Abstract
Proteomics based biological research is greatly expanded by high quality mass spectrometry studies, which are themselves enabled by access to quality mass spectrometry resources, such as high-quality curated proteome data repositories. We present a PeptideAtlas for the domestic chicken, containing an extensive and robust collection of chicken tissue and plasma samples with substantial value for the chicken proteomics community for protein validation and design of downstream targeted proteome quantitation. The Chicken PeptideAtlas contains 6,646 canonical proteins at a protein FDR of 1.3%, derived from ~100,000 peptides at a peptide level FDR of 0.1%. The rich collection of readily accessible data is easily mined for the purposes of data validation and experimental planning, particularly in the realm of developing proteome quantitation workflows. Herein we demonstrate the use of the atlas to mine information on common chicken acute phase proteins and biomarkers for cancer detection research, as well as their localization and polymorphisms. This wealth of information will support future proteome-based research using this highly important agricultural organism in pursuit of both chicken and human health outcomes.
Keywords: Biomarkers, Chicken, Ovarian Cancer, PeptideAtlas
Graphical Abstract
INTRODUCTION
The domestic laying hen (Gallus gallus) is the most abundant avian species on the planet1 and a cornerstone species in food production and classical embryological study.2 This importance was underscored by its selection as the first classical “production” organism to be fully genome sequenced, in 2004.3 With the establishment of functional genomic tools for the chicken, it has officially entered the post-genomic era dominated by proteomic studies.4
Mass spectrometry (MS) based proteomics has reached widespread acceptance as a powerful tool to study organisms and protein products through multiple stages of disease, development, and degradation.5 While the experimental techniques are well developed and robust, the application of proteomic methods to non-classical model organisms remains nascent. This is particularly true for organisms of predominantly agricultural interest, such as chickens, whose investigators frequently lack access to advanced MS resources available to human health researchers.6 The result is that only ~10% of the chicken genome is well annotated7,8 in spite of the enormous potential power of the chicken as a biological model system and its substantive importance as an agricultural product. The chicken was a classic model organism for embryological development in the pre-genomic era and the development of genetic tools have returned it to the fore.2,9 Further, the hen ovary exhibits many similar genetic and physiological traits to humans and is the only non-human animal known to spontaneously develop ovarian cancer.10,11
Clearly there is enormous medical and economic potential in proteomics studies involving Gallus gallus that is limited by access to MS resources and the incomplete nature of chicken proteomic/genomic databases. Successful mass spectrometry experiments are greatly enabled by access to publicly available databases of validated data, such as those found in PeptideAtlas12 or the ProteomeXchange13 consortium database, PRIDE;14,15 however, these databases are currently devoid of substantial information regarding the domestic hen. The completion of a reference database for chicken proteomics would substantially advance experimental planning and data validation for global and targeted analysis in this model system.
PeptideAtlas (http://www.peptideatlas.org) curates and compiles mass spectrometry data derived from a variety of experiments through the reprocessing of available MS data using the Trans-Proteomic Pipeline (TPP), a freely available open source suite of tools for tandem MS experiments.16–18 The resulting proteome builds contain high stringency (<1.5% false discovery rate (FDR)) protein-level identifications and peptide observations from numerous experiments in a readily accessible interface, one that is particularly well suited to the evaluation of candidate peptides for targeted proteomics via SRM or data-independent approaches such as DIA.19 More recently, the TPP has also incorporated the iterative reSpect algorithm to assess spectral chimeras and identify peptides that would otherwise be missed.20 PeptideAtlas contains numerous well studied proteomes and model organisms such as Saccharomyces cerevisiae with > 70% proteome coverage,21 and includes classical agriculturally important organisms, such as the pig (Sus scrofa domesticus)22 and the co (Bos Taurus)23.
Here we present an initial build of a chicken PeptideAtlas, containing a variety of structurally complex ovarian tissue types, as well as diagnostically relevant plasma samples. Currently, the chicken PeptideAtlas is directed toward the hen as an ovarian cancer model organism, but data from liver tissue and plasma are included as a useful baseline, and additional tissue types will be incorporated into later builds. We demonstrate the current utility of the atlas through the mining of reported ovarian cancer biomarkers, as well as general inflammatory response proteins, and the selection of candidate peptides for quantitative SRM analysis.
EXPERIMENTAL PROCEDURES
Sample Collection and Processing
The biological samples included in the chicken PeptideAtlas were collected from two cohorts of Bovan’s white leghorn commercial laying hens in the Poultry Science department at North Carolina State University. Birds were managed in accordance with the Institute for Laboratory Animal Research Guide with all of the husbandry practices being approved by and under the oversight of North Carolina State University Institutional Animal Care and Use Committee (IACUC). Plasma and tissue from the initial cohort of B-strain hens were obtained in a previous study,24 with samples collected from a second cohort of 300 C-strain hens similarly. In brief, ~2mL of blood was collected from each bird every four months beginning at an age of 26 weeks. The collected blood was centrifuged at 3000 × g at 4 °C and the plasma was collected in cryogenic tubes for long term storage at −80 °C. At an age of 136 weeks the birds were sacrificed by cervical dislocation and dissected to ascertain gross internal pathology. Tissue was collected from all birds presenting neoplastic lesions on the ovary, oviduct, duodenum, pancreas, GI tract, and/or liver as well as from a selected number of “healthy” birds with no visible distress. Tissues were preserved for proteomic study by snap freezing in liquid nitrogen and stored at −80 °C.
Whole tissue samples were lysed using a 1:5 ratio of tissue to buffer containing 50 mM Tris pH = 7.8, 8 M urea, 2 M thiourea, 10 mM EDTA, 10 mM DTT, and 0.001 % sodium azide and homogenized using an OMNI TIP Homogenizing kit, as described previously.25 A subset of liver and ovary tissues were instead subjected to laser microdissection to isolate yolk, ovarian stroma, and follicular wall tissue, which were subsequently lysed in a buffer of 100 mM Tris-HCl, pH 7.5, 100 mM dithiothreitol (DTT), and 4% sodium dodecyl sulfate.26 Plasma samples were used as collected.
Cell lysates and plasma were processed for LC-MS/MS analyses using either a 1D SDS-PAGE in-gel digestion protocol24,25 or filter aided sample preparation (FASP)27 as described previously26. A subset of the FASP prepared samples were de-glycosylated on-filter using peptide-N-glycosidase F (PNGase F) according to a combined glycomics and proteomics protocol28.
