exposed, allowing it to interact with Glu-255 in the carboxyl-terminal domain, leading to a compact structure. In apoE2 and apoE3, however, Arg-61 is not exposed, and thus apoE4 domain interaction (interaction between Arg-61 and Glu-255) does not occur in these isoforms. Domain interaction in human apoE4 has been suggested to represent a major underlying factor contributing to the adverse effects of this isoform (19). Mouse apoE contains the equivalent of Arg-112 and Glu-255 but lacks the equivalent of Arg-61 and would be expected to resemble apoE3 in terms of lipoprotein-binding preference and metabolism. All other mammalian species examined also have Thr-61 and, therefore, lack the critical Arg-61 equivalent (8).
In this study, we sought to introduce apoE4 domain interaction into mouse apoE by supplying the critical Arg-61. Our goal was to generate an in vivo model suitable for testing the hypothesis that domain interaction contributes to the effects of apoE4 in plasma lipoprotein metabolism and neurodegeneration. Here, we report the successful application of gene targeting in embryonic stem (ES) cells to generate mice displaying apoE4 domain interaction in vivo.
Materials and Methods
Expression and Purification of Wild-Type (wt) and Arg-61 Mouse ApoE.
A cDNA clone encoding mouse apoE (obtained from Aldons Lusis, University of California, Los Angeles) was subcloned into the glutathione S-transferase fusion protein expression vector pGEX3X (20). Mouse Arg-61 apoE was generated by substituting arginine for threonine at the equivalent of position 61 in human apoE by PCR with mutagenic oligonucleotide primers (21). Both Arg-61 and wt mouse apoE were expressed as glutathione S-transferase fusion proteins in Escherichia coli, cleaved from glutathione S-transferase, and purified as described (19).
Distribution of ApoE Among Plasma Lipoproteins and Fractionation of Mouse Plasma.
ApoE was iodinated with the Bolton–Hunter reagent (DuPont) (22) and incubated with normal human plasma at 37°C for 2 h. The plasma was fractionated into various lipoprotein classes by FPLC on a Superose 6 column (Amersham Pharmacia), as described (19). The 125I content was determined in a Beckman 8000 counter (Beckman Coulter). Mouse plasma was fractionated by FPLC, and the total cholesterol level in each fraction was determined with a colorimetric assay (Spectrum; Abbott).
Generation of an Arg-61 Apoe Allele.
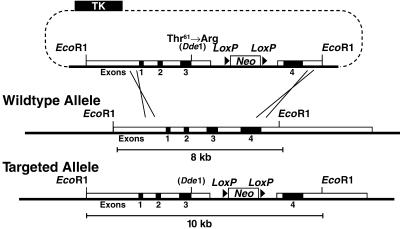
A sequence-replacement, gene-targeting vector was constructed from subclones of an 8.3-kb EcoRI fragment spanning exons 1–4 of the mouse Apoe gene isolated from a 129/SvJae mouse genomic bacterial artificial chromosome library (Research Genetics, Huntsville, AL) (Fig. 1). Site-directed mutagenesis by PCR substituted a G for a C at the last nucleotide position of exon 3 that codes for Thr-61 to create an arginine codon and a unique DdeI restriction site.
Figure 1.
Generation and characterization of an Arg-61 Apoe allele. (A) The sequence-replacement, gene-targeting strategy. A 7-kb EcoRI-BstB1 fragment containing exons 1, 2, and 3 and 5′ flanking sequences was inserted upstream of a neo cassette flanked by loxP sites. A 1.3-kb fragment containing exon 4 and 3′ flanking sequences was amplified by PCR and cloned into the Asc1 site downstream of the neo cassette. Homologous recombination between the gene-targeting vector and the Apoe locus in ES cells introduced a G-for-C change in the second position of codon 61 at the 3′ end of exon 3 and introduced a unique DdeI restriction site. A neo cassette flanked by loxP sites was placed in close proximity to the novel Arg-61 codon in intron 3 to monitor the correct integration of the point mutation. Targeted ES cell clones and mice were identified by digestion of genomic DNA with EcoRI and Southern blotting with an Apoe exon 4 probe, which revealed an expanded 10.3-kb fragment vs. a wt 8.3-kb fragment. Cre-mediated excision of the neo cassette produces a mutant Apoe allele (designated Arg-61 Apoe).
Generation of Homozygous Arg-61 Apoe Mice.
