Abstract
Endohyphal bacteria (EHB), dwelling within fungal hyphae, markedly affect the growth and metabolic potential of their hosts. To date, two EHB belonging to the family Burkholderiaceae have been isolated and characterized as new taxa, Burkholderia rhizoxinica (HKI 454T) and Mycoavidus cysteinexigens (B1-EBT), in Japan. Metagenome sequencing was recently reported for Mortierella elongata AG77 together with its endosymbiont M. cysteinexigens (Mc-AG77) from a soil/litter sample in the USA. In the present study, we elucidated the complete genome sequence of B1-EBT and compared it with those of Mc-AG77 and HKI 454T. The genomes of B1-EBT and Mc-AG77 contained a higher level of prophage sequences and were markedly smaller than that of HKI 454T. Although the B1-EBT and Mc-AG77 genomes lacked the chitinolytic enzyme genes responsible for invasion into fungal cells, they contained several predicted toxin-antitoxin systems including an insecticidal toxin complex and PIN domain imposing an addiction-like mechanism essential for endohyphal growth control during host colonization. Despite the different host fungi, the alignment of amino acid sequences showed that the HKI 454T genome consisted of 1,265 (32.6%) and 1,221 (31.5%) orthologous coding sequences (CDSs) with those of B1-EBT and Mc-AG77, respectively. This comparative study of three phylogenetically associated endosymbionts has provided insights into their origin and evolution, and suggests the later bacterial invasion and adaptation of B1-EBT to its host metabolism.
Keywords: genome, endohyphal symbiont, interaction, M. cysteinexigens, B. rhizoxinica
‘The swollen hypha with two characteristic filiform appendages at its tip’ was the first report of the presence of an endobacterium in the cytosol of Glomeromycotina spores (44). Further morphological confirmation was obtained in subsequent studies (6, 7), and the presence of endobacteria then became widely evident in fungal lineages (14, 60). Recent studies revealed that bacteria and fungi interact at different levels in the bacterial-fungal interface, such as the bacterial slack attachment with the hyphal surface or metabolic complementation with associates and alterations to partners of various forms ranging from antibiosis and signaling to genetic exchange (20, 25, 57). However, endohyphal symbionts residing inside fungal cells exemplify the most intimate illustration of congenial interactions. Despite their genetic and functional diversities, bacterial-fungal interactions have become a new interest in microbiology with significance for microbial ecology (20).
Bacterial endohyphal symbionts are best characterized by the family Burkholderiaceae (Burkholderia-related) in the class Betaproteobacteria and frequently occur in the fungal phyla of Mucoromycota (63). Ascomycota and Basidiomycota are also known to host gammaproteobacteria (2) and alphaproteobacteria (61), respectively. Two endohyphal bacterium (EHB) belonging to the family Burkholderiaceae were isolated as new taxa, Burkholderia rhizoxinica (HKI 454T) and B. endofungorum (HKI 456T); however, only B. rhizoxinica HKI 454T has been characterized in detail (53). An interesting multilateral microbial interaction involving B. rhizoxinica HKI 454T has been shown to cause rice seedling blight and the causative agent is the Mucoromycotinan fungus Rhizopus microsporus. Phytopathogenic R. microsporus used an antimitotic agent, rhizoxin, as a virulence factor (59, 67). It later became evident that this virulence factor was not produced by R. microsporus itself, but by the EHB, B. rhizoxinica HKI 454T (53, 54). B. rhizoxinica controls the vegetative reproduction of the host (31). A previous study isolated B. rhizoxinica HKI 454T together with its host strain R. microsporus van Tieghem var. chinensis (ATCC 62417) from rice seedlings in Japan (54). Recent findings have characterized several strains of the non-pathogenic fungus Mortierella elongata (Mortierellomycotina) hosting a Burkholderia-related obligate endohyphal symbiont named B1-EBT (58), which was recently characterized as Mycoavidus cysteinexigens, gen. nov., sp. nov., based on its phenotypic, chemotaxonomic, and phylogenetic features (49). The basic function and characteristics of B1-EBT as a fungal endosymbiont currently remain unknown. An isolate of M. elongata (AG77) from a soil/litter sample close to a poplar tree in Duke forest, the USA was revealed to host M. cysteinexigens and it was attained through metagenome sequencing of the host M. elongata AG77 together with the endobacterium M. cysteinexigens (67). A large and growing endobacterial population of M. cysteinexigens AG77 has been observed under nitrogen limiting conditions, whereas M. elongata AG77 exhibited decreased growth (38).
Bacterial endosymbionts of fungi are widespread, and extremely different microbial populations may co-exist in the same host fungal strain. For example, rod-shaped Gramnegative ‘Candidatus Glomeribacter gigasporarum’ (CaGg) and coccoid Gram-positive Mollicutes-related endobacteria (Mre) co-exist in a single spore of the arbuscular mycorrhizal fungus Gigaspora margarita (13). Diversity in microbes is influenced by many different factors such as vertical and horizontal gene transfer, the activity of mobile genetic elements, the recombination machinery, and mutation rate (46, 47, 66). B. rhizoxinica HKI 454T is the rhizoxin-producing EHB of the phytopathogen R. microsporus and causes blight symptoms in rice seedlings, whereas M. cysteinexigens B1-EBT is the endohyphal symbiont of the non-pathogenic fungus M. elongata. Despite diverse host specificity and modes of interaction with respective hosts, B. rhizoxinica and M. cysteinexigens have a close phylogenetic relationship (49, 67). Although CaGg is the most phylogenetically related to M. cysteinexigens, the CaGg genome was incompletely sequenced, which may have been due to approximately 7% DNA contamination by the fungal host G. margarita (22). In addition, the coexisting endofungal bacteria Mre and other CaGg species were potentially contained in the spore lysates of G. margarita used to concentrate the endobacterial fraction (13, 42), which produced a number of contigs or induced big gaps in the subsequent genome assembly. Although the draft genome of M. cysteinexigens B1-EBT was sequenced in our previous study (21), information regarding structure, mobile genetic elements, rRNA operons, and tRNAs was still inconclusive. Therefore, in the present study, we elucidated the complete genome sequences of M. cysteinexigens B1-EBT by sequencing the DNA extract from culture collection and comparing it with the complete genome of the uncultured endosymbiont of M. elongata AG77 (named here as Mc-AG77), and also with that of the cultured B. rhizoxinica HKI 454T as a reference. Comparative genomic analyses of the three phylogenetically associated endosymbionts; M. cysteinexigens B1-EBT, Mc-AG77, and B. rhizoxinica HKI 454T, in two different fungal hosts may provide insights into their origin and evolution as well as potential host-bacterium interactions.
Materials and Methods
DNA extraction and genome sequencing
Strain B1-EBT was incubated on cysteine-containing buffered charcoal-yeast extract agar (B-CYEα) as described in our previous study (49). Colonies were suspended in sterile water and recovered by centrifuging at 12,000×g for 2 min. The DNA of strain B1-EBT was extracted using the lysozyme buffer method (69). Genomic DNA (21.8 μg) was used to prepare a sequencing library with a SMRTbell Template Prep Kit (Pacific Biosciences of California, Menlo Park, CA, USA). Genome sequencing was performed by Macrogen Korea using PacBio RSII single-molecule real-time (SMRT) sequencing technology (Pacific Biosciences of California).