LC-MS/MS Analyses
Peptide samples were analyzed by LC-MS/MS using one of three sets of conditions:
Separation was performed on a nanoLC-2D (Eksigent Technologies) coupled to an LTQ-FT-Ultra with a 7T FT-ICR Mass Analyzer (Thermo Scientific) in a vented column configuration29. The column consisted of a 75 μm × 5 cm Integrafrit trap and 75 μm × 15cm Picofrit column (New Objective) self-packed with 5μm Magic C18AQ stationary phase. Mobile phases A and B consisted of 98/2/0.2 and 2/98/0.2 water/ACN/formic acid, and the separation was performed as a 60 minute gradient from 2–60% B at a flow rate of 350 nL/min. Data was collected in Top-8 data-dependent mode over a mass range of 400 – 1600 m/z with a full scan resolving power of 100,000 FWHM @ 400 m/z using IT-CID and an AGC target of 1e6. Dynamic exclusion windows of 3 minutes were applied to each MS/MS precursor mass.
Separation was conducted with an EASY nLC II (Thermo Scientific) was coupled to a Q Exactive benchtop mass spectrometer (Thermo Scientific). The trap and column were arranged in the vented column configuration as in (1), and used a mobile phase gradient of 2%–5% B (2 min), 5%–40% B (200 min) at a flow rate of 300 nL/min. Parameters for the mass spectrometer were set according to optimums derived for proteomics experiments on the Q Exactive.30 Briefly, data was collected in Top-12 data-dependent MS/MS mode using HCD with a full scan mass range of 400 – 1600 m/z, a full scan resolving power of 70,000 and MS/MS resolving power of 17,500 FWHM @ 200 m/z, and a dynamic exclusion window of 30 s. AGC targets were set at 1e6 and 2e5 for full scan and MS/MS scan respectively, with complementary maximum injections times of 30ms and 250ms.
Chromatographic separation was carried out with an EASY nLC-1000 (Thermo Scientific) coupled to a Q Exactive HF (Thermo Scientific). A 75 μm × 5cm trap was prepared with a frit synthesized as described by Meiring et al.31 and coupled to a 75 μm × 30 cm Picofrit emitter (New Objective). Both were self-packed with 2.6 mm, 100 Å particle size C18 stationary phase (Phenomenex) and assembled in the same column setup as in (1) and (2). Peptides were separated by a gradient of 2 – 20% B (100 min), 20 – 32% B (30 min) at 300 nL/min. MS parameters were independently optimized for proteomics experiments on the Q Exactive HF and are reported in Hecht et al.28 The data was collected in Top 20 data-dependent MS/MS mode using HCD with a 15 s dynamic exclusion window, a full scan mass range of 375–1500 m/z and resolving power of 120,000 and 15,000 FWHM @ 200 m/z for the MS1 and MS2 scans respectively. The full scan AGC and injection were set to 1e6 and 30ms for the MS1 scan and 1e5 and 30ms for the MS2 scans.
Data Processing and PeptideAtlas Assembly
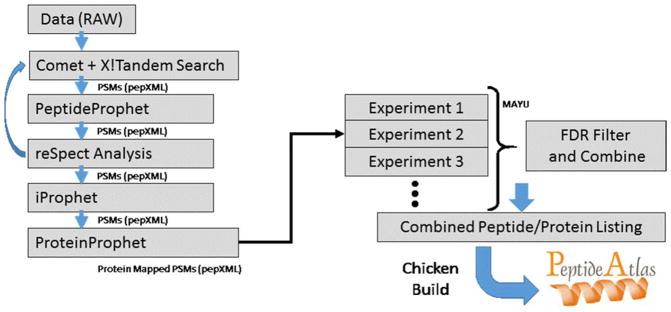
The processing of collected MS data followed the general workflow of Farrah et al.32 with the Trans-Proteomic Pipeline18 as the main component (Figure 1).
Figure 1.
Schematic workflow of the PeptideAtlas build process.
First, vendor .RAW files were converted to the mzML format33 using the ProteoWizard34 msConvert tool version 3.0.794. Next, the mzML files were searched using both Comet35 and X!Tandem36 with the hrk-score plugin. The chicken search database was compiled as a nonredundant union of 24,231 chicken sequences from UniProtKB/TrEMBL, 16,354 sequences from Ensembl release 82,37 and the cRAP database. For Q Exactive files, parent mass errors were set to ± 20 ppm with product ion mass tolerances of ± 20 ppm for X!Tandem and ± 0.03 Da for Comet. LTQ-FT data used a parent mass error of ± 50 ppm and product mass tolerance of ± 0.4 Da for both X!Tandem and Comet. Searching allowed semi-tryptic peptides and a maximum of two missed cleavages. A static modification of carbamidomethyl on C, and a variable modification for oxidation on M, acetylation of protein N-termini, and pyro-glu on Q, E, and C were also used. For samples processed with deglycosylation, a variable modification of deamidation on N was also searched.
The search results were processed with the TPP 18,38 version 4.8.1 tools. The pepXML17 output for Comet was used directly, and the TPP tool Tandem2XML was used to convert the native X!Tandem output to pepXML. For each experiment, the set of pepXML files from the two search outputs were processed together with PeptideProphet.39 The two output files were then combined via iProphet.40
The iProphet output was further processed using the reSpect algorithm20 to access spectral chimeras. During reSpect searching the precursor mass tolerance was increased to match the MS isolation window. For LTQ-FT data the parent mass tolerance was increased to 2.0 Da and likewise increased to 1.4 Da for Q Exactive data. The new set of mzML files generated by reSpect were searched and processed using the TPP as for the initial files. For Q Exactive data, this process was repeated an additional time, for two complete rounds of reSpect in total. The spectra used for reSpect were filtered for a minimum iProphet probability ≥ 0.0 during the initial round, and ≥ 0.5 for the second round of analysis.
Using the PeptideAtlas processing pipeline, all the iProphet results from standard and reSpect searches were filtered at a variable probability threshold to maintain a constant FDR threshold for each experiment. The filtered data was assessed with the MAYU software41 to calculate decoy-based FDRs at the peptide-spectrum match (PSM), distinct peptide, and protein levels. All results were collated in the Chicken PeptideAtlas, available at http://www.peptideatlas.org.
RESULTS & DISCUSSION
Overview of the PeptideAtlas
The chicken PeptideAtlas represents the most extensive collection of publicly accessible proteomic data for Gallus gallus, an important economic organism and resource tool. The database incorporates data from individual birds in various cancerous disease states, and fully samples the reproductive tissue, including differentiated portions of the ovary (i.e. small white follicles, ovarian stroma, etc.), which are nearly unexplored (Table 1).26 The extensive sampling is enhanced by high resolution MS and MS/MS IT-CID and HCD data collected from both LTQ-FT-ICR and Orbitrap instruments, providing the highest quality mass spectra in order to support further study of reproduction and cancer biology in hen based systems.
