Abstract
Many thermophiles thriving in a natural high-temperature environment remain uncultivated, and their ecophysiological functions in the biogeochemical cycle remain unclear. In the present study, we performed long-term continuous cultivation at 65°C and 70°C using a microbial mat sample, collected from a subsurface geothermal stream, as the inoculum, and reconstructed the whole genome of the maintained populations using metagenomics. Some metagenome-assembled genomes (MAGs), affiliated into phylum-level bacterial and archaeal clades without cultivated representatives, contained genes involved in nitrogen metabolism including nitrification and denitrification. Our results show genetic components and their potential interactions for the biogeochemical nitrogen cycle in a subsurface geothermal environment.
Keywords: thermophiles, metagenomics, long-term continuous cultivation, nitrogen cycle, subsurface geothermal groundwater
Phylogenetically diverse, uncultivated thermophiles are present in natural high-temperature environments, such as terrestrial hot springs and deep-sea hydrothermal vents, as suggested by 16S rRNA gene analyses (3, 16, 32). Culture-independent single-cell genomics and metagenomics are powerful techniques and have become standards in microbial ecology (7, 13, 23, 31). Previous studies using single-cell genomics and metagenomics revealed the metagenome-assembled genomes (MAGs) or single amplified genomes (SAGs) of uncultivated thermophiles, and suggested their metabolic potential (4, 27, 30, 34). However, their physiological functions and ecological roles remain unclear because of the absence of cultivated representatives or the availability of partial genome sequences only. One of the research approaches to overcome this difficulty is a genomic analysis based on temperature-controlled laboratory cultivation, which has provided direct evidence for growth temperatures and metabolism (10, 11).
Nitrogen is one of the key elements for life in anabolism and catabolism as well as for the biogeochemical cycle. Uncultivated thermophiles detected in a high-temperature environment potentially play significant roles in the biogeochemical nitrogen cycle, as indicated by their metabolic potential (4–6, 27). In order to obtain a better understanding of the mechanisms by which as well as what thermophilic microbial populations contribute to the nitrogen cycle in the natural environment, we collected a microbial mat sample in an ammonia-rich geothermal groundwater stream in Japan and performed continuous cultivation at 65°C and 70°C using flow-through bioreactors with the mat sample as the inoculum as previously reported (25). At the original sampling site, discharged groundwater has constantly shown a high temperature (69–72°C), circumneutral pH (6.1–6.4), low salinity (0.1%), and high concentrations of NH4+ (>100 μM) and Fe2+ (>70 μM) (14, 17, 24, 33). Previous studies demonstrated the presence of uncultivated microorganisms affiliated with Candidatus (Ca.) “Nitrosocaldus”, “Aigarchaeota”, and “Acetothermia” in the stream and inoculated mat sample (14, 24–27, 34). The MAGs of “Aigarchaeota” and “Acetothermia” in the stream have already been reported (27, 34). Although no MAGs/SAGs of “Nitrosocaldus” have been described to date, “Nitrosocaldus” spp. have predominated in microbial communities as shown by a 16S rRNA gene analysis (24, 26). Geochemical measurements and a nitrogen isotopic analysis of mat samples have provided evidence for the microbiological oxidation of ammonia and nitrite (24). Furthermore, the oxidation of ammonia to nitrite by “Nitrosocaldus” spp. at 70°C was demonstrated using bioreactors for more than 2 years (25). These findings have suggested that the nitrogen cycle, such as nitrification, is driven by these uncultivated microorganisms in this high-temperature environment.
We herein demonstrated the metabolic potential of maintained populations in high-temperature bioreactors, which operated for more than 4 years, as revealed by metagenomics. Details of the experimental procedures employed are described in Supplementary Information. In brief, DNA was extracted from samples in a bioreactor (Reactor-65), which operated at 65°C for 349 d after the inoculation, and in another bioreactor (Reactor-70A), which operated at 70°C for 45 and 1,579 d after the inoculation. On day 159, a subsample from Reactor-70A was inoculated into a new bioreactor (Reactor-70B), which operated at 70°C, in order to assess the reproducibility of cultivation, and a sample was obtained from Reactor-70B 1,420 d after the re-initiation of cultivation. DNA extraction was performed in duplicate, except for Reactor-70A on day 45. Extracted DNA from seven samples was applied to shotgun metagenomic sequencing as previously described (12). All reads were cleaned up (Table S1) and co-assembled. This resulted in 26,561 contigs (≥1 kbp) with an N50 value of 10,355 bp. Binning using longer contigs (≥2.5 kbp) and manual curation resulted in a total of 41 MAGs (Fig. S1). The MIMAG (Minimum Information about a Metagenome-Assembled Genome) developed by the Genomic Standards Consortium (8) as a standard for reporting MAGs, such as estimates of genome completeness and contamination, was summarized in Table S2. Phylogenetic trees were constructed for 16S rRNA gene sequences (Fig. S2) and for the concatenated protein sequences of 43 single-copy marker genes (Fig. 1A).