The gene-targeting vector was electroporated into strain 129/SVJae ES cells as described (23), and drug-resistant clones were screened by Southern blot analysis. Several targeted clones were microinjected into C57BL/6 blastocysts to generate chimeric mice harboring a mutant Apoe allele, Arg-61 neo+, in which Thr-61 was converted to arginine and intron 3 contained the neo cassette. Male chimeras were crossed with C57BL/6 females to generate heterozygous neo+/wt mice. The neo+/wt mice were intercrossed to generate neo+/neo+ mice and were crossed with a Cre-deleter transgenic mouse (24) to remove the neo cassette and generate heterozygous Arg-61/wt mice. Heterozygous mice were backcrossed to C57BL/6 to eliminate the Cre transgene and intercrossed to generate homozygous Arg-61 apoE mice. All of the mice had a mixed genetic background (≈87% C57BL/6). The mice were weaned at 21 days of age, housed in a barrier facility with a 12-h light/12-h dark cycle, and fed a chow diet containing 4.5% fat (Ralston Purina).
Northern Blot Analysis of Total RNA in Various Tissues.
Total RNA was extracted from several tissues and organs according to the method of Chomczynski and Sacchi (25) by using Triazol reagent (GIBCO/BRL). Total RNA (≈20 μg) was electrophoresed in a 1% agarose gel containing 20% formaldehyde, transferred to Hybond membrane (Amersham Pharmacia), and hybridized to a mouse apoE cDNA probe labeled with [32P]dCTP in Quickhyb solution (Stratagene) at 65°C overnight. The blot was washed in 0.3× standard sodium citrate (150 mM NaCl/15 mM sodium citrate) and 0.1% SDS at 55°C for 1 h and exposed to x-ray film overnight. A second blot of identical samples run on the same gel was hybridized with a mouse β-actin probe.
Isolation of Primary Hepatocytes from Mouse Liver.
Primary mouse hepatocytes were isolated and prepared as described (26). A confluent layer of hepatocytes was plated in each well of six-well plates coated with collagen type I (Becton Dickinson). Cells were incubated at 37°C under 5% CO2 with 2 ml of culture medium consisting of DMEM (GIBCO/BRL) supplemented with 1% (vol/vol) glutamine, 1% (vol/vol) 10 mM nonessential amino acids, and 10% (vol/vol) FBS.
Isolation of Cerebrospinal Fluid.
Cerebrospinal fluid (CSF) was isolated by perfusing anesthetized mice with PBS, pH 7.4, for 3 min at a flow rate of 3 ml/min. The skull then was exposed and punctured with a 28-gauge needle. CSF was retracted slowly into a 1-ml syringe connected to the needle with 0.012-inch siliconized tubing.
Isoelectric-Focusing (IEF)–PAGE.
Primary hepatocyte culture medium and plasma FPLC fractions were dialyzed against deionized water and lyophilized. Serum samples and CSF (5 μl) were directly lyophilized. Freeze-dried material was resuspended into sample buffer (10% sucrose/0.1% decyl sulfate/0.2 mM Tris, pH 9.0), incubated for 30 min at 37°C, and run on a 5% polyacrylamide gel containing 8 M urea and 200 mM ampholites, pH 3.5–6.5 (Amersham Pharmacia). The gel was run at 200 V overnight at 4°C and was transferred to a nitrocellulose membrane for immunodetection.
Determination of Plasma Lipid Levels.
Plasma lipid measurements were performed on 8- to 15-week-old mice, which were fasted for 4 h, anesthetized, and bled by retroorbital puncture. Total levels of cholesterol and triglycerides in whole plasma or in FPLC fractions were determined with colorimetric assays (Spectrum; Abbott; and Triglycerides; Boehringer Mannheim). Statistical analysis was performed by using Student's t test.
Generation of Polyclonal Antiserum Against Mouse ApoE.
Polyclonal antiserum specific to mouse apoE was raised by immunizing New Zealand White rabbits with a 100-μg mixture of pure recombinant wt and Arg-61 mouse apoE emulsified in complete Freund's adjuvant. Rabbits were boosted twice with the antigens emulsified in incomplete Freund's adjuvant. The presence of apoE in mouse plasma or FPLC fractions was detected by Western blot analysis with a horseradish peroxidase-conjugated anti-rabbit antibody (GIBCO/BRL) and chemiluminescent reagent (Amersham Pharmacia) and quantified with a PhosphorImager (Bio-Rad) and quantification software (Bio-Rad quantity one).