Table 1.
Summary of sample types included in the chicken PeptideAtlas, including disease status of epithelial ovarian cancer (EOC) and/or oviductal cancer (OVD). A complete list of included files can be found in the Table S-1.
| ID | Tissue | Disease State | Instrument Protocol | # Spectra ID’d | # Distinct | # Proteins |
|---|---|---|---|---|---|---|
| 6757 | Liver | Healthy | 2 | 659875 | 34645 | 3616 |
| 6773 | Ovary | Healthy | 1 | 163185 | 15048 | 2725 |
| 6758 | Ovary | Late State EOC | 1 | 170087 | 17160 | 3149 |
| 6762 | Ovary | Early Stage EOC | 1 | 200708 | 18906 | 3446 |
| 6754 | Ovary | Late State EOC-OVD | 1 | 1234 | 671 | 317 |
| 6772 | Oviduct | Healthy | 1 | 55924 | 8888 | 1232 |
| 6761 | Oviduct | Early Stage EOC | 1 | 64427 | 10978 | 1665 |
| 6765 | Oviduct | Late Stage EOC | 1 | 43347 | 10418 | 1633 |
| 6764 | Oviduct | Late Stage EOC-OVD | 1 | 24018 | 8716 | 1566 |
| 6774 | Ovarian Granulosa | Healthy | 2 | 69787 | 16457 | 2779 |
| 6770 | Ovarian Stroma | Healthy | 2 | 77159 | 19709 | 2915 |
| 6756 | Small White Follicle | Healthy | 2 | 4938 | 2329 | 576 |
| 6771 | Ovary | Healthy | 2 | 91301 | 19561 | 2928 |
| 6760 | Ovarian Follicle Yolk | Healthy | 2 | 56365 | 13006 | 2178 |
| 6766 | Plasma | Early Stage EOC | 1 | 161840 | 3594 | 228 |
| 6767 | Plasma | Late Stage EOC | 1 | 111422 | 3849 | 245 |
| 6753 | Plasma | Late Stage EOC-OVD | 1 | 57269 | 3150 | 206 |
| 6763 | Plasma | Healthy | 1 | 188913 | 4277 | 239 |
| 6755 | Plasma | Healthy | 3 | 133106 | 6751 | 226 |
| 6768 | Plasma | Late Stage EOC | 3 | 144047 | 7121 | 279 |
| 6801 | Ovary | Healthy | 3 | 269263 | 29960 | 4149 |
| 6797 | Ovary | Late Stage EOC | 3 | 292654 | 35261 | 4310 |
| 6795 | Oviduct | Late Stage EOC | 3 | 331682 | 36078 | 4728 |
| 6799 | Oviduct | Healthy | 3 | 187123 | 18221 | 2968 |
| 6798 | Plasma | Healthy | 3 | 255303 | 9250 | 248 |
| 6796 | Ovary | Healthy | 3 | 482706 | 32718 | 3990 |
| 6800 | Ovary | Late Stage EOC | 3 | 507992 | 33611 | 4151 |
| 6794 | Oviduct | Healthy | 3 | 381373 | 26717 | 3033 |
| 6802 | Oviduct | Late Stage EOC | 3 | 379049 | 36305 | 4461 |
Accessing the PeptideAtlas is accomplished through a readily accessible web GUI, providing observation information at both the protein and individual peptide level. Proteins and peptides of interest can be accessed using a sequence and/or list of accession number derived from Uniprot or Genbank This information can be readily used for comparisons between individual proteins contained in a single PeptideAtlas build, across species within the broader PeptideAtlas project, or generated from other sources such as KEGG or Reactome pathways.42,43
The chicken PeptideAtlas incorporates mass spectral data from over 800 individual biological experiments containing over 8 million spectra. The current build includes 5 million PSMs with an FDR threshold of 0.01% and over one hundred thousand distinct peptides at a false-discovery rate of 1%. This translates into 6,646 canonical proteins at a protein FDR of 1.3%. A substantial number of additional proteins are at least partially distinguishable, meaning that they possess at least one unique, well detected peptide within the chicken PeptideAtlas; the total number of observed proteins is 11,370, corresponding to 47% of Gallus gallus’ 23,997 predicted gene products based on the UniProtKB/TrEMBL database. A sizeable fraction of these detected species exist at the single peptide level, which is to be expected based on the reproductive system focus of the current datasets. Future inclusion of additional sample types is expected to increase the overall gene product coverage of the chicken PeptideAtlas.
Chimeric MS/MS reSpect Analysis
One of the basic assumptions of MS/MS data processing is that the fragment ion spectra obtained are derived from an individual precursor molecule isolated by MS. In spite of continued advancement in MS, precursor isolation windows remain relatively large to the achievable mass accuracy of the analyzers. Further, the complexity of whole proteomes remains well beyond the separation capacity of current chromatographic systems. The end result is that fragment spectra are frequently “chimera” containing peptide fragments from multiple precursors. While this is sometimes an intentional byproduct of the acquisition method, as in data-independent analyses, the collection of chimera can significantly reduce identifications in data-dependent sampling of complex samples due to the algorithmic MS/MS interpretation limitations44,45. This build of this initial chicken PeptideAtlas incorporates data processed with the newly developed reSpect tool,20 which enables multiple processing of complex MS/MS spectra to isolate co-fragmented chimeric peptides and increase overall peptide/protein IDs.
Examining the output of the reSpect reveals the effect of its inclusion on the chicken PeptideAtlas coverage. New peptide identifications derived from the reSpect output account for 825,135/5,125,800 = 16.1% of the total PSMs included in the chicken PeptideAtlas, only five specific instances of which corresponded to the identification that would originally have been assigned by a standard search. This frequency of spectral chimeras is in concordance with previously reported values.45 Within the chimeric group, 653,874 (79.2%) of the identifications have precursor masses falling outside the m/z tolerances of the initial search, and are therefore unintentional precursor inclusions as the result of wide precursor isolation windows. Interestingly, 378,194 (45.8%) of the PSMs were obtained from spectra that were assigned a ≥ 90% probability in the initial search, while a roughly equivalent number, 387,714 (47.0%), were from spectra whose assignment was given a probability of < 50%. Therefore, the chimera spectra seem to be equally likely to be minor components of an otherwise identifiable spectrum or major interferents who impinge spectral assignment.