PacBio read assembly and genome completing
The long single pass reads (122,714 reads with 1,069,198,690 bases in total) obtained from 1 SMRT cell were assembled using a PacBio de novo assembler optimized for the speed Hierarchical Genome Assembly Process (HGAP, v3.0) (9). Three credible contigs were generated: contig 1, 1,355,844 bp with 282-fold coverage; contig 2, 1,037,856 bp with 293-fold coverage; and contig 3, 398,059 bp with 102-fold coverage. These three contigs were combined with the previous draft genome contigs sequenced by the Roche 454 Titanium system (21) by aligning them using the sequence assembler ATGC ver. 4.2 (Genetyx Co., Tokyo, Japan), resulting in a completely circular genome sequence. The positions of 454 contigs in the complete genome were visualized using a CIRCOS circular visualization data tool (ver. 0.69) (30).
Multilocus sequence analysis
Five housekeeping genes (atpD, gyrB, lepA, recA, and rpoB) and the 16S rRNA gene sequence were selected for a multilocus sequence analysis (MLSA) as described previously (17), but with slight modifications. In the present study, nine major members (Cupriavidus necator N-1T, C. pinatubonensis JMP134, Pandoraea apista DSM 16535, P, pulmonicola DSM 16583, B. diffusa RF2- non-BP9, B. cepacia ATCC 25416, B. psudomallei K96243, and B. thailandensis E264T) belonging to the family Burkholderiaceae with completely sequenced genomes, and two species (Ralstonia pickettii 12DT and R. solanacearum GMI1000) within the family Ralstoniaceae, were selected as the interspecific references and Polynucleobacter necessarius subsp. necessaries STIR1 as outgroup, respectively, in the MLSA. Briefly, multiple sequence alignments were inferred for each dataset using MAFFT (ver. 7.222). MEGA (ver. 7) was used to modify the alignment scale and change the PHYLIP multiple sequence alignment format. The multiple alignment file and module selection of nucleotide substitution were combined using Kakusan version 4. Among-site rate variations were modeled by a GAMMA distribution with four rate categories. Tree searches were initiated using Randomized Axelerated Maximum Likelihood (RAxML) based on the concatenated dataset (12,492 and 11,023 positions in total with/without 16S rRNA, respectively) of the five housekeeping genes and 16S rRNA. Only strains with a full set of sequenced genes were compared in this analysis. Multilocus phylogenetic trees were visualized using the program FigTree (ver. 1.4.3) (49).
Genome annotation and general comparison
The complete genome sequence of strain B1-EBT was annotated automatically using Microbial Genome Annotation Pipeline (MiGAP) including MetaGeneAnnotator (23) for identifying protein coding sequences (CDSs), tRNAscan-SE (39) for tRNAs, and RNAmmer (35) for rRNAs. GC-skew and GC-content were calculated using a custom Perl script. In order to confirm the annotation of CDSs, the predicted protein sequences were annotated manually using BLASTP with the NCBI-nr database and KEGG Orthology and Links Annotation (KOALA) (26). PHAge Search Tool Enhanced Release (PHASTER) was used to identify prophage sequences within the genome (3, 71). Detailed pan-genome analyses were performed at the protein level using a bidirectional best-hit (BDBH) algorithm implemented in GET_HOMOLOGUES (ver. 2.0.23), imposing a minimum pairwise alignment coverage of 75.0% with default parameters (11) and visualized using the CIRCOS circular visualization data tool (ver. 0.69) (30).
Nucleotide sequence accession number
The genome sequence of M. cysteinexigens B1-EBT has been deposited in the DDBJ/EMBL/GenBank databases under the accession number AP018150. The complete genome sequences of B. rhizoxinica HKI 454T and M. cysteinexigens AG77 were obtained from NCBI RefSeq and GenBank (Supplemental Table 7, Reference 68), respectively.
Results and Discussion
General characteristics of the complete M. cysteinexigens B1-EBT genome
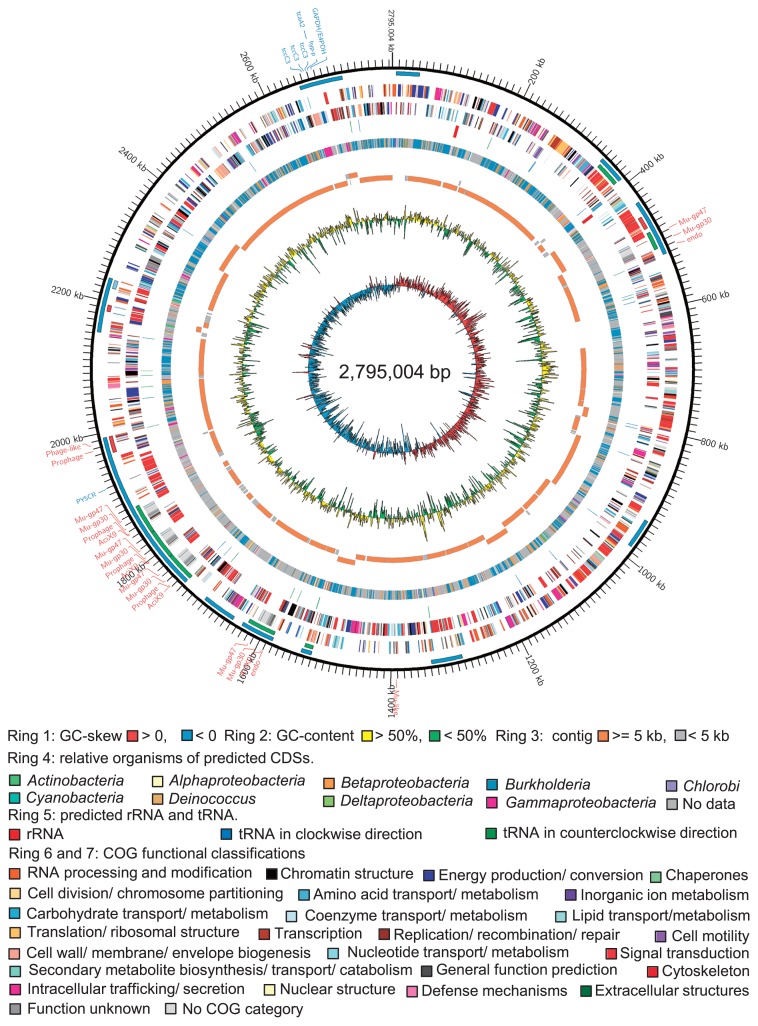
The complete B1-EBT genome consisted of a 2,795,004-bp circular chromosome with a G+C content of 48.9%, which contained two sets of ribosomal RNA (rrn) operons, 41 tRNAs, and 2,317 predicted CDSs (Fig. 1). This result was approximately in accordance with our studied draft genome of strain B1-EBT (21), revealing a genome size of 2,653,566 bp with a G+C content of 46.1%, 1 set of rrn operons, 39 tRNAs, and 2,390 CDSs. Most of the CDSs were identified as well conversed with CDSs in bacteria within the genus Burkholderia (Fig. 1). Of note, the CDSs encoding insecticidal toxin proteins, which are of potential benefit to the fungal host, were close to those conserved by Xenorhabdus (Gammaproteobacteria), a genus living symbiotically with soil entomopathogenic nematodes.
Fig. 1.
Schematic spherical illustration of the B1-EBT genome. Circles display from the inside to the outside as follows: (ring 1) GC-skew (G-C/G+C ratio) using a 1-kb window; (ring 2) GC-content using a 1-kb window; (ring 3) 454-contigs; (ring 4) relative bacteria of predicted CDSs; (ring 5) rRNAs and tRNAs; (rings 6 and 7) predicted CDSs transcribed in a counterclockwise/clockwise direction; (ring 8) intact, questionable, and incomplete prophage sequences are indicated in blue, green, and red solid bars, respectively; (ring 9) transposons; and (ring 10) scale in kb. Red and blue text outside shows the positions of phage-related genes and insecticidal toxin proteins, respectively.