Results
Introduction of ApoE4 Domain Interaction into Mouse ApoE.
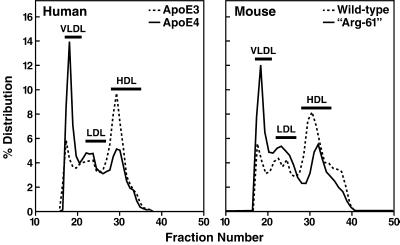
Site-directed mutagenesis was used to substitute arginine for threonine at the position equivalent to 61 in human apoE. The effect of the mutation on the lipoprotein-binding preference was examined in an in vitro human plasma assay. Recombinant wt mouse apoE, like human apoE3, bound preferentially to HDL; in contrast, mouse Arg-61 apoE, like human apoE4, displayed a preference for VLDL (Fig. 2), indicating that domain interaction could be introduced into mouse apoE.
Figure 2.
Plasma lipoprotein-binding preferences of human apoE and mouse apoE. Recombinant mouse apoE and human apoE isoforms were radiolabeled, incubated with human plasma, and fractionated into various lipoprotein classes by size-exclusion FPLC.
Arg-61 Apoe Gene-Targeted Mice.
We next sought to extend this observation in vivo. Mice expressing Arg-61 apoE were generated by gene targeting in ES cells (Fig. 1), and homozygous gene-targeted mice were identified by Southern blot analysis of tail DNA. Digestion of genomic DNA with EcoRI revealed an expanded Apoe locus due to presence of the neo cassette (not shown). A DdeI digest revealed the mutated Arg-61 codon in exon 3 (data not shown). Homozygous Arg-61 apoE gene-targeted mice were mated with Cre-deleter transgenic mice, in which Cre recombinase expression in germ cells causes organwide recombination of DNA flanked by loxP sites (24). Removal of the neo cassette resulted in normal levels of the Arg-61 apoE mRNA compared with wt in all tissues examined (Fig. 3). An identical Northern blot was hybridized to a β-actin probe to control for levels of total mRNA in the tissue samples. Levels of β-actin mRNA were similar in all pairs of tissue (Fig. 3).
Figure 3.
Comparison of apoE mRNA levels in various tissues and organs from wt (+) mice and Arg-61-targeted mice crossed with Cre-deleter mice (R). (A) Total RNA from various tissues and organs was isolated from wt and Agr-61 mice and subjected to Northern blot analysis with an Apoe exon 4 probe. Tissues and organs included: B, brain; Li, liver; Lu, lung; M, muscle; Si, small intestine; T, testis; H, heart; St, stomach; K, kidney; Sk, skin; Sp, spleen; and E, eye. (B) A control mouse β-actin hybridization is shown.
Analysis of Arg-61 ApoE Protein in Mouse Plasma.
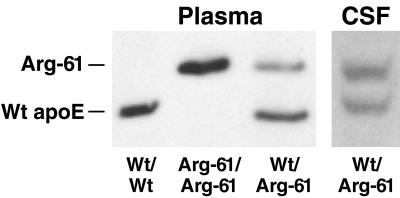
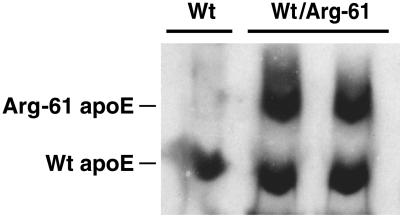
IEF–PAGE/Western blot analysis of mouse plasma demonstrated that Arg-61 apoE migrated with one additional positive charge unit relative to wt mouse apoE (Fig. 4). This finding is consistent with the substitution of arginine for Thr-61 and confirms that the mutation is carried in the expressed protein. As shown in plasma from heterozygous Arg-61/wt mice, the Arg-61 apoE levels were consistently 60–70% (n = 30) lower than the levels of wt apoE (Fig. 4). This finding indicates that Arg-61 apoE is catabolized more rapidly in plasma, because homozygous gene-targeted mice had Arg-61 apoE mRNA levels identical to those of apoE in wt mice (Fig. 3). To exclude the possibility that the lower levels of Arg-61 apoE in plasma resulted from reduced Arg-61 apoE secretion by the liver, cultures of primary hepatocytes from two Arg-61/wt heterozygous mice were examined. The culture medium contained equivalent levels of both apoE isoforms, demonstrating that the Arg-61 Apoe allele retained full transcriptional and translational activity (Fig. 5).