Regardless of the source or intensity of the chimeras, the reSpect step was able to improve the depth of PSM and peptide identification by an appreciable margin. For the most part, the additional PSMs correspond with peptides that are already included as the primary ID from another spectrum. However, 8,976 unique peptides were identified only as the result of reSpect processing, corresponding to ~ 8% of the total peptide IDs in the chicken PeptideAtlas. The inclusion of additional unique peptide IDs is obviously of significant importance given the goal of complete organism coverage, but additional validation of already identified peptides is likewise important for increasing the confidence of spectral assignment when combining the results of multiple searches in iProphet.40
Ovarian Cancer Biomarkers in the Chicken PeptideAtlas
Ovarian cancer (OVC) is the most lethal of the gynecological cancers, with the single most important prognostic marker being clinical stage at diagnosis.46,47 Currently, the vast majority of OVC cases present in later stages47,48 and, as consequence, there is substantial interest in discovering biomarkers for early stages of the disease.49,50 To this end, the domestic fowl has rapidly emerged as a uniquely suited organism for the study of spontaneous ovarian cancer.51–54 Throughout the past 75 years, pathological examination of the hen has noted that spontaneous ovarian tumor formation occurs in up to 35% of the population and presents with the same histopathological subtypes seen in human disease.55–58 An extensive degree of molecular investigation has identified both human similar molecular expression and novel markers using mRNA expression analysis and immunohistochemistry (IHC) based assays.59–65 One major limitation of the model is the lack of commercially available chicken specific reagents for IHC and Western Blot analysis; the use of cross-reactive human antibodies has yielded inconsistent results, especially in the case of CA125, the primary prognostic marker for human OVC.66–68 The PeptideAtlas is a powerful resource for mining of proteotypic peptides to develop SRM-based methods69 that can augment or replace inconsistent mRNA and IHC approaches.70–72 Further, the current sample set is an empirical examination of the existence of many biomarkers previously unexplored by LC-MS/MS. Several examples of chicken biomarkers will be discussed. An extensive, but by no means complete, list of ovarian cancer associated proteins can be found in Table 2 and in a previous review by Hawkridge et al.53
Table 2.
A list of ovarian cancer associated protein biomarkers detected in the PeptideAtlas.
| Protein | Uniprot Accession | # Peptide Observations | # Distinct Peptides | Protein Group(s) | # Unique Proteins | Sample Type | Ref. |
|---|---|---|---|---|---|---|---|
| alpha-2-macroglobulin (A2M) | E1C544 | 48220 | 440 | E1C344 | 4 | P, T | [118] |
| Angiotensin-converting enzyme | F1NYM0 | 399 | 45 | F1NYM0 | 2 | T | [119] |
| Apolipoprotein A-1 | P08250 | 34918 | 242 | P08250 | 1 | P, T | [120] |
| Apolipoprotein B | F1NV02 | 103453 | 1236 | F1NV02 | 17 | P, T | [121] |
| B-2 Microglobulin | P21611 | 525 | 9 | P21611 | 6 | P, T | [122] |
| Cadherin-1 | P08641 | 791 | 42 | E1C6M9 | 4 | T | [123] |
| ERBB2/HER2 Analog | Q2EJ72 | 244 | 20 | E1C0S3 | 12 | P, T | [124] |
| Fibronectin | P11722 | 10241 | 117 | F1NJT3 | 6 | P, T | [125] |
| Fibulin-1 | F1NX60 | 1613 | 40 | F1NX60 | 3 | P, T | [126] |
| Hras / Kras | P08642 | 533 | 12 | F5ANT1 | 4 | T | [11] |
| Metalloproteinase inhibitor 2 | R4GIL5 | 46 | 4 | R4GIL5 | 2 | P, T | [127] |
| Mucin-6 | R9PXQ5 | 1488 | 121 | R9PXQ5 | 2 | T | [128] |
| Mucin-5B | Q98UI9 | 7483 | 273 | Q98UI9 | 2 | T | [128] |
| Ovalbumin | P01012 | 117199 | 528 | R9XP5 | 2 | P, T | [129] |
| Ovostatin | P20740 | 3570 | 123 | F1NU63 | 4 | T | [130] |
| OVOSTL | F1NEW8 | 1829 | 69 | F1NEW8 | 1 | P, T | [130] |
| SERPINA1 / Alpha-1-antitrypsin | E1C7T1 | 2158 | 48 | E1C7T1 | 1 | P, T | [84] |
| SERPINA3 / Alpha-1-antichymotrypsin | F1NPN5 | 1813 | 44 | F1NPN5 | 1 | P, T | [85] |
| SERPINB11 | P01013 | 1357 | 66 | IOJ178 | 4 | T | [59] |
| SERPINB2 / Plasminogen activator inhibitor 2 | E1BTH3 | 190 | 15 | E1BTH3 | 1 | T | [131] |
| Transglutaminase | Q01841 | 16255 | 102 | Q01841 | 3 | T | [129] |
| Transthyretin | P27731 | 5864 | 59 | P27731 | 1 | P, T | [132] |
| Vimentin | P09654 | 33522 | 128 | F1NJ08 | 22 | T | [133] |
Localization of Serine Protease Inhibitors
Serine protease inhibitors (Serpins) are a superfamily of protease inhibiting proteins ubiquitous to eukaryotic organisms whose regulatory members share a unique mechanism of irreversible “suicide inhibition” of target preoteases.73–75 Because protein homeostasis is controlled by proteinases and their inhibitors, this superfamily has attracted attention in the field of cancer biology, where tumor progression necessarily requires disruption of homeostasis.75,76 Within the family, specific proteins have been identified as prognostic biomarkers,77,78 tumor tissue markers,79 and potential diagnostic markers for a range of cancers,80–83 examples of which will be discussed further. Searching the chicken PeptideAtlas for proteins that have a description that contains “SERPIN” yields 18 unique protein groups from multiple subgroups of the serpin family, including many of interest in cancer research.
Serpin class “A” molecules are one of the largest serpin groups in humans, consisting of extracellular molecules.75 Five chicken orthologs of this class (E1C7T1, F1NPN5, E1BS56, E1C206, and F1NPN4) were identified in the chicken PeptideAtlas with an average coverage of 70%. As expected, excreted species are observed in plasma and liver tissue. Two of the species however, are also observed in hen reproductive tissues. SERPINA1 (E1C7T1, alpha-1-antitrypsin) is the most common serum serpin and its misregulation has been associated with endometrial and cervical cancer in humans.84 SERPINA3 (F1NPN5, alpha-1-antichymotrypsin) has likewise been associated with the promotion of endometrial cancer cells, as well as numerous other cancer types.85 The detection of these potential biomarkers in endometrial tissue of the chicken ovary is not unexpected, but cancerous associations derived from human studies have not been confirmed in the chicken. This relationship is worthy of further MS studies in the chicken model system, with particular emphasis on the translation of disease behavior between species.