Nevertheless, the annotation of the complete genome indicated that B1-EBT lacks cysteine biosynthetic pathways, as reported previously (21). In addition, the B1-EBT genome had conserved 11 transposons, 7 intact prophages, and 1 questionable and 3 incomplete prophages. Six of these transposons overlapped with the 11 prophage sequences (Fig. 1) encoding prophage-related proteins such as phage integrase and phage transposase (Fig. 2A and S1), suggesting that these regions were acquired by prophage insertion. Besides transposons, there were several retron-type reverse transcriptase and phage proteins. The genetic evidence supporting the introduction to foreign DNA contrasts endosymbionts such as Buchnera sp. APS, which was harbored by Acyrthosiphon pisum (pea aphid) (43, 62). The B1-EBT genome also contained several predicted toxin-antitoxin systems including the insecticidal toxin complex RelE/StbE, HipA, MazF, and PIN domain proteins encoding genes. These features imposed an addiction-like mechanism on the prokaryotic host genome and arbitrated the stability of mobile elements (1). In addition to their genetic addiction, these factors have been anticipated to facilitate growth retardation under stress conditions (51). Therefore, toxin-antitoxin systems may be essential for endohyphal growth control during host colonization. The B1-EBT genome also included 358 hypothetical proteins. We hypothesized that some strain-specific genomic components of unknown function may correlate with the evolution of the endofungal lifestyle.
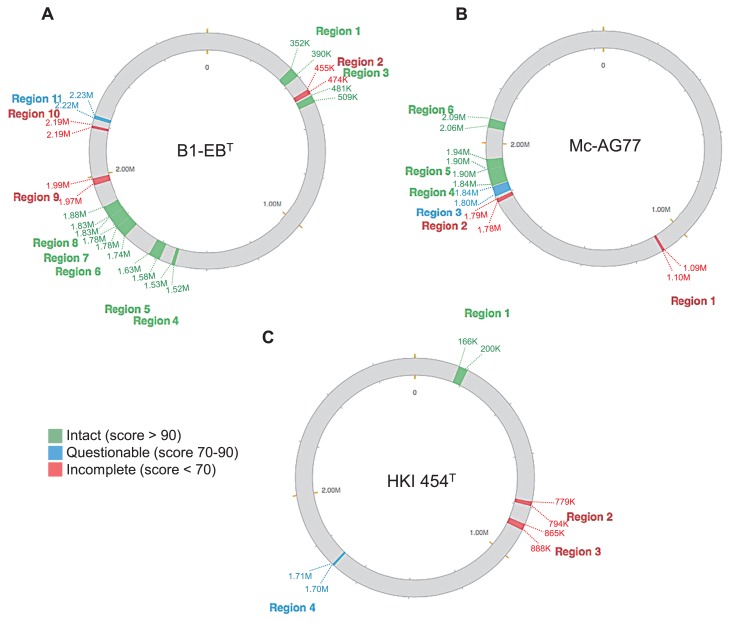
Fig. 2.
Prophage sequences within B1-EBT (A), Mc-AG77 (B), and HKI 454T (C) genomes. The details of each region are shown in Fig. S1. Position-based identification of prophage region is shown in colors. The scale bar is in bp.
Phylogenetic analysis of M. cysteinexigens and related taxa within the family Burkholderiaceae
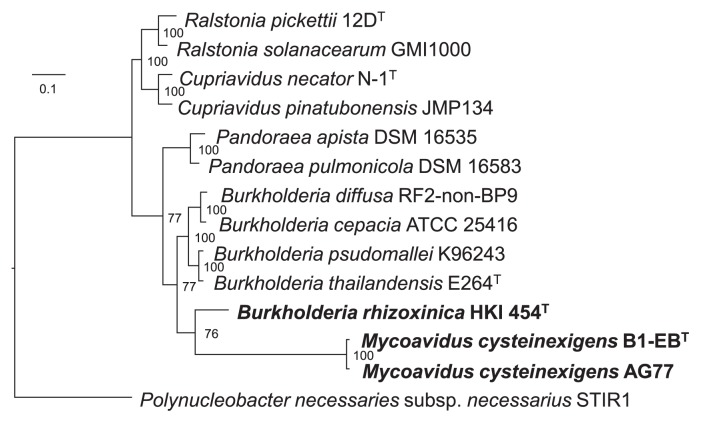
In the present study, we generated a multilocus phylogenetic tree of 14 reference strains (including 4 type strains) of Burkholderiaceae using the atpD, gyrB, lepA, recA, and rpoB genes in combination with the 16S rRNA gene sequence and employed RAxML with support values estimated with 100 bootstrap replicates for the evolution of the genus Mycoavidus as a sister clade to Burkholderia (Fig. 3 and S2). The output of this analysis also showed a distinct clade for B. rhizoxinica, which was positioned within the genus Burkholderia. Based on phylogenomics and the separation of fungal hosts, two alternative scenarios, ‘early’ and ‘late’ bacterial invasion, were hypothesized (5). Regarding Burkholderia-related endobacteria, early bacterial invasion estimated the existence of an ancestral free-living Burkholderia-related endobacteria that invaded the common ancestor of Glomeromycotina and Mortierellomycotina (5, 33). As discussed by Bonfante & Desirò (2017) (5), late bacterial invasion may have occurred when the evolutionary lines leading to Glomeromycotina and Mortierellomycotina had already separated. However, B. rhizoxinica appeared to have an independent origin shared with the free-living Burkholderia-related endobacteria that diverged from the ancestral lineage (34). This hypothetical scenario may lead to further discussions on whether specific bacterial-fungal interactions occurred in the common ancestor only (not separated) and not in separated Glomeromycotina and Mortierellomycotina in which obligate endobacteria essentially lack the ability to grow independently of their fungal hosts.
Fig. 3.
Randomized axelerated maximum likelihood (RAxML) tree based on concatenated sequences (11,023 positions in total) of five housekeeping genes (atpD, gyrB, lepA, recA, and rpoB) indicating the relative placement of the three endofungal bacteria (bold) and other genera in the families Burkholderiaceae and Ralstoniaceae. Pairwise distance between B1-EBT and Mc-AG77 is 0.013. The horizontal lines show genetic distances, which are supported by values estimated with 100 bootstrap replicates. The scale bar indicates the number of substitutions per nucleotide position.