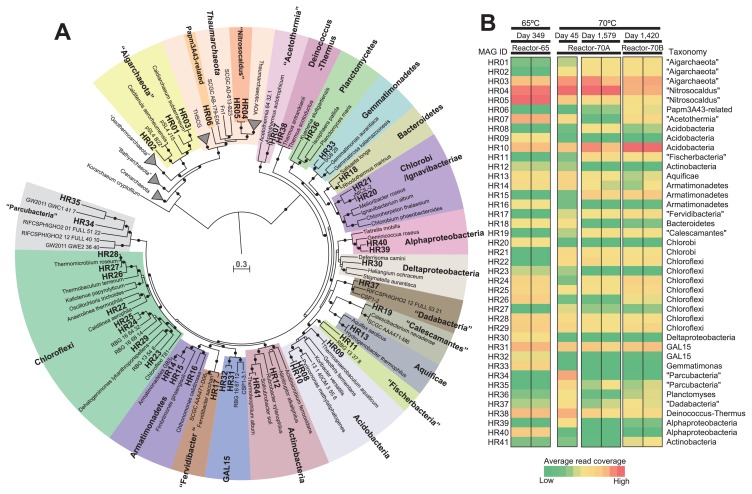
Fig. 1.
Phylogeny and relative abundance of cultivates based on the analysis of MAGs. (A) Maximum likelihood tree of concatenated amino acid sequences of 43 conserved single-copy marker proteins. The MAGs obtained in this study are shown in bold. The tree was rooted at the midpoint between Archaea and Bacteria. Filled and open circles at branches indicate more than 70% and 50–70% of bootstrap values (1,000 replicates), respectively. The scale bar represents 0.3 amino acid substitutions per sequence position. (B) Heat-map representing the relative abundance of MAGs in each metagenome (a total of seven) based on average read coverages.
These MAGs were assigned to 19 bacterial clades including “Calescamantes”, “Fervidibacteria”, and “Parcubacteria” (30), “Dadabacteria” (15), “Fischerbacteria” (1), and GAL15 defined as a phylum-level clade in the Silva database (29) without cultivated representatives, and two archaeal clades including “Aigarchaeota” and Thaumarchaeota at the phylum level (see Supplementary Information for details on the taxonomic affiliations of MAGs). Thaumarchaeotic MAGs were classified into two lineages—”Nitrosocaldus” and a deeper lineage most closely related (16S rRNA gene similarity, 90.3%) to the environmental clone Papm3A43 recovered from deep-sea hydrothermal fluid (18). Based on cultivation temperatures and periods, all MAG-derived cultivates were thermophilic or thermo-tolerant.
In order to assess the relative abundance of these MAG populations in the bioreactor, read coverages for MAGs were displayed as a heat-map figure (Fig. 1B; Table S2). Several MAGs in “Aigarchaeota”, “Nitrosocaldus”, Acidobacteria, and GAL15 were relatively abundant in all samples from the bioreactors at 65°C and 70°C over time. Some MAGs in Armatimonadetes, “Fischerbacteria”, “Calescamantes”, Chloroflexi, and “Aigarchaeota” were less abundant at 65°C, but were relatively abundant at 70°C, which suggests that they were adapted to the higher temperature.