Serpin class “B” molecules are generally intracellular proteins and similarly to class A are frequently associated with regulation and disease;76 the current chicken PeptideAtlas build contains three such proteins. SERPINB11 (P01013) is an important regulator of estrogen expression in chicken oviductal tissue; it is implicated in both normal oviduct development in the chicken and the development of ovarian endometrial cancer.59,79 Peptides from this protein were identified with 100% sequence coverage across healthy and cancerous oviduct tissues, consistent with its specific role in that tissue type. Interestingly, while identifications in plasma or liver tissue are limited to single observations, SERPINB11 peptides can be identified at consistent low levels in ovary samples from some healthy individuals, as well as in cancerous samples. This is consistent with the low levels of expression seen in RT-PCR experiments with developing ovarian follicles79 but the potential for ovarian tumors to originate from oviduct or mullerian cell incursion is considered quite high.86 The monitoring of cellular migration in the hen reproductive system offers the potential for understanding the biological origin and progression of ovarian cancer, which will aid in our understanding, detection, and treatment of the disease.
Additional serpins common in specific sample types are associated with multiple disease states and biological stress responses. SERPINC1 (F1NLP7, Antithrombin-III) and SERPINH1 (P13731, Hsp47) are among the most abundant proteins detected in plasma samples within the PeptideAtlas, as befitting their roles in blood. Antithrombin-III is an anticoagulant shown to be effective prognostic markers for some cancer types due to the systemic response to carcinogenesis,87 but more commonly used as a protein therapeutic.88 Hsp47 meanwhile, is of interest due to its relationship with thermal regulation and stress.89 It is therefore apparent that in addition to its use as an ovarian cancer model system, the included samples allow for the creation of experiments addressing additional diseases and biology of interest in chickens.
Acute Phase Proteins in the Chicken PeptideAtlas
One of the broadest diagnostic tools for the investigation of systemic response to stressors is the acute phase response (APR). The APR is a generalized systematic response of the immune system to numerous events, including trauma, inflammation, infection, stress, or carcinogenesis, with the goal of restoring organism homeostasis and promoting healing.90 Acute Phase Proteins (APPs) are hepatic proteins produced as the result of pro-inflammatory cytokines during trauma and are a powerful tool for examining immune and stress response.91 The initial description of APPs involved the changing concentrations of plasma C-reactive protein in response to pneumococcal infection92 but has since expanded to encompass numerous plasma proteins in both human and non-human animal systems.93,94 Generally, the APPs are divided into classes based on whether they increase or decrease in concentration in response to the APR, as well as the magnitude of the change. This expression can be highly species specific, requiring the development of unique assays and the selection of unique assay targets for different study organisms.93
APP profiles in many species have been used as diagnostic tools and as a mechanism to understand the biological response to system challenge, such as infection.95 Unsurprisingly, various APPs in the chicken (Table 2) have likewise been thoroughly classified by response and magnitude.96 Albumin is the only notable negative APP (relative abundance decreasing upon immune challenge) in the chicken, but with plasma concentrations approaching 20 mg/mL it is a trivial target for proteomic analysis.97 The positive APPs range in concentration from ng/mL – μg /mL quantities and their responses can be categorized as major (≥10-fold abundance increase), moderate (4–10 fold), and minor (≤3 fold).96–98 Monitoring APPs at these levels has previously relied on enzyme-linked immunoassays (ELISA), often using partially cross-reactive murine or human anitbodies.99 This method is undesirable for the discovery of avian APPs, due to cost, laboriousness, and frequent lack of similarity with more well understood mammalian systems. For example, haptoglobin is an very common APP in a wide range of species,91 but it was not until the chicken genome was sequenced that it was determined that no homologous protein exists in G. gallus;100 an antioxidant protein from gene PIT54 fills the hemoglobin binding role in chickens instead.100,101 Invaluable information on novel APP proteins, as well as thorough investigation of existing APPs will be greatly enhanced by the application of quantitative mass spectrometry techniques enabled, in part, by quality peptide and spectral libraries.
The chicken PeptideAtlas contains extensive liver and plasma samples from both healthy and diseased chickens, and contains all well-studied APPs in the organism (Table 3). Due to the underannoted nature of the chicken proteome, many APPs contain multiple potential variants in the global UniProtKB/TrEMBL database that are grouped by the chicken PeptideAtlas under the most likely canonical form. The PeptideAtlas can be used to explore the existence of one or more protein isoforms, as well as a starting point for the development of distinguishing assays.
Table 3.
Acute phase proteins of the chicken identified in the chicken PeptideAtlas. A highly redundant number of spectra were collected for each peptide, with numerous unique peptide IDs allowing for nearly complete coverage of many APPs.