Comparative genomics of EHB belonging to M. cysteinexigens and B. rhizoxinica
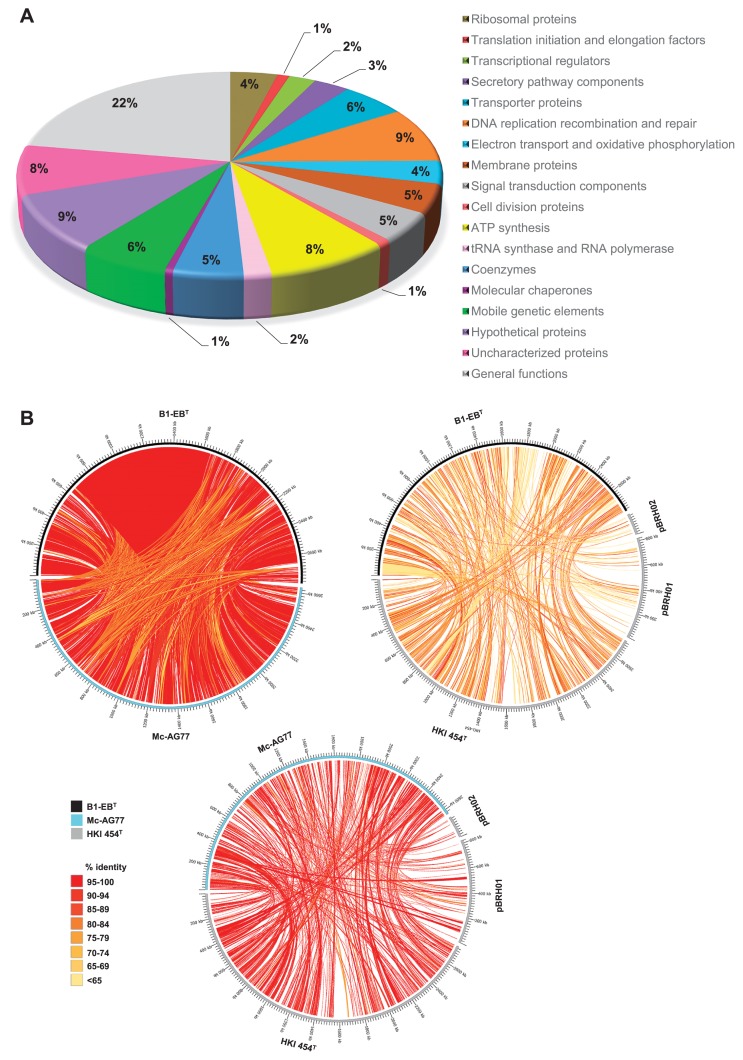
In order to obtain insights into the origin and evolution of gene families characteristic of the phylogenetically associated endohyphal symbionts in diverse fungal hosts, we conducted comparative genome analyses on M. cysteinexigens B1-EBT, Mc-AG77, and B. rhizoxinica HKI 454T by the computation of pairwise homologous gene clusters. We excluded CaGg from this comparative analysis because of its controversial genome sequence, which we clarified earlier in this study. A previous study reported that the total genome of B. rhizoxinica HKI 454T is 3,750,139 bp, consisting of the chromosome and two plasmids, pBRH01 and pBRH02 (33). The entire genome comprises 3,878 CDSs, 2,437 of which were estimated to have biological functions, 1,441 had no known function, 897 appeared to be similar to other database entries, and 544 remained without function and similarity (34). The genome of Mc-AG77 is 2,638,116 bp and contained 2,255 CDSs (68). Both bacterial genomes were markedly smaller than that of B. rhizoxinica HKI 454T and harbored fewer transcriptional regulator genes (Table 1). Pan-genome GET_HOMOLOGUES analyses at the protein level using the BDBH algorithm resulted in the extraction of 1,266 and 1,221 ortholog clusters with shared homology to the HKI 454T genome based on the % identity of amino acid pairs with B1-EBT and Mc-AG77, respectively, whereas 1,788 homologous regions with high % identity were predicted between B1-EBT and Mc-AG77. We herein summarized the distribution (% shared proteomes) of ortholog clusters based on the % identity among the proteomes of Mycoavidus and B. rhizoxinica genomes (Fig. 4A). Pairwise comparative mapping of the B1-EBT, Mc-AG77, and HKI 454T genomes displayed regions of shared homology (Fig. 4B). Although the MLSA of 14 reference strains of Burkholderiales using five housekeeping genes and the 16S rRNA gene revealed that M. cysteinexigens B1-EBT and Mc-AG77 both had a similar distance with B. rhizoxinica HKI 454T (Fig. 3 and S2), pairwise comparative mapping displayed the high homology of Mc-AG77 with B. rhizoxinica HKI 454T. The B1-EBT genome was disrupted by a large amount of mobile genetic elements, with 146 CDSs (6.3%) encoding proteins with similarities to transposases or inactivated derivatives, plasmid stability proteins, reverse transcriptases, and phage proteins, which was similar to HKI 454T (Table S1); however, it is currently unclear how many of these were active insertion elements. We also predicted 271 (11.6%) characterized proteins, 137 hypothetical proteins, and 51 transposase and inactivated derivatives in B1-EBT, which were non-homologous with Mc-AG77 and HKI 454T (Table S2). Since the annotations of Mc-AG77 CDSs are not yet known, we attempted to predict their functions from homology shared with the B1-EBT genome.
Table 1.
Comparative genome analysis of M. cysteinexigens B1-EBT, Mc-AG77, and B. rhizoxinica HKI 454T.
| Organism | Genome size (bp) | G+C content (%) | Coding sequences (CDSs) | Mobile genetic, elements, (% for all CDSs) | Transcriptional, regulators, (% for all CDSs) | References |
|---|---|---|---|---|---|---|
| M. cysteinexigens B1-EBT | 2,795,004 | 48.9 | 2,317 | 146 (6.3) | 39 (1.68) | This research |
| M. cysteinexigens AG77 | 2,638,116 | 49.0 | 2,255 | 71 (3.14)** | 39 (1.72)*** | Uehling et al., 2017 (68) |
| B. rhizoxinica HKI 454T | 3,750,139* | 60.7 | 3,870 | 255 (6.6) | 193 (5.0) | Lackner et al., 2011 (34) |
Includes a 2,755,309-bp chromosome, 822,304-bp mega plasmid, and 172,525-bp plasmid.
Genes coding for mobile genetic elements are predicted from the homologous sequence of the B1-EBT genome.
Genes coding for transcriptional regulators are predicted from the homologous sequence of the B1-EBT genome.
Fig. 4.
Genome comparison of three beta-proteobacterial endohyphal symbionts. (A) Pie chart showing the distribution (% shared proteomes) of ortholog clusters (BDBH) based on % identity among the proteomes of Mycoavidus and B. rhizoxinica genomes. (B) Comparative mapping showing links among homologous sequences. The circular exterior displays the genomes of organisms clockwise. Ribbons between genomes indicate regions of shared homology based on the % identity of amino acid pairs between each pair of organisms. Darker colors correspond to higher identity links and the lighter color corresponds to a lower identity (see legends).
Prophage sequences, horizontal gene transfer, and genome reductions in M. cysteinexigens and B. rhizoxinica
Genomes of obligate intracellular bacteria are often categorized by high levels of genetic drift, genome streamlining, and a lack of mobile DNA (plasmid, transposon, and prophage), which emerge with prominent degrees of stability and gene synteny (8, 36). However, a number of studies have showed that the smaller genomes of obligate intracellular species comprise a level of prophage sequences (e.g., the prophage sequence of Wolbachia pipientis is less than 2.0% of the total genome) similar to the lower end of the range found in facultative intracellular species (8). We herein identified the prophage sequence in the genomes of EHB M. cysteinexigens B1-EBT, Mc-AG77, and B. rhizoxinica HKI 454T by PHASTER. Of note, the chromosomes of B1-EBT, Mc-AG77 comprised a markedly higher level of prophage sequences than those in the HKI 454T chromosome. The prophages and their remnants in B1-EBT and Mc-AG77 accounted for 11.5% (11 regions with 267 phage-related CDSs) and 7.2% (6 regions with 133 phage-related CDSs) of the total lengths of their chromosomes, respectively (Fig. 2A and B), whereas that of HKI 454T was only 2.9% (4 regions with 79 phage-related CDSs, Fig. 2C). A longer association with a host resulted in the endobacterial genomes undergoing a mutational bias toward a lower GC content (12, 40, 43). The GC contents of prophage sequences in B1-EBT and Mc-AG77 chromosomes (prophage GC contents, 49.2% and 48.4%, respectively) were closer to those of their chromosomes (Table 2), whereas an apparently lower GC content (58.6%) was observed for the prophage sequences in the HKI 454T chromosome (chromosome GC content, 61.2%) (Fig. 2C). This result suggests a relatively longer association of prophages with M. cysteinexigens hosts than those with B. rhizoxinica HKI 454T.
Table 2.