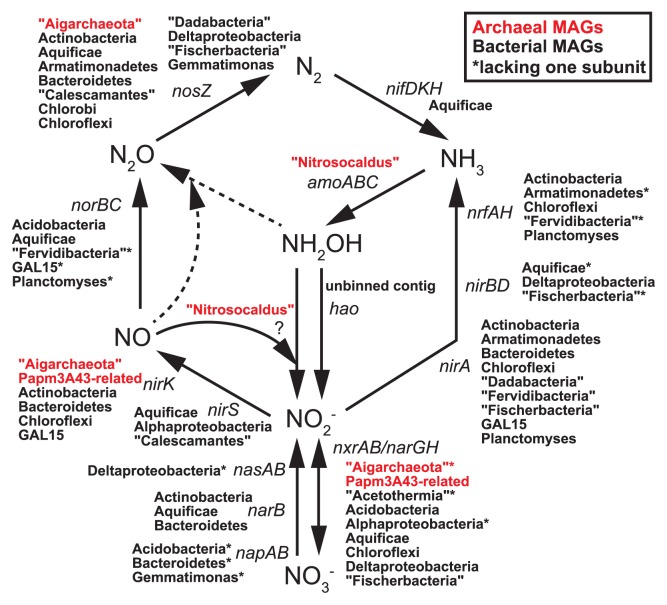
In order to clarify the ecological roles of the maintained populations in the nitrogen cycle, such as nitrification catalyzed by ammonia monooxygenase (Amo), hydroxylamine oxidoreductase (Hao), and nitrite oxidoreductase (Nxr), assimilatory and dissimilatory nitrate reduction by nitrate and nitrite reductases (Nap/Nar/Nas/Nir), denitrification by nitric oxide reductase (Nor) and nitrous oxide reductase (Nos), and nitrogen fixation by nitrogenase (Nif), we focused on genes for the key subunits of the above enzymes (Fig. 2).
Fig. 2.
Potential contribution of MAG-derived cultivates in the biogeochemical nitrogen cycle. Taxonomic names for the archaeal and bacterial MAGs are colored in red and black, respectively. Solid and dashed arrows indicate enzymatic and abiotic reactions, respectively. Asterisks indicate MAGs lacking a gene for one of the key subunits of the enzymes (see Table S3 for details).
The amoABC genes were found in the “Nitrosocaldus” MAGs (HR04 and HR05), but not in any of the other MAGs or unbinned contigs. These amo genes have also been detected in enrichment cultures of “Nitrosocaldus yellowstonii” (10); however, its entire genome has not been reported. Although the oxidation of NH3 to NO2− by the “Nitrosocaldus”-related populations in our bioreactors has been demonstrated previously (25) and in the present study (Fig. S3), the enzymes that catalyze NH2OH to NO2− in archaeal ammonia-oxidizers have yet to be identified (20). Since a hao gene was detected in an unbinned contig, we were unable to exclude the possibility that NH2OH oxidation was catalyzed by an undefined cultivate. The amo genes were not found in Papm3A43-related MAG (HR06), and were also absent in the previously reported MAGs and SAGs of deeper-branching thaumarchaeotic lineages than “Nitrosocaldus” (4, 22, 30). As suggested previously (4, 22), the HR06-derived archaeon may not be an ammonia oxidizer.
The narGH/nxrAB and nirK genes were found in Papm3A43-related MAGs, in addition to some MAGs of “Aigarchaeota” and several bacterial MAGs. We performed a phylogenetic analysis of the subunit NarH/NxrB (Fig. S4), which has been used in an Nar/Nxr analysis (28), indicating that the NarH/NxrB sequences obtained were distantly related to previously identified NxrB proteins. Thus, a phylogenetic analysis was not sufficient to identify which gene products were realistically involved in microbial NO2− oxidation. Nevertheless, NO3− production was observed in the bioreactors (up to 150 μM in the effluent) operated at 70°C for more than 4 years (Fig. S3), indicating that some genes for Nar/Nxr were involved in NO2− oxidation to NO3−. Nearly half of the MAGs (19 out of 41) including “Aigarchaeota”, “Fischerbacteria”, and “Dadabacteria” contained nosZ. Thus, they may produce N2 from N2O, which may be produced abiotically from NH2OH and NO (21) or biotically from NO via NorBC.
Our results provide insights into previously unrecognized physiological and metabolic properties, particularly in the nitrogen cycle, of thermophilic microbial populations in high-temperature bioreactors, some of which represent phylum-level clades without cultivated representatives. nosZ has not been found in the MAGs/SAGs of “Aigarchaeota” reported to date. However, HR02, one of the “Aigarchaeota” MAGs found in this study, contained the gene. Although the contribution to the nitrogen cycle of deep-branching thaumarchaeotic lineages including the MAG named Fn1 (22) is unknown, the HR06-derived archaeon with nar/nxr may be involved in nitrite oxidation or nitrate reduction. No MAGs/SAGs of “Dadabacteria”, “Fischerbacteria”, or GAL15 have been obtained from any geothermal environment to date. Thus, HR11, HR31, HR32, and HR37 are the first thermophilic MAGs in these groups, and contain several genes involved in the nitrogen cycle, similar to previously reported MAGs from a nongeothermal environment (1, 15). Several MAGs/SAGs of “Calescamantes” and “Fervidibacteria” have been reported from geothermal environments, and contain genes involved in the nitrogen cycle, such as narG, nirS, and nosZ (6, 30), which is consistent with our results.