Figure 4.
IEF and Western blot of mouse plasma and CSF apoE. Mouse apoE isoforms in plasma and CSF were monitored by IEF and Western blot detection. Mice were fasted for 4 h before bleeding.
Figure 5.
IEF and Western blot of apoE secreted from primary hepatocytes into the culture medium. Primary hepatocytes were isolated from wt and Arg-61/wt heterozygous mice and placed into culture for 2 days, and the medium was collected. Mouse wt and Arg-61 apoE in medium were assessed by IEF and Western blot detection.
Plasma Lipid Levels.
Levels of plasma cholesterol in homozygous Arg-61 mice were not significantly different from those of wt mice (60.5 ± 9.1 vs. 59.7 ± 8.0 mg/dl, n = 16, P = 0.8). Plasma triglyceride levels in Arg-61 mice also were not significantly different from those of wt mice (20.5 ± 8.2 vs. 24.1 ± 12.1 mg/dl, n = 8, P = 0.5). Fractionation of mouse plasma also showed no marked change in the lipid distribution of plasma lipoproteins (data not shown).
Distribution of Arg-61 and Wt ApoE in Plasma Lipoproteins.
On a chow diet, mice normally have low levels of circulating VLDL, reflecting low levels of hepatic apoB100 expression. To study the distribution of Arg-61 apoE relative to wt apoE in plasma lipoproteins in vivo, the plasma VLDL level in Arg-61/wt mice was raised by feeding them a cholesterol-rich (Paigen) diet (ICN, Costa Mesa, CA) for 6 days (Fig. 6). The lipoprotein-binding preferences of Arg-61 and wt apoE were then examined by fractionating mouse plasma by size-exclusion FPLC chromatography and IEF–PAGE. Relative to wt apoE, Arg-61 apoE bound preferentially to lower density lipoproteins (Fig. 6), and little, if any, Arg-61 apoE was found in the HDL fraction. This finding confirms the shift in lipoprotein-binding preference observed in vitro (Fig. 2).
Figure 6.
Distribution of wt and Arg-61 apoE in plasma from a heterozygous, gene-targeted mouse on a high-cholesterol diet. Arg-61/wt apoE heterozygous mice were fed the Paigen diet for 6 days to increase VLDL levels. Nonfasted plasma was fractionated by size-exclusion FPLC. (Upper) Fractions corresponding to the different lipoprotein classes were resolved by IEF, and apoE was detected by Western blotting.
These results are consistent with the notion that the catabolism of Arg-61 apoE, relative to wt apoE, is enhanced because it binds preferentially with lipoproteins of lower density, which turn over more rapidly in plasma. Similar levels of Arg-61 and wt apoE were present in CSF (Fig. 4), suggesting an equal production and similar metabolism of both apoE isoforms in CSF.
Discussion
This study demonstrates that apoE4 domain interaction can be introduced into mouse apoE by supplying the critical equivalent of Arg-61. Both in vitro and in vivo, Arg-61 mouse apoE, like human apoE4, bound preferentially to VLDL. In contrast, wt mouse apoE (Thr-61) displayed an apoE3-like preference for HDL. Thus, although it has the equivalent of Arg-112 found in apoE4, wt mouse apoE (and presumably all other mammalian forms of apoE that also have Thr-61) behaves functionally like apoE3, reflecting the absence of apoE4 domain interaction. In parallel with human apoE4, the introduction of apoE4 domain interaction into mouse apoE likely involves the interaction between Arg-61 and the Glu-255 equivalent. However, it is possible that another glutamic acid residue in the vicinity of the Glu-255 equivalent participates. Establishing this point will require additional mutagenesis studies. The Arg-61 mouse apoE model reported here will facilitate studies of the role of apoE4 domain interaction in lipoprotein metabolism, atherosclerosis, and neurodegeneration.