Phage-related CDSs in genomes of M. cysteinexigens B1-EBT, Mc-AG77, and B. rhizoxinica HKI 454T.
| Bacterial host of relative phage/prophage in | B1-EBT | Mc-AG77 | HKI 454T | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| CDS count | Percentage | CDS count | Percentage | CDS count | Percentage | |
| Actinobacteria | 2 | 0.75 | 1 | 0.75 | 0 | — |
| Bacteroidetes | 4 | 1.50 | 2 | 1.50 | 0 | — |
| Cyanobacteria | 2 | 0.75 | 4 | 3.01 | 0 | — |
| Firmicutes | 9 | 3.37 | 7 | 5.26 | 4 | 5.06 |
| Alphaproteobacteria | 13 | 4.87 | 3 | 2.26 | 0 | — |
| Betaproteobacteria | 39 | 14.61 | 21 | 15.79 | 57 | 72.15 |
| Gammaproteobacteria | 197 | 73.78 | 95 | 71.43 | 18 | 22.78 |
| Tenericutes (Mollicutes) | 1 | 0.37 | 0 | — | 0 | — |
|
| ||||||
| Total | 267 | 100.00 | 133 | 100.00 | 79 | 100.00 |
As an important means for gene-gain events, prophages horizontally transfer genes from one bacterial genome to another. More than 70% of phage-related CDSs in the genomes of M. cysteinexigens were close to those CDSs of the bacteriophages infecting Gammaproteobacteria such as Aggregatibacter actinomycetemcomitans, Edwardsiella tarda, and Escherichia coli, followed by those of phage-infecting bacteria of Betaproteobacteria, Alphaproteobacteria, Firmicutes, Cyanobacteria, Bacteroidetes, Actinobacteria, and Tenericutes (Mollicutes) (Table 2 and S3). In contrast, most phage-related CDSs in the B. rhizoxinica HKI 454T chromosome (72.2%) were similar to those found in the phages infecting betaproteobacteria, particularly B. cenocepacia, B. cepacia, and B. thailandensis, followed by the CDSs related to the phages infecting bacteria of Gammaproteobacteria (22.8%) and Firmicutes (5.0%) (Table 2 and S3). This phage originated by distinction, suggesting a difference in horizontal gene transformation between the two lineages. However, many researchers have shown that the smaller genomes of obligate intracellular bacteria preferred the inactivation and deletion of prophage sequences over their insertion and duplication (8, 36). An analysis using PHASTER showed that only one prophage region in the B. rhizoxinica HKI 454T chromosome was intact, which was consistent with the deletion of the prophage in the intracellular bacterial genome. Nevertheless, M. cysteinexigens had more intact prophages in their chromosomes (7 in B1-EBT, and 3 in Mc-AG77), indicating that prophages in M. cysteinexigens genomes were undergoing deletion and inactivation processes. In consideration of the relatively longer association of prophages with M. cysteinexigens, the deletion rate of prophages in the genomes of M. cysteinexigens appeared to be slower than those proceeding in the genome of B. rhizoxinica, suggesting that the prophages still possess physiological functions in M. cysteinexigens. However, the lengths of non-prophage regions in the chromosomes of B1-EBT, Mc-AG77, and HKI 454T were 2,469,125 bp, 2,447,528 bp, and 2,674,052 bp, respectively. Therefore, the primordial genetic elements in the M. cysteinexigens genome were presumed to be more reduced than those in the HKI 454T chromosome if the two endohyphal bacteria were derived from a common ancestor bacterium.
Metabolic potential and nutrient uptake in M. cysteinexigens and B. rhizoxinica
There has always been a primary interest in the metabolic potential of EHB, whether they are nutrient producers or consumers and how they exchange nutrients and cofactors with their fungal hosts. We compared the primary metabolism, cofactor biosynthesis, and membrane transport of B1-EBT and Mc-AG77 with HKI 454T (Table 3) and created a model of metabolic processes in M. cysteinexigens (Fig. 5). The deduction of primary metabolism and transporters suggested that M. cysteinexigens (B1-EBT and Mc-AG77) possess genes for glucose/maltose/N-acetylglucosamine- and mannose/fructose-specific phosphotransferase system components, but do not utilize glucose, arabinose, mannose, mannitol, N-acetyl-d-glucosamine, maltose, or gluconate. It was already known that HKI 454T prefers glycerol over glucose as a carbon source (53). HKI 454T may consume organic acids such as citrate, malate, glycerol, and ethanol from Rhizopus (41), whereas no genes encoding ethanol assimilation or organic acid importers were detected in the B1-EBT genome (Table 3 and S1), suggesting that B1-EBT cannot utilize these organic acids from its host.
Table 3.
Comparison of metabolic potentials, nutrient uptake and defense responses in B1-EBT, Mc-AG77, and HKI 454T.
| Attributes | B1-EBT | Mc-AG77 | HKI 454T |
|---|---|---|---|
|
| |||
| Primary metabolism | |||
| Gluconeogenesis* | − | − | + |
| Ethanol assimilation* | − | − | + |
| Import of organic acids | − | − | + |
| Amino acid metabolism | + | + | + |
| Biosynthetic routes for proteinogenic amino acids* | + | + | + |
| Dipeptide and oligopeptide import system* | + | + | + |
|
| |||
| Cofactor biosynthesis | |||
|
| |||
| Biosynthesis of pyridoxine*, heme*, flavin*, biotin, and thiamine* | + | + | + |
|
| |||
| Membrane transport | |||
|
| |||
| Putative cobalamin transport system* | + | + | + |
| Fe2+, Mg2+, Co2+, Zn2+, K+ uptake system | + | + | + |
| Na+/Ca2+ antiporter* | + | + | + |
| Na+/citrate symporter* | + | + | + |
| NRPS/PKS** system for rhizoxin biosynthesis | − | − | + |
| Lantibibiotic biosynthesis* | − | − | + |
| Chitinolytic enzymes and chitin-binding proteins* | − | − | + |
|
| |||
| Amino acid metabolism | |||
|
| |||
| Branched-chain amino acid | + | + | + |
| Aromatic amino acid | + | + | + |
| Histidine | + | + | + |
| Glutamate/aspartate | + | + | + |
| Glycine | + | + | + |
| Citrate | + | + | + |
| Biosynthetic routes for proteinogenic amino acids | + | + | + |
| Efflux systems for arginine, histidine, lysine | + | + | + |
| Efflux systems for cysteine | − | − | + |
|
| |||
| Polar lipids | |||
|
| |||
| Phosphatidylethanolamine* | + | + | − |
| Phosphatidylglycerol* | + | + | − |
| Diphosphatidylglycerol* | + | + | − |
| Aminophospholipid* | + | + | − |
| Aminolipids* | + | + | − |
|
| |||
| Fatty acid synthesis | |||
|
| |||
| Acetyl-CoA carboxylase components | + | + | + |
| Saturated/unsaturated synthase components* | + | + | + |
| Peripheral enzymes (ACP synthase and biotin ligase) * | + | + | + |
| Glycerophospholipid metabolism* | + | + | + |
|
| |||
| Lipopolysaccharide gene cluster | |||
|
| |||
| Lipopolysaccharide | − | − | + |
| O-antigen* | − | − | + |
| Inner membrane ABC transporter, MsbA* | + | + | + |
| Transport complex (LptA, LptAB, LptC, LptD, LptE, LptG) * | − | − | + |
|
| |||
| Exopolysaccharide/capsular polysaccharide biosynthesis | |||
|
| |||
| Antigen flippase and polymerase* | − | − | + |
| Trans-envelope transport complex (Wza/Wzb/Wzc) * | − | − | + |
| Glycosyltransferases gene* | + | + | + |
| Mannosyltransferase gene* | − | − | + |
Genes listed in Mc-AG77 are predicted from the homologous sequence of the B1-EBT genome.