In addition to nitrogen metabolism, other potential metabolic functions of MAGs, such as carbon fixation and energy acquisition, are summarized in Table S3 and Supporting Information. In the bioreactors, the energy source for sustaining the microcosm was limited to NH3 added to media at a final concentration of approximately 200 μM (12; Supporting Information). The ammonia-oxidizing archaeal populations derived from the abundant “Nitrosocaldus” MAG (HR04) may be primary producers in the microcosm as suggested previously (12). NO2− produced from the oxidation of NH3 may be used by nitrite-oxidizing autotrophs potentially derived from some of the MAGs containing nxr/nar genes. However, the carbon fixation pathways present have yet to be identified. Unexpectedly, Fe(II), which was in media at a final concentration of approximately 100 μM to simulate the chemical composition of geothermal groundwater at the sampling site, may be another potential energy source for the population. The “Fischerbacteria” and Chlorobi MAGs (HR11 and HR21) contained a homolog for Cyc2 (Fig. S5), an outer membrane c-type cytochrome that may catalyze Fe(II) oxidation in several iron oxidizers, such as Acidithiobacillus spp., Ferriphaselus spp., Gallionella spp., and Mariprofundus spp. (2, 9, 19). Moreover, a near-complete gene set including rbcSL in the Calvin cycle for carbon fixation was found in HR11. These results imply that the bacterium derived from the “Fischerbacteria” MAG is potentially an iron-oxidizing autotroph and has a role as a primary producer in the microcosm.
The present study revealed the unique physiological functions and ecological roles of diverse, previously uncultivated thermophiles in the nitrogen cycle in a subsurface geothermal environment. These results provide important information for the isolation of novel MAG-derived thermophiles, and will contribute to the characterization and elucidation of their ecophysiologies. Further cultivation-based metagenomics for a number of high-temperature environments with different pH, salinities, redox potentials, and energy sources may reveal uncultivated thermophiles and unidentified thermophilic microbial ecosystems.
Supplementary Material
Acknowledgements
We would like to thank Dr. Takuro Nunoura (JAMSTEC) for his helpful comments. We would like to thank Editage for the English language review. We also would like to thank the two anonymous reviewers for their helpful comments. This work was supported by JSPS KAKENHI Grant Numbers JP16H06180 and JP16K14663 (to S.K.), and JP22540499 (to M.N.), and partially by the Cabinet Office, Government of Japan, through the Next-generation Technology for Ocean Resources Exploration (called as Zipangu-in-the-ocean project) in the Cross-ministerial Strategic Innovation Promotion Program (SIP).
Footnotes
Accession number
The sequence data set obtained in this study was deposited into DDBJ (DNA Data Bank of Japan) under the accession number DRA006147, and linked to the BioProject with the accession number PRJDB6348.