| Protein | PeptideAtlas Protein Name (UniProt Accession Number(s)) | PeptideAtlas Group(s) | Spectra | Unique IDs | % Cover | Ref |
|---|---|---|---|---|---|---|
| Ceruloplasmin | F1N9R5, F1NPE0, Q5ZIT6, F1NY12 | F1N9R5 | 2111 | 83 | 68.4 | [134] |
| Fibronectin | P11722, F1NJT4, P11722, O57403 | F1NJT3 | 10241 | 117 | 94.5 | [125] |
| Alpha-1-acid glycoprotein | Q8JIG5, A7UEB0 | Q8JIG5 | 17665 | 117 | 87.6 | [97] |
| Ovotransferrin | P02789, E1BQC2, F1P2F0, P02789, P15989, Q4ADJ6, Q4ADJ7 // F1NVN3, Q92062 | E1BQC2 // Q92062 | 159189 | 668 | 97.5 | [135] |
| Serum Amyloid A | F1NW65, D6P887, Q9PSM7 | F1NW65 | 42 | 5 | 46.7 | [98] |
| PIT54 | Q98TD1, H9KZK6 | Q98TD1 | 13572 | 144 | 97.6 | [136] |
| Mannan-binding lectin | G3FGP5, O57451, Q98TA4, F1NKG3, U6A6U9, F1NKG3 | G3FGP5 | 628 | 19 | 70.1 | [137] |
| Hemopexin | H9L385, Q90WR3 // P20057 | H9L385 // P20057 | 17639 | 134 | 98.5 | [138] |
| Serum Albumin | P19121, F2Z4L6, R4GFD0 | P19121 | 194669 | 819 | 97.0 | [139] |
| Fibrinogen | F1P4V1, P14448 // F1NUL9, F1P5F9, Q02020 // E1BV78, O93568 | F1P4V1 // F1NUL9 // E1BV78 | 114957 | 981 | 87.2 | [140] |
Hemopexin Polymorphism
Hemopexin is an acute phase serum glycoprotein with enormous binding affinity for free heme, and is found across multiple animal families.102 The protein was initially isolated on the basis of its heme-affinity103 and has multifunctional properties in iron-metabolism.104 However, it has been shown to be APP based on regulated expression in response to hormonal stimuli,105 and infection with various pathogens,106–108 which is believed to be its primary biological role.104
The chicken PeptideAtlas contains multiple entries for chicken Hemopexin proteins derived from multiple sources. The manually annotated UniProtKB/Swiss-Prot database contains chicken protein P20057, which is an N-terminal fragment rather than a complete protein,109 while the UniProtKB/TrEMBL database contains an entry from the chicken genome (ENSGALG00000022586 / H9L385) as well as a non-genomic sequence fragment from a partial cDNA library (Q90WR3). Mutual alignment of the sequences revealed virtually no similarity between the manually determined N-terminal sequence and either of the other forms, while H9L385 and Q90WR3 possess 99% sequence similarity within the fragment region (Figure S-1 and S-2). The observed peptides within the chicken PeptideAtlas yield over 98% sequence coverage for the genetically derived data, while no portion of the manually determined sequence could be identified, casting doubt on its veracity. Within the two genetic sequences, three polymorphisms can be observed, two of which (p.Arg282Met and p.Lys148Arg) are not addressable because the small peptides constitute the unobserved 2% of the sequence. However, variants of the remaining polymorphism (p.Arg235His) were detected with unique peptides. The peptides CSGEPFQAITSDDSGR from H9L385 and CSGEPFQAITSDDSGHIYAFR from Q90WR3 (polymorphic site noted) were each observed in several hundred quality spectra across experiments, many with nearly full y-ion series, lending credence to the existence of the non-genomic protein sequence. Disaggregation of the samples to examine individual organisms from a published study24 reveals that specific chickens can present with either one or both of the sequences. Chickens #602, 612, 630, 639, and 666 exhibit peptides from both sequences, while #600, 601, and 650 exhibit only the unique peptide from H9L385 and #620 exhibits the form from Q90WR3, implying both hetero- and homozygosity of the polymorphism occur in the population. The chicken PeptideAtlas thus reveals the existence of multiple forms of a common chicken APP and provides peptides suitable for quantification of either individual polymorphisms or total protein.
Peptide Deglycosylation
N-linked glycosylation is an extremely common post-translational modification consisting of potentially diverse oligosaccharide structures linked to asparagine residues at a conserved sequence motif (N-X-(S/T), X≠P) and which affects the majority of gene products.110 The modification that occurs is a non-template driven process and due to the availability of N-glycan precursor, the accessibility of the glycosylation site, and the variable transit time of proteins, the majority of glycosylation motifs are not modified by sugars; others exhibit partial or complete occupancy, which can have significant implications for their biological function.111,112
During sample processing for the chicken PeptideAtlas a representative number of samples were deglycosylated using the enzyme peptide-N-glycosidase F (PNGase F) to cleave N-glycans and generate a detectable asparagine deamidation (mass shift: +0.9848 Da). This processing has a two-fold implication on the resulting peptides: increased peptide coverage from detection of newly deglycosylated peptides, and identification of potential glycosylation sites for future study based on the co-occurrence of the glycosylation motif and asparagine deamidation. The latter is usually performed with 18O water to provide unambiguous identification of enzymatic deamidation,113 but this is unfeasible for a project of this size. Nevertheless, a processing method designed to reduce non-specific deamidation combined with high resolution mass spectrometry enables accurate site determination sufficient for initiating deeper study.28,114
The protein prothrombin is an important coagulation precursor with multiple known glycosylation sites in different species.115 The glycosylation of these sites is important to protein function and processing, but different species exhibit different numbers of active glycosylation sites.116,117 The protein sequence in the chicken (UniProt Accession: F1NXV6) is 607 amino acids long and contains four potential N-glycosylation motifs at Asn122, Asn144, Asn161, and Asn403 which are homologous with the four sites observed in the human form of the protein (Figure S-3). Specific deamidated versions of each of the four potential glycopeptides (GTInYTK, FnASIYPDLTENYCR, NPDnNSEGPWcYTR, and nLTTNDILVR) were able to be observed, as well as longer fragments containing miss cleavages. Of the tryptic peptides, three of the four were observed only in deglycoslated samples, and were observed only in their deamidated forms; this implies that these sites are wholly glycosylated and the identification of these peptides is entirely related to the PNGase F processing. In contrast, one of the peptides (NPDNNSEGPWCYTR) was observed in both deamidated and non-deamidated forms in the deglycosylated samples, as well as being identified in the PNGase F free preparations, implying only partial site occupancy. Direct comparison of biological samples 6755 and 6798 (Table 1) showed a 70% increase in the number of peptide observations as the result of the inclusion of deamidated, and therefore deglycosylated, peptides. Homology comparison to humans would predict that the three wholly glycosylated sites would be the only occupied sites.115 However, the rigorous identification of a partially occupied site could point to a different structural or processing role in the chicken than the human, which could be worthy of further study.
CONCLUSION
The initial build of the chicken PeptideAtlas represents over 100,000 high confidence peptide identifications from MS/MS spectra corresponding to nearly 50% of the translated chicken proteome. The collection is rich in representative proteins of substantial interest in future health research in both chicken and human. Herein we have demonstrated the coverage of the chicken PeptideAtlas for an expansive number of proteins relevant to the chicken as a model for human ovarian cancer, important for investigations of systemic inflammatory responses in a veterinary context, and of general biological and agricultural interest to the study of the chicken organism. The chicken PeptideAtlas provides a readily accessible repository of information on the MS observable chicken proteome, providing a basis for proteomic experiments in the assay of known proteins, useful for discovery of novel biomarkers, and validation of genomic information at the proteomic level. We have demonstrated the use of this information for the isolation of single amino acid polymorphisms in inflammatory proteins as well as localization of tissue specific markers at the sample level within the chicken PeptideAtlas. This powerful resource is freely available to mine data for efforts such as in biologically driven hypotheses of both human health and agricultural interest and for approaches in biomarker discovery for quantitative MS assays.
Supplementary Material
Figure S-1: BLAST sequence alignment of P20057 and H9L385
Figure S-2: BLAST sequence alignment of Q90WR3 and H9L385
Figure S-3: BLAST sequence alignment of F1NXV6 and P00734 with annotated glycosylation sites (sequence motif N-X-(S/T), X≠P).