Non-ribosomal peptide synthetase/polyketide synthase
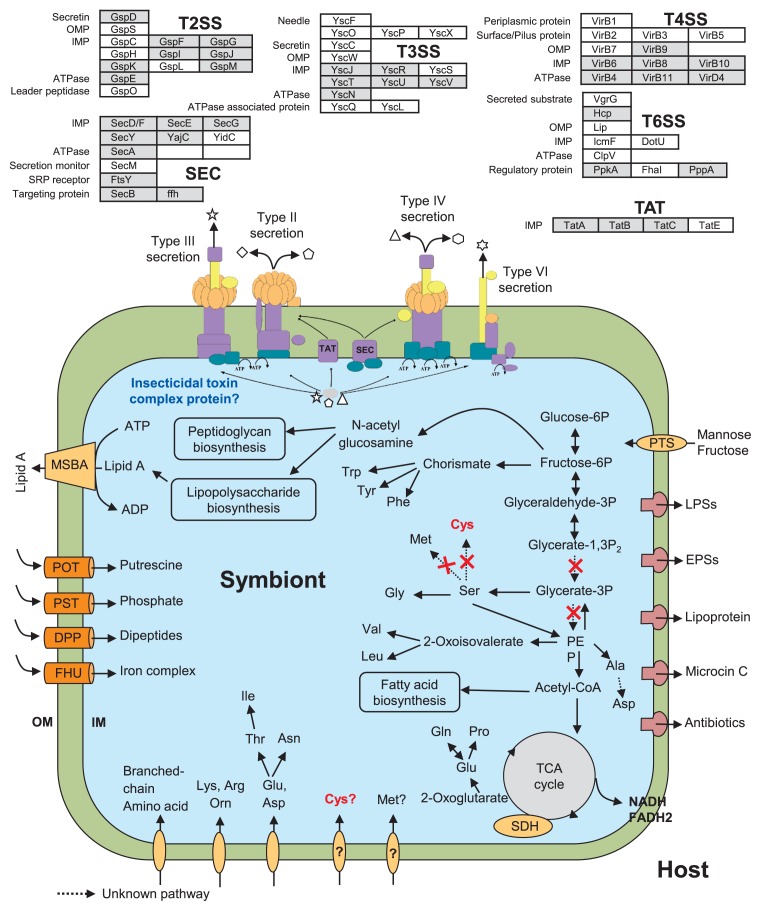
Fig. 5.
Model representing metabolic processes and the secretion system assumed from the B1-EBT genome. Putative type II, III, IV, and VI secretory proteins are highlighted in grey. POT, polyamine transport system; PST, phosphate-specific transport; DPP, dipeptide permease; FHU, ferrichrome-uptake; EPSs, exopolysaccharides; LPSs, lipopolysaccharides; IM, inner membrane; OM, outer membrane; OMP, outer membrane protein; IMP, inner membrane protein; SEC, general secretion pathway; TAT, twin arginine translocation pathway.
M. cysteinexigens and B. rhizoxinica both harbor genes responsible for importing branched-chain and aromatic amino acids, histidine, acidic amino acids, and glycine; the biosynthetic routes for all proteinogenic amino acids and efflux systems for basic amino acids (Table S4) (68). Additionally, both endosymbionts had genes for dipeptide and oligopeptide import systems. However, B1-EBT and Mc-AG77 do not possess efflux systems for cysteine. B1-EBT possesses genes encoding biosynthesis enzymes for essential cofactors such as pyridoxine, heme, flavin, biotin, and thiamine, which were similar to those in HKI 454T. HKI 454T may provide some amino acids and cofactors to its host (34), which suggests that amino acid metabolism and transportation systems adapted during endobacterium-host symbiosis in B1-EBT and HKI 454T.
Secondary metabolites, lipopolysaccharides, exopolysaccharides, and efflux pumps in M. cysteinexigens and B. rhizoxinica
The genomic locus for antimitotic rhizoxin biosynthesis in HKI 454T revealed a hybrid system of non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) (52). The NRPS/PKS system architecture in HKI 454T possessed a phosphopantetheinyl transferase to activate the acyl and peptidyl carrier protein domains, 14 NRPS multimodular enzymes, and macrolide-specific ABC-transporters for toxin delivery to the host fungus (34). Since no peptides corresponding to these gene clusters were isolated from HKI 454T, it was hypothesized that some NRPS products may function as siderophores or antibiotics (34). M. cysteinexigens B1-EBT contained a NRPS module (MCB1EB_1072), which was an ortholog of the adenylation domain in the first NRPS module of bacillibactin present in pBRH01 (RBRH_00588) in B. rhizoxinica HKI 454T. On the other hand, this NRPS module in B1-EBT shared strong identity (97%) with Glomeribacter sp. 1016415. However, its function and biosynthetic potential remain unclear. Virulence factors help bacteria to invade their hosts, which were completely functional in EHB HKI 454T. Therefore, the mechanisms by which B1-EBT invaded its host fungus without a complete virulence system have not yet been elucidated. Although bacterial intrusion often depends on genetic characteristics, the invasion mechanism strictly differed with the environment encountered by the bacterium (56) and with different life history policies such as avirulent microorganisms, latent pathogens, and virulent pathogens in the early stages of infection or at a certain phase of their development (28). A single mutation was shown to result in the loss of the virulence factor, transforming the pathogenic fungus, Colletotrichum magna, into an endophyte (19). Therefore, virulence-related genes may be lost during bacterial genome reduction in B1-EBT. We speculated how M. cysteinexigens may invade host fungal tissues, such as when fungal hyphae are damaged by other fungi or by grazing soil fauna. Subsequent invasion occurred through breaks in the fungal cell wall. In contrast, B. rhizoxinica HKI 454T invades the host, even under laboratory conditions (34).
Lipopolysaccharides (LPSs) and exopolysaccharides (EPSs) play key roles in the recognition and stability of the symbiosis of B. rhizoxinica HKI 454T within its host and a gene cluster involved in LPS biosynthesis in HKI 454T (37). Besides LPS biosynthesis, genome sequencing revealed genes that enabled HKI 454T to also produce EPSs (34). The biosynthesis of the homopolymeric O-antigen LPS depends on an ABC transporter (70). B1-EBT and Mc-AG77 genomes identified an inner membrane ABC transporter, MsbA, similar to the macrolidespecific ABC transporter in HKI 454T and a glycosyltransferase gene (Table S1), which indicates that only these two components of the carbohydrate-derived putative LPS/EPS system of the recognition process are present in M. cysteinexigens.
Secretion systems and insecticidal toxin complexes in M. cysteinexigens and B. rhizoxinica
Secretory proteins pass through the cell membrane in bacteria, either via the general secretion pathway (4, 15) or twin arginine translocation pathway (50), and regulate host control mechanisms. Therefore, in the present study, we analyzed the genes coding for putative protein secretion systems. The Mc-AG77 genome acquired genes for type II (T2SS), III (T3SS), and IV (T4SS) secretion systems, and these contributed to the exchange of proteins and DNA with its host (68). Additionally, the B1-EBT genome possesses a few components of a type VI secretion system (T6SS).