References
- 1.Anantharaman K., Brown C.T., Hug L.A., et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barco R.A., Emerson D., Sylvan J.B., Orcutt B.N., Jacobson Meyers M.E., Ramirez G.A., Zhong J.D., Edwards K.J. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol. 2015;81:5927–5937. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns S.M., Delwiche C.F., Palmer J.D., Pace N.R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beam J.P., Jay Z.J., Kozubal M.A., Inskeep W.P. Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J. 2014;8:938–951. doi: 10.1038/ismej.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beam J.P., Jay Z.J., Schmid M.C., et al. Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous ‘streamer’ community. ISME J. 2016;10:210–224. doi: 10.1038/ismej.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becraft E.D., Dodsworth J.A., Murugapiran S.K., et al. Single-cell-genomics-facilitated read binning of candidate phylum EM19 genomes from geothermal spring metagenomes. Appl Environ Microbiol. 2016;82:992–1003. doi: 10.1128/AEM.03140-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blainey P.C. The future is now: Single-cell genomics of bacteria and archaea. FEMS Microbiol Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers R.M., Kyrpides N.C., Stepanauskas R., et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotech. 2017;35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castelle C., Guiral M., Malarte G., Ledgham F., Leroy G., Brugna M., Giudici-Orticoni M.-T. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem. 2008;283:25803–25811. doi: 10.1074/jbc.M802496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Torre J.R., Walker C.B., Ingalls A.E., Konneke M., Stahl D.A. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 11.Elkins J.G., Podar M., Graham D.E., et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai M., Nishi S., Tsuda M., Sunamura M., Takaki Y., Nunoura T. Library construction from subnanogram DNA for pelagic sea water and deep-sea sediment. Microbes Environ. 2017;32:336–343. doi: 10.1264/jsme2.ME17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraoka S., Yang C.-c., Iwasaki W. Metagenomics and bioinformatics in microbial ecology: Current status and beyond. Microbes Environ. 2016;31:204–212. doi: 10.1264/jsme2.ME16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirayama H., Takai K., Inagaki F., Yamato Y., Suzuki M., Nealson K.H., Horikoshi K. Bacterial community shift along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles. 2005;9:169–184. doi: 10.1007/s00792-005-0433-8. [DOI] [PubMed] [Google Scholar]
- 15.Hug L.A., Thomas B.C., Sharon I., et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–173. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz P., Pitulle C., Hershberger K.L., Pace N.R. Novel division level bacterial diversity in a yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki F., Takai K., Hirayama H., Yamato Y., Nealson K.H., Horikoshi K. Distribution and phylogenetic diversity of the subsurface microbial community in a Japanese epithermal gold mine. Extremophiles. 2003;7:307–317. doi: 10.1007/s00792-003-0324-9. [DOI] [PubMed] [Google Scholar]
- 18.Kato S., Yanagawa K., Sunamura M., et al. Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough. Environ Microbiol. 2009;11:3210–3222. doi: 10.1111/j.1462-2920.2009.02031.x. [DOI] [PubMed] [Google Scholar]
- 19.Kato S., Ohkuma M., Powell D.H., Krepski S.T., Oshima K., Hattori M., Shapiro N., Woyke T., Chan C.S. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol. 2015;6:1265. doi: 10.3389/fmicb.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerou M., Offre P., Valledor L., Abby S.S., Melcher M., Nagler M., Weckwerth W., Schleper C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2016;113:E7937–E7946. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlowski J.A., Stieglmeier M., Schleper C., Klotz M.G., Stein L.Y. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Handley K.M., Gilbert J.A., Kostka J.E. Metabolic potential of fatty acid oxidation and anaerobic respiration by abundant members of Thaumarchaeota and Thermoplasmata in deep anoxic peat. ISME J. 2015;9:2740–2744. doi: 10.1038/ismej.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narihiro T., Kamagata Y. Genomics and metagenomics in microbial ecology: Recent advances and challenges. Microbes Environ. 2017;32:1–4. doi: 10.1264/jsme2.ME3201rh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa M., Koba K., Makabe A., et al. Nitrification-driven forms of nitrogen metabolism in microbial mat communities thriving along an ammonium-enriched subsurface geothermal stream. Geochim Cosmochim Acta. 2013;113:152–173. [Google Scholar]
- 25.Nishizawa M., Sakai S., Konno U., et al. Nitrogen and oxygen isotope effects of ammonia oxidation by thermophilic thaumarchaeota from a geothermal water stream. Appl Environ Microbiol. 2016;82:4492–4504. doi: 10.1128/AEM.00250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunoura T., Hirayama H., Takami H., et al. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ Microbiol. 2005;7:1967–1984. doi: 10.1111/j.1462-2920.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- 27.Nunoura T., Takaki Y., Kakuta J., et al. Insights into the evolution of archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pester M., Maixner F., Berry D., et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol. 2014;16:3055–3071. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinke C., Schwientek P., Sczyrba A., et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 31.Stepanauskas R. Single cell genomics: An individual look at microbes. Curr Opin Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Takai K., Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai K., Hirayama H., Sakihama Y., Inagaki F., Yamato Y., Horikoshi K. Isolation and metabolic characteristics of previously uncultured members of the order Aquificales in a subsurface gold mine. Appl Environ Microbiol. 2002;68:3046–3054. doi: 10.1128/AEM.68.6.3046-3054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takami H., Noguchi H., Takaki Y., et al. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One. 2012;7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.