Heterozygous Arg-61/wt apoE mice displayed a lipoprotein metabolism phenotype resembling that in human apoE4/3 subjects. Like apoE4/3 subjects, who have lower levels of apoE4 than of apoE3 (15), Arg-61/wt mice had lower levels of Arg-61 apoE than wt apoE. In fact, the relative levels of the mouse apoE isoforms mirrored that in apoE4/3 subjects (15). This finding is consistent with the more rapid catabolism, and lower plasma levels, of apoE4 than of apoE3 or apoE2 (13, 27). Other findings provided additional evidence for enhanced catabolism of Arg-61 apoE. In vivo, the level of Arg-61 apoE mRNA in the liver (the primary source of plasma apoE) was normal. In vitro, primary hepatocytes from heterozygous Arg-61/wt mice secreted equal amounts of each isoform. Another similarity to human apoE4 was the preference of Arg-61 apoE for lower-density apoB-containing lipoproteins, which turn over more rapidly than HDL in plasma. In CSF, which lacks apoB-containing lipoproteins, the levels of Arg-61 and wt apoE were equal. Taken together, these results demonstrate that apoE4 domain interaction was introduced in vivo into Arg-61 apoE mice.
ApoE4 is associated with elevated plasma LDL levels and with a predisposition to atherosclerosis (28). Recently, the effects of apoE4 on the metabolism of apoB48 and apoB100 were determined with a stable isotope approach in humans (29). The presence of one APOE4 allele was associated with higher LDL levels, owing to lower fractional catabolism of LDL and a 33% increase in the conversion of VLDL to LDL.
Because Arg-61 mice reproduce key features of apoE4 metabolism in plasma, they will be invaluable for understanding how apoE4 domain interaction contributes to increased plasma LDL levels and atherosclerosis in humans. However, in mice, plasma cholesterol is transported mostly by HDL, and plasma LDL levels are intrinsically low because of low apoB100 expression. In contrast, human plasma cholesterol is transported largely by LDL, which contain apoB100. The lower levels of plasma apoB in mice likely explain the absence of detectable differences in plasma and VLDL lipid levels in Arg-61 mice, despite a marked alteration in apoE catabolism. To reproduce more closely the human apoE4 phenotype in human lipoprotein metabolism, we will cross Arg-61 mice with transgenic mice expressing high levels of human apoB100 (30).
The APOE4 allele is a major susceptibility gene associated with neurodegenerative disorders, including Alzheimer's disease (31), and with poor clinical outcome after head trauma and stroke (32, 33). The isoform-specific effects of apoE in the human nervous system are thought to include effects on lipid transport (34), on the metabolism of tau protein and amyloid β-peptide, and on susceptibility to oxidation (35, 36). The precise role of apoE4 in neurodegenerative disorders remains unclear; however, apoE4 is less able than apoE2 or apoE3 to promote the growth and repair of neurons after injury (7, 37). Studies of transgenic mice expressing apoE isoforms in neurons have provided evidence that, relative to apoE3, apoE4 predisposes to neuronal loss and hyperphosphorylation of tau protein and exerts a dominant-negative effect in neurodegeneration (38–40). These mice also suffer from a loss of cognitive function (41). The Arg-61 mouse apoE model will allow the determination of which of the above effects results directly from apoE4 domain interaction.
The Arg-61 apoE mouse model has unique features that avoid some potential complications of other mouse models of human apoE isoforms. First, the “humanized” Arg-61 mouse Apoe gene is expressed under the control of the endogenous, tissue-specific control elements. Thus, site-of-integration effects on protein expression levels, species effects, and the use of nonnatural apoE promoters are eliminated. Recent mouse models of apoE isoforms have been generated by gene targeting in ES cells to replace the murine Apoe locus with human APOE alleles (42–45), and some lines displayed unexpected lipoprotein metabolism phenotypes. For example, gene-targeted mice expressing human apoE3 showed increased diet-induced hyperlipidemia, demonstrating that, in mice, human apoE3 is less efficient at clearing lipoproteins than wt mouse apoE (42). Similarly, one line of gene-targeted mice expressing human apoE4, unlike human apoE4 subjects, had normal levels of plasma cholesterol and LDL and retarded clearance of VLDL (44). However, elevated levels of plasma cholesterol were found in human apoE4 gene-targeted mice created by using a different gene-targeting strategy (45). These observations demonstrate that several factors complicate the development of faithful mouse models for human proteins, including apoE isoforms. In addition, the presence of the neo cassette in a knock-in allele can result in a phenotype that is independent of the proposed contribution of the targeted allele (46).