T2SS is known to be involved in the secretion of toxins and lytic proteins (e.g., chitinolytic enzymes, chitin-binding proteins) outside the cell in various bacteria (10, 18). In addition, microbial chitinases are presumed to degrade hyphal walls and allow bacteria to invade fungi. The betaproteobacterium Nitrosomonas europea appears to have significant, but incomplete T2SS core components (10). B. rhizoxinica HKI 454T possessed a complete T2SS, whereas B1-EBT possessed a partial T2SS constituent, which is homologous with that in HKI 454T (Table S5). It currently remains unclear whether T2SS homologs in diverse bacteria are expressed and functional. B. rhizoxinica and other free-living, pathogenic Burkholderia species (e.g. B. thailandensis and B. pseudomallei) contain hrp T3SS gene clusters and these are known to play a significant role in symbiosis through the translocation of effector proteins with their hosts (32, 64). B1-EBT possessed a putative T3SS constituent with partial homology with the HKI 454T T3SS pathway (Table S6). In contrast, the T3SS components of Mc-AG77 shared strong homology with Gammaproteobacteria such as Salmonella and Yersinia (68).
The T4SS system was shown to be involved in the plant pathogenicity of B. cenocepacia (16). A partial T4SS was found in the B1-EBT genome and in the plasmid pBRH02 of HKI 454T (Table S1). In the B1-EBT genome, we recognized a hypothetical protein homolog type 4 major prepilin protein, pilus assembly pathway ATPase PulE/Tfp homolog type 4 pilus biogenesis protein, a prepilin signal peptidase, and ATPase pilus assembly distributed throughout the chromosome in HKI 454T (Table S1). Adhesive structures, particularly type 4 bacterial pili, are generally major adhesion factors for host surfaces in Gram-negative bacteria (27). However, the pilus structure in B1-EBT lacked several key genes e.g., secretin, motor protein, and other accessory proteins. Moreover, hemolysin gene products are known to be involved in plant pathogenicity (24), and the hemagglutinin-like proteins, putative hemolysin/phospholipase/lecithinase, were also found in the B1-EBT genome and had sequence similarities with RBRH_02330, RBRH_00133, and RBRH_01303 of HKI 454T. Additionally, a gene cluster coding for an insecticidal toxin protein (coding genes: CSV86_11535, tccC3, PCH70_41490, and tcaA2) was found in the homologs of Xenorhabdus bovienii SS-2004 and Photorhabdus asymbiotica (45, 55). The insecticidal toxin proteins present in the HKI 454T genome were completely different from those present in the B1-EBT genome.
Conclusions
Compared with animals and plants, the huge genetic diversity of the bacterial biosphere makes it an outstanding resource for studying genetics and evolution. The bacteria that dwell inside eukaryotes contain some of the smallest, most stable, highly repetitive, and recombined bacterial genomes sequences currently known (40, 48). Although bacterial-fungal symbiosis has important implications for agriculture, biotechnology, and food safety, this interaction has long been overlooked (29, 65). The genome of B1-EBT offers an extraordinary picture of an obligate symbiont that has most likely lost its capacity to grow outside of its fungal host (i.e., cannot grow in conventional nutrient media) (12, 21, 49). In the present study, we found that the B1-EBT and Mc-AG77 genomes exhibited amino acid similarities between ~20–84% and ~18–100%, respectively, with HKI 454T (Table S1). Obligate endosymbiotic bacteria were anticipated to have lost many essential genes as a result of genome reduction during evolution (51). In contrast to other fungal endosymbionts in the family Burkholderiaceae, both strains of M. cysteinexigens lack many genes associated with host invasion such as chitinolytic enzymes (chitinase, chitosanase), chitin-binding proteins, and several T2SS components, combined with sugar importers and the ATP/ADP antiporter, during bacterial genome reduction, suggesting later bacterial invasion and adaptation to its host metabolism.
Supplementary Material
Acknowledgements
We thank Drs. Reiko Fujimura and Kenshiro Oshima from The University of Tokyo for their help with the data analysis. We are grateful to Hikaru Nabatame and Nanami Konuma for their technical assistance. This research was supported by the Institute for Fermentation, Osaka, JSPS KAKENHI Grant Number 17K07695, and the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO).
References
- 1.Arcus V.L., Rainey P.B., Turner S.J. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005;13:360–365. doi: 10.1016/j.tim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Arendt K.R., Hockett K.L., Araldi-Brondolo S.J., Baltrus D.A., Arnold A.E. Isolation of endohyphal bacteria from foliar Ascomycota and in vitro establishment of their symbiotic associations. Appl Environ Microbiol. 2016;82:2943–2949. doi: 10.1128/AEM.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks B.C., Palmer T., Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Bonfante P., Desirò A. Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. ISME J. 2017;11:1727–1735. doi: 10.1038/ismej.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonfante-Fasolo P., Scannerini S. The ultrastructure of the zygospore in Endogone flammicorona Trappe and Gerdemann. Mycopathologia. 1976;59:117–123. [Google Scholar]
- 7.Bonfante-Fasolo P., Scannerini S. Cytological observations on the mycorrhiza Endogone flammicorona-Pinus strobus. Allionia. 1977;22:23–34. [Google Scholar]
- 8.Bordenstein S.R., Reznikoff W.S. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- 9.Chin C.-S., Alexander D.H., Marks P., et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 10.Cianciotto N.P. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Contreras-Moreira B., Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale C., Moran N.A. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Desirò A., Salvioli A., Ngonkeu E.L., et al. Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J. 2014;8:257–270. doi: 10.1038/ismej.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desirò A., Faccio A., Kaech A., Bidartondo M.I., Bonfante P. Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytol. 2015;205:1464–1472. doi: 10.1111/nph.13136. [DOI] [PubMed] [Google Scholar]
- 15.Driessen A.J.M., Manting E.H., van der Does C. The structural basis of protein targeting and translocation in bacteria. Nat Struct Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
- 16.Engledow A.S., Medrano E.G., Lipuma J.J., Gonzalez C.F., Mahenthiralingam E. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J Bacteriol. 2004;186:6015–6024. doi: 10.1128/JB.186.18.6015-6024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrada-De Los Santos P., Vinuesa P., Martínez-Aguilar L., Hirsch A.M., Caballero-Mellado J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol. 2013;67:51–60. doi: 10.1007/s00284-013-0330-9. [DOI] [PubMed] [Google Scholar]
- 18.Francetic O., Belin D., Badaut C., Pugsley A.P. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 2000;19:6697–6703. doi: 10.1093/emboj/19.24.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman S., Rodriguez R.J. Genetic conversion of a fungal plant pathogen to a non-pathogenic, endophytic mutualist. Science. 1993;260:75–78. doi: 10.1126/science.260.5104.75. [DOI] [PubMed] [Google Scholar]
- 20.Frey-Klett P., Burlinson P., Deveau A., Barret M., Tarkka M., Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimura R., Nishimura A., Ohshima S., Sato Y., Nishizawa T., Oshima K., Hattori M., Narisawa K., Ohta H. Draft genome sequence of the betaproteobacterial endosymbiont associated with the fungus Mortierella elongata FMR23-6. Genome Announc. 2014;2:e01272–14. doi: 10.1128/genomeA.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghignone S., Salvioli A., Anca I., et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 2012;6:136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon D., Desmarais C., Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilhabert M.R., Kirkpatrick B.C. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute a biofilm maturation to X. fastidios and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005;18:857–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman M.T., Arnold A.E. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol. 2010;76:4063–4075. doi: 10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Kline K.A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi D.Y., Palumbo J.D. Bacterial endophytes and their effects on plants and uses in agriculture. In: Bacon C.W., White J.F., editors. Microbial Endophytes. Marcel Dekker; New York: 2000. pp. 199–236. [Google Scholar]