Table S-1: Complete list of experimental data incorporated into the PeptideAtlas
Acknowledgments
Funding Sources
This research was generously funded by the National Institutes of Health NCI IMAT Program Grant R33 (CA147988-02), the US Dept. of Education GAANN Fellowship Program in Molecular Biotechnology at NC State Grant (P200A140020), the W.M. Keck Foundation, and North Carolina State University and also funded in part by the National Institute of General Medical Sciences (NIGMS) grant R01 GM087221 and 2P50 GM076547/Center for Systems Biology.
Hen plasma and tissue samples were obtained with the generous assistance of Dr. James N. Petitte and Rebecca Wysocky of the NC State University Department of Poultry Science and Elizabeth Hecht of the NC State University Chemistry Department. Additional thanks are extended to Angelito Nepomuceno and Adam Hawkridge for their assistance in collecting MS data for inclusion.
Footnotes
Links to the GUI front-end of the chicken PeptideAtlas are available for reviewers at: https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/main.cgi?SBEAMSentrycode=G71UmfA
The related SRMAtlas, containing a reference library of SRM transitions derived in these experiments, will be available in 2017.
Author Contributions
All authors have given approval to the final version of the manuscript.
References
- 1.Food and Agriculture Organization of the United Nations. http://faostat3.fao.org/
- 2.Tickle C. The Contribution of Chicken Embryology to the Understanding of Vertebrate Limb Development. Mech Dev. 2004;121(9):1019–1029. doi: 10.1016/j.mod.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME, et al. Sequence and Comparative Analysis of the Chicken Genome Provide Unique Perspectives on Vertebrate Evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 4.Cogburn LA, Porter TE, Duclos MJ, Simon J, Burgess SC, Zhu JJ, Cheng HH, Dodgson JB, Burnside J. Functional Genomics of the Chicken—A Model Organism. Poult Sci. 2007;86(10):2059–2094. doi: 10.1093/ps/86.10.2059. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Fonslow BR, Shan B, Baek M-C, Yates JR. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem Rev. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida AM, Bassols A, Bendixen E, Bhide M, Ceciliani F, Cristobal S, Eckersall PD, Hollung K, Lisacek F, Mazzucchelli G, et al. Animal Board Invited Review: Advances in Proteomics for Animal and Food Sciences. Animal. 2015;9(01):1–17. doi: 10.1017/S1751731114002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ensembl genome browser 82: Gallus gallus - Description. http://useast.ensembl.org/Gallus_gallus/Info/Annotation.
- 8.organism:gallus in UniProtKB. http://www.uniprot.org/uniprot/?query=Organism%3Agallus&sort=score.
- 9.Vergara MN, Canto-Soler MV. Rediscovering the Chick Embryo as a Model to Study Retinal Development. Neural Develop. 2012;7:22. doi: 10.1186/1749-8104-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredrickson TN. Ovarian Tumors of the Hen. Environ Health Perspect. 1987;73:35–51. doi: 10.1289/ehp.877335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakim AA, Barry CP, Barnes HJ, Anderson KE, Petitte J, Whitaker R, Lancaster JM, Wenham RM, Carver DK, Turbov J, et al. Ovarian Adenocarcinomas in the Laying Hen and Women Share Similar Alterations in p53, Ras, and HER-2/Neu. Cancer Prev Res (Phila Pa) 2009;2(2):114–121. doi: 10.1158/1940-6207.CAPR-08-0065. [DOI] [PubMed] [Google Scholar]
- 12.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, Aderem A, Boyle R, Brunner E, Donohoe S, et al. Integration with the Human Genome of Peptide Sequences Obtained by High-Throughput Mass Spectrometry. Genome Biol. 2005;6(1):R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange Provides Globally Coordinated Proteomics Data Submission and Dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas Project. Nucleic Acids Res. 2006;34(suppl 1):D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vizcaino JA, Cote R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A Guide to the Proteomics Identifications Database Proteomics Data Repository. Proteomics. 2009;9(18):4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, et al. A Guided Tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller A, Eng J, Zhang N, Li X, Aebersold R. A Uniform Proteomics MS/MS Analysis Platform Utilizing Open XML File Formats. Mol Syst Biol. 2005;1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutsch EW, Mendoza L, Shteynberg D, Slagel J, Sun Z, Moritz RL. Trans-Proteomic Pipeline, a Standardized Data Processing Pipeline for Large-Scale Reproducible Proteomics Informatics. Proteomics Clin Appl. 2015;9(7–8):745–754. doi: 10.1002/prca.201400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: A Resource for Target Selection for Emerging Targeted Proteomics Workflows. EMBO Rep. 2008;9(5):429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shteynberg D, Mendoza L, Hoopmann MR, Sun Z, Schmidt F, Deutsch EW, Moritz RL. reSpect: Software for Identification of High and Low Abundance Ion Species in Chimeric Tandem Mass Spectra. J Am Soc Mass Spectrom. 2015;26(11):1837–1847. doi: 10.1007/s13361-015-1252-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King NL, Deutsch EW, Ranish JA, Nesvizhskii AI, Eddes JS, Mallick P, Eng J, Desiere F, Flory M, Martin DB, et al. Analysis of the Saccharomyces Cerevisiae Proteome with PeptideAtlas. Genome Biol. 2006;7:R106. doi: 10.1186/gb-2006-7-11-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hesselager MO, Codrea MC, Sun Z, Deutsch EW, Bennike TB, Stensballe A, Bundgaard L, Moritz RL, Bendixen E. The Pig PeptideAtlas: A Resource for Systems Biology in Animal Production and Biomedicine. Proteomics. 2016;16(4):634–644. doi: 10.1002/pmic.201500195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bislev SL, Deutsch EW, Sun Z, Farrah T, Aebersold R, Moritz RL, Bendixen E, Codrea MC. A Bovine PeptideAtlas of Milk and Mammary Gland Proteomes. Proteomics. 2012;12(18):2895–2899. doi: 10.1002/pmic.201200057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkridge AM, Wysocky RB, Petitte JN, Anderson KE, Mozdziak PE, Fletcher OJ, Horowitz JM, Muddiman DC. Measuring the Intra-Individual Variability of the Plasma Proteome in the Chicken Model of Spontaneous Ovarian Adenocarcinoma. Anal Bioanal Chem. 2010;398(2):737–749. doi: 10.1007/s00216-010-3979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepomuceno AI, Shao H, Jing K, Ma Y, Petitte JN, Idowu MO, Muddiman DC, Fang X, Hawkridge AM. In-Depth LC-MS/MS Analysis of the Chicken Ovarian Cancer Proteome Reveals Conserved and Novel Differentially Regulated Proteins in Humans. Anal Bioanal Chem. 2015;407(22):6851–6863. doi: 10.1007/s00216-015-8862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nepomuceno AI, Muddiman DC, Petitte JN. Global Proteomic Analysis of Functional Compartments in Immature Avian Follicles Using Laser Microdissection Coupled to LC-MS/MS. J Proteome Res. 2015;14(9):3912–3923. doi: 10.1021/acs.jproteome.