In summary, we report the development of an in vivo model designed to test the role of apoE4 domain interaction in lipoprotein metabolism, atherosclerosis, and neurodegeneration. Gene-targeted mice expressing Arg-61 apoE display two key phenotypes associated with apoE4 in humans. Arg-61 apoE has a preference for lower density lipoproteins, and it is less abundant than wt mouse apoE in plasma likely because of a more rapid catabolism. Taken together, these results indicate that Arg-61 gene-targeted mice will serve as a model to study the effect of domain interaction in apoE4 as a cause for disease in humans, opening the possibility of developing isoform-specific therapeutic strategies.
Acknowledgments
We thank Heather Myers for help with ES cell culture, Bryan Tow for blastocyst injections, Dr. Murielle Véniant for help with hepatocyte cell culture, Dr. Edward Kim for help in assembling the gene-targeting vector, and Dr. Gail Martin for providing Cre-deleter transgenic mice. We also thank Brian Auerbach for manuscript preparation, Gary Howard and Stephen Ordway for editorial assistance, and Jack Hull and John Carroll for preparation of figures. This work was supported by National Institutes of Health Grants HL47660 and NS35939 (to K.H.W.) and a fellowship from the Heart and Stroke Foundation of Canada (to R.L.R.).
Abbreviations
- apo
apolipoprotein
- LDL
low density lipoprotein(s)
- VLDL
very low density lipoprotein(s)
- HDL
high density lipoprotein
- IEF–PAGE
isoelectric-focusing PAGE
- CSF
cerebrospinal fluid
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mahley R W. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Reyland M E, Williams D L. J Biol Chem. 1991;266:21099–21104. [PubMed] [Google Scholar]
- 3.Ishigami M, Swertfeger D K, Granholm N A, Hui D Y. J Biol Chem. 1998;273:20156–20161. doi: 10.1074/jbc.273.32.20156. [DOI] [PubMed] [Google Scholar]
- 4.Avila E M, Holdsworth G, Sasaki N, Jackson R L, Harmony J A K. J Biol Chem. 1982;257:5900–5909. [PubMed] [Google Scholar]
- 5.Hui D Y, Harmony J A K, Innerarity T L, Mahley R W. J Biol Chem. 1980;255:11775–11781. [PubMed] [Google Scholar]
- 6.Pepe M G, Curtiss L K. J Immunol. 1986;136:3716–3723. [PubMed] [Google Scholar]
- 7.Weisgraber K H, Mahley R W. FASEB J. 1996;10:1485–1494. doi: 10.1096/fasebj.10.13.8940294. [DOI] [PubMed] [Google Scholar]
- 8.Weisgraber K H. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 9.Mahley R W, Rall S C., Jr . In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Childs B, Kinzler K W, Vogelstein B, editors. Vol. 2. New York: McGraw–Hill; 2001. pp. 2835–2862. [Google Scholar]
- 10.Saunders A M, Strittmatter W J, Schmechel D, St. George-Hyslop P H, Pericak-Vance M A, Joo S H, Rosi B L, Gusella J F, Crapper-MacLachlan D R, Alberts M J, et al. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 11.Strittmatter W J, Roses A D. Proc Natl Acad Sci USA. 1995;92:4725–4727. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davignon J, Cohn J S, Mabile L, Bernier L. Clin Chim Acta. 1999;286:115–143. doi: 10.1016/s0009-8981(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 13.Gregg R E, Zech L A, Schaefer E J, Stark D, Wilson D, Brewer H B., Jr J Clin Invest. 1986;78:815–821. doi: 10.1172/JCI112645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinmetz A, Jakobs C, Motzny S, Kaffarnik H. Arteriosclerosis. 1989;9:405–411. doi: 10.1161/01.atv.9.3.405. [DOI] [PubMed] [Google Scholar]
- 15.Weisgraber K H. J Lipid Res. 1990;31:1503–1511. [PubMed] [Google Scholar]
- 16.Aggerbeck L P, Wetterau J R, Weisgraber K H, Wu C-S C, Lindgren F T. J Biol Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 17.Wetterau J R, Aggerbeck L P, Rall S C, Jr, Weisgraber K H. J Biol Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 18.Dong L-M, Wilson C, Wardell M R, Simmons T, Mahley R W, Weisgraber K H, Agard D A. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 19.Dong L-M, Weisgraber K H. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 20.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innerarity T L, Pitas R E, Mahley R W. J Biol Chem. 1979;254:4186–4190. [PubMed] [Google Scholar]