- 29.Kobayashi D.Y., Crouch J.A. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu Rev Phytopathol. 2009;47:63–82. doi: 10.1146/annurev-phyto-080508-081729. [DOI] [PubMed] [Google Scholar]
- 30.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lackner G., Hertweck C. Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog. 2011;7:5–8. doi: 10.1371/journal.ppat.1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lackner G., Moebius N., Hertweck C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2011;5:252–261. doi: 10.1038/ismej.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lackner G., Moebius N., Partida-Martinez L., Hertweck C. Complete genome sequence of Burkholderia rhizoxinica, an endosymbiont of Rhizopus microsporus. J Bacteriol. 2011;193:783–784. doi: 10.1128/JB.01318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lackner G., Moebius N., Partida-Martinez L.P., Boland S., Hertweck C. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence J.G., Hendrix R.W., Casjens S. Where are the pseudogenes in bacterial. Trends Microbiol. 2001;9:535–540. doi: 10.1016/s0966-842x(01)02198-9. [DOI] [PubMed] [Google Scholar]
- 37.Leone M.R., Lackner G., Silipo A., Lanzetta R., Molinaro A., Hertweck C. An unusual galactofuranose lipopolysaccharide that ensures the intracellular survival of toxin-producing bacteria in their fungal host. Angew Chem, Int Ed. 2010;49:7476–7480. doi: 10.1002/anie.201003301. [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Yao Q., Dearth S.P., et al. Integrated proteomics and metabolomics suggests symbiotic metabolism and multimodal regulation in a fungal-endobacterial system. Environ Microbiol. 2017;19:1041–1053. doi: 10.1111/1462-2920.13605. [DOI] [PubMed] [Google Scholar]
- 39.Lowe T.M., Eddy S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1996;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCutcheon J.P., McDonald B.R., Moran N.A. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millati R., Edebo L., Taherzadeh M.J. Performance of Rhizopus, Rhizomucor, and Mucor in ethanol production from glucose, xylose, and wood hydrolyzates. Enzyme Microb Technol. 2005;36:294–300. [Google Scholar]
- 42.Mondo S.J., Toomer K.H., Morton J.B., Lekberg Y., Pawlowska T.E. Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution. 2012;66:2564–2576. doi: 10.1111/j.1558-5646.2012.01611.x. [DOI] [PubMed] [Google Scholar]
- 43.Moran N.A., McCutcheon J.P., Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 44.Mosse B. Honey-coloured, sessile Endogone spores. Arch Microbiol. 1970;70:167–175. [Google Scholar]
- 45.Mulley G., Beeton M.L., Wilkinson P., et al. From insect to man: photorhabdus sheds light on the emergence of human pathogenicity. PLoS One. 2015;10:e0144937. doi: 10.1371/journal.pone.0144937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naito M., Pawlowska T.E. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mobile Genet Elem. 2015;6:e1136375. doi: 10.1080/2159256X.2015.1136375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naito M., Pawlowska T.E. Defying muller’s ratchet: ancient heritable endobacteria escape extinction through retention of recombination and genome plasticity. mBio. 2016;7:1–8. doi: 10.1128/mBio.02057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M. The 169-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2008;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 49.Ohshima S., Sato Y., Fujimura R., Takashima Y., Hamada M., Nishizawa T., Narisawa K., Ohta H. Mycoavidus cysteinexigens gen. nov., sp. nov., an endohyphal bacterium isolated from a soil isolate of the fungus Mortierella elongata. Int J Syst Evol Microbiol. 2016;66:2052–2057. doi: 10.1099/ijsem.0.000990. [DOI] [PubMed] [Google Scholar]
- 50.Osborne A.R., Rapoport T.A., van den Berg B. Protein translocation by the Sec61/Secy channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 51.Pandey D.P., Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Partida-Martinez L.P., de Looß C.F., Ishida K., Ishida M., Roth M., Buder K., Hertweck C. Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl Environ Microbiol. 2007;73:793–797. doi: 10.1128/AEM.01784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Partida-Martinez L.P., Groth I., Schmitt I., Richter W., Roth M., Hertweck C. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporous. Int J Syst Evol Microbiol. 2007;57:2583–2590. doi: 10.1099/ijs.0.64660-0. [DOI] [PubMed] [Google Scholar]
- 54.Partida-Martinez L.P., Hertweck C.A. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chem Bio Chem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 55.Proschak A., Zhou Q., Schöner T., Thanwisai A., Kresovic D., Dowling A., Proschak E., Bode H.B. Biosynthesis of the insecticidal xenocyloins in Xenorhabdus bovienii. Chem Bio Chem. 2014;15:369–372. doi: 10.1002/cbic.201300694. [DOI] [PubMed] [Google Scholar]
- 56.Ribet D., Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Herrera J., León-Ramírez C., Vera-Nuñez A., Sánchez-Arreguín A., Ruiz-Medrano R., Salgado-Lugo H., Sánchez-Segura L., José Peña-Cabriales J. A novel intracellular nitrogen-fixing symbiosis made by Ustilago maydis and Bacillus spp. New Phytol. 2015;207:769–777. doi: 10.1111/nph.13359. [DOI] [PubMed] [Google Scholar]
- 58.Sato Y., Narisawa K., Tsuruta K., Umezu M., Nishizawa T., Tanaka K., Yamaguchi K., Komatsuzaki M., Ohta H. Detection of betaproteobacteria inside the mycelium of the fungus Mortierella elongata. Microbes Environ. 2010;25:321–324. doi: 10.1264/jsme2.me10134. [DOI] [PubMed] [Google Scholar]
- 59.Sato Z., Noda T., Matsuda I., Iwasaki S., Kobayashi H., Furukawa J., Okuda S. Studies on rhizoxin, a phytotoxin produced by Rhizopus chinensis causing rice seedling blight. Ann Phytopathol Soc Jpn. 1983;49:128. [Google Scholar]
- 60.Scannerini S., Bonfante-Fasolo P. Bacteria and bacterialike objects in endomycorrhizal fungi (Glomeceae) In: Margulis L., Fester R., editors. Symbiosis as source of evolutionary innovation: speciation and morphogenesis. The MIT Press; London: 1991. pp. 273–287. [PubMed] [Google Scholar]
- 61.Sharma M., Schmid M., Rothballer M., et al. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol. 2008;10:2235–2246. doi: 10.1111/j.1462-5822.2008.01202.x. [DOI] [PubMed] [Google Scholar]
- 62.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 63.Spatafora J.W., Chang Y., Benny G.L., et al. A phylum-level phylogenetic classification of zygomycete fungi based on genomescale data. Mycologia. 2016;108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens M.P., Haque A., Atkins T., et al. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology. 2004;150:2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 65.Tarkka M.T., Sarniguet A., Frey-Klett P. Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr Genet. 2009;55:233–243. doi: 10.1007/s00294-009-0241-2. [DOI] [PubMed] [Google Scholar]
- 66.Toomer K.H., Chen X., Naito M., Mondo S.J., Bakker H.C., VanKuren N.W., Lekberg Y., Morton J.B., Pawlowska T.E. Molecular evolution patterns reveal life history features of mycoplasma-related endobacteria associated with arbuscular mycorrhizal fungi. Mol Ecol. 2015;24:3485–3500. doi: 10.1111/mec.13250. [DOI] [PubMed] [Google Scholar]
- 67.Tsuruo T., Oh-hara T., lida H., et al. Rhizoxin, a macrocylic lactone antibiotic, as a new antitumor agent against huamn and murine tumor cells and their vincristine-resistant sublines. Cancer Res. 1986;46:381–385. [PubMed] [Google Scholar]
- 68.Uehling J., Gryganskyi A., Hameed K., et al. Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ Microbiol. 2017;19:2964–2983. doi: 10.1111/1462-2920.13669. [DOI] [PubMed] [Google Scholar]
- 69.Wang G.C.Y., Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:347–352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.