- 23.Farese R V, Jr, Ruland S L, Flynn L M, Stokowski R P, Young S G. Proc Natl Acad Sci USA. 1995;92:1774–1778. doi: 10.1073/pnas.92.5.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers E N, Lewandoski M, Martin G R. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, Cham C M, Véniant M M, Ambroziak P, Young S G. J Clin Invest. 1998;101:1468–1477. [PMC free article] [PubMed] [Google Scholar]
- 27.Utermann G. In: Diabetes, Obesity and Hyperlipidemias III. Crepaldi G, Tiengo A, Baggio G, editors. Amsterdam: Elsevier Science; 1985. pp. 1–28. [Google Scholar]
- 28.Davignon J, Gregg R E, Sing C F. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Welty F K, Lichtenstein A H, Barrett P H R, Jenner J L, Dolnikowski G G, Schaefer E J. Arterioscler Thromb Vasc Biol. 2000;20:1807–1810. doi: 10.1161/01.atv.20.7.1807. [DOI] [PubMed] [Google Scholar]
- 30.Borén J, Lee I, Zhu W, Arnold K, Taylor S, Innerarity T L. J Clin Invest. 1998;101:1084–1093. doi: 10.1172/JCI1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corder E H, Saunders A M, Strittmatter W J, Schmechel D E, Gaskell P C, Small G W, Roses A D, Haines J L, Pericak-Vance M A. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 32.Nicoll J A R, Roberts G W, Graham D I. Ann NY Acad Sci. 1996;777:271–275. doi: 10.1111/j.1749-6632.1996.tb34431.x. [DOI] [PubMed] [Google Scholar]
- 33.Slooter A J C, Tang M-X, van Duijn C M, Stern Y, Ott A, Bell K, Breteler M M B, Van Broeckhoven C, Tatemichi T K, Tycko B, et al. J Am Med Assoc. 1997;277:818–821. doi: 10.1001/jama.277.10.818. [DOI] [PubMed] [Google Scholar]
- 34.LaDu M J, Gilligan S M, Lukens J R, Cabana B G, Reardon C A, Van Eldik L J, Holtzman D M. J Neurochem. 1998;70:2070–2081. doi: 10.1046/j.1471-4159.1998.70052070.x. [DOI] [PubMed] [Google Scholar]
- 35.Strittmatter W J, Weisgraber K H, Huang D Y, Dong L-M, Salvesen G S, Pericak-Vance M, Schmechel D, Saunders A M, Goldgaber D, Roses A D. Proc Natl Acad Sci USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaDu M J, Falduto M T, Manelli A M, Reardon C A, Getz G S, Frail D E. J Biol Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- 37.Mahley R W, Huang Y. Curr Opin Lipidol. 1999;10:207–217. doi: 10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Am J Pathol. 2000;156:951–964. doi: 10.1016/S0002-9440(10)64963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buttini M, Orth M, Bellosta S, Akeefe H, Pitas R E, Wyss-Coray T, Mucke L, Mahley R W. J Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buttini M, Akeefe H, Lin C, Mahley R W, Pitas R E, Wyss-Coray T, Mucke L. Neuroscience. 2000;97:207–210. doi: 10.1016/s0306-4522(00)00069-5. [DOI] [PubMed] [Google Scholar]
- 41.Raber J, Wong D, Buttini M, Orth M, Bellosta S, Pitas R E, Mahley R W, Mucke L. Proc Natl Acad Sci USA. 1998;95:10914–10919. doi: 10.1073/pnas.95.18.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan P M, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick R L, Quarfordt S H, Maeda N. J Biol Chem. 1997;272:17972–17980. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan P M, Mezdour H, Quarfordt S H, Maeda N. J Clin Invest. 1998;102:130–135. doi: 10.1172/JCI2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knouff C, Hinsdale M E, Mezdour H, Altenburg M K, Watanabe M, Quarfordt S H, Sullivan P M, Maeda N. J Clin Invest. 1999;103:1579–1586. doi: 10.1172/JCI6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamanaka H, Katoh-Fukui Y, Suzuki K, Kobayashi M, Suzuki R, Motegi Y, Nakahara Y, Takeshita A, Kawai M, Ishiguro K, et al. Hum Mol Genet. 2000;9:353–361. doi: 10.1093/hmg/9.3.353. [DOI] [PubMed] [Google Scholar]
- 46.Kaul A, Köster M, Neuhaus H, Braun T. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]