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal Sample Preparation Method for Proteome Analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 28.Hecht ES, McCord JP, Muddiman DC. A Quantitative Glycomics and Proteomics Combined Purification Strategy. J Vis Exp. 2016;(109) doi: 10.3791/53735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews GL, Shuford CM, Burnett JC, Hawkridge AM, Muddiman DC. Coupling of a Vented Column with Splitless NanoRPLC-ESI-MS for the Improved Separation and Detection of Brain Natriuretic Peptide-32 and Its Proteolytic Peptides. J Chromatogr B. 2009;877(10):948–954. doi: 10.1016/j.jchromb.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randall SM, Cardasis HL, Muddiman DC. Factorial Experimental Designs Elucidate Significant Variables Affecting Data Acquisition on a Quadrupole Orbitrap Mass Spectrometer. J Am Soc Mass Spectrom. 2013;24(10):1501–1512. doi: 10.1007/s13361-013-0693-y. [DOI] [PubMed] [Google Scholar]
- 31.Meiring HD, van der Heeft E, ten Hove GJ, de Jong APJM. Nanoscale LC–MS(n): Technical Design and Applications to Peptide and Protein Analysis. J Sep Sci. 2002;25(9):557–568. [Google Scholar]
- 32.Farrah T, Deutsch EW, Omenn GS, Sun Z, Watts JD, Yamamoto T, Shteynberg D, Harris MM, Moritz RL. State of the Human Proteome in 2013 as Viewed through PeptideAtlas: Comparing the Kidney, Urine, and Plasma Proteomes for the Biology- and Disease-Driven Human Proteome Project. J Proteome Res. 2014;13(1):60–75. doi: 10.1021/pr4010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, et al. mzML--a Community Standard for Mass Spectrometry Data. Mol Cell Proteomics. 2011;10(1):R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat Biotechnol. 2012;30(10):918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng JK, Jahan TA, Hoopmann MR. Comet: An Open-Source MS/MS Sequence Database Search Tool. Proteomics. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 36.Craig R, Beavis RC. TANDEM: Matching Proteins with Tandem Mass Spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 37.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 39.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 40.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: Multi-Level Integrative Analysis of Shotgun Proteomic Data Improves Peptide and Protein Identification Rates and Error Estimates. Mol Cell Proteomics MCP. 2011;10(12):M111.007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein Identification False Discovery Rates for Very Large Proteomics Data Sets Generated by Tandem Mass Spectrometry. Mol Cell Proteomics MCP. 2009;8(11):2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: A Knowledgebase of Biological Pathways. Nucleic Acids Res. 2005;33(suppl 1):D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houel S, Abernathy R, Renganathan K, Meyer-Arendt K, Ahn NG, Old WM. Quantifying the Impact of Chimera MS/MS Spectra on Peptide Identification in Large-Scale Proteomics Studies. J Proteome Res. 2010;9(8):4152–4160. doi: 10.1021/pr1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoopmann MR, Finney GL, MacCoss MJ. High-Speed Data Reduction, Feature Detection and MS/MS Spectrum Quality Assessment of Shotgun Proteomics Data Sets Using High-Resolution Mass Spectrometry. Anal Chem. 2007;79(15):5620–5632. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McPhail S, Johnson S, Greenberg D, Peake M, Rous B. Stage at Diagnosis and Early Mortality from Cancer in England. Br J Cancer. 2015;112(s1):S108–S115. doi: 10.1038/bjc.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Facts & Figures. [accessed May 12, 2016];2016 http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2016/
- 48.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian Cancer. The Lancet. 2014;384(9951):1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 49.Clarke-Pearson DL. Screening for Ovarian Cancer. N Engl J Med. 2009;361(2):170–177. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 50.Buys SS, Partridge E, Black A, et al. Effect of Screening on Ovarian Cancer Mortality: The Prostate, Lung, Colorectal and Ovarian (Plco) Cancer Screening Randomized Controlled Trial. J Am Med Assoc. 2011;305(22):2295–2303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 51.Giles JR, Elkin RG, Trevino LS, Urick ME, Ramachandran R, Johnson PA. The Restricted Ovulator Chicken: A Unique Animal Model for Investigating the Etiology of Ovarian Cancer. Int J Gynecol Cancer. 2010;20(5):738–744. doi: 10.1111/igc.0b013e3181da2c49. [DOI] [PubMed] [Google Scholar]
- 52.Johnson PA, Giles JR. The Hen as a Model of Ovarian Cancer. Nat Rev Cancer. 2013;13(6):432–436. doi: 10.1038/nrc3535. [DOI] [PubMed] [Google Scholar]
- 53.Hawkridge AM. The Chicken Model of Spontaneous Ovarian Cancer. Proteomics - Clin Appl. 2014;8(9–10):689–699. doi: 10.1002/prca.201300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson PA, Giles J. Use of Genetic Strains of Chickens in Studies of Ovarian Cancer. Poult Sci. 2006;85(2):246–250. doi: 10.1093/ps/85.2.246. [DOI] [PubMed] [Google Scholar]
- 55.McGowan JP. The Biological Significance of Ovarian Tumors in the Fowl. J Cancer Res. 1930;14(4):527–535. [Google Scholar]
- 56.Papasolomontos P, Appleby E, Mayor O. Pathological Findings in Condemned Chickens: A Survey of 1,000 Carcasses. Vet Rec. 1969;85(17):459–465. doi: 10.1136/vr.84.17.459. [DOI] [PubMed] [Google Scholar]
- 57.Benedet JL, Bender H, Jones H, Ngan HY. Pecorelli SFIGO Staging Classifications, Clinical Practice Guidelines in the Management of Gynecologic Cancers, FIGO Committee on Gynecologic Oncology. Int J Gynaecol Obstet. 2000;70(2):209–262. [PubMed] [Google Scholar]
- 58.Barua A, Bitterman P, Abramowicz JS, Dirks AL, Bahr JM, Hales DB, Bradaric MJ, Edassery SL, Rotmensch J, Luborsky JL. Histopathology of Ovarian Tumors in Laying Hens, a Preclinical Model of Human Ovarian Cancer. Int J Gynecol Cancer. 2009;19(4):531–539. doi: 10.1111/IGC.0b013e3181a41613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim W, Kim J-H, Ahn SE, Jeong W, Kim J, Bazer FW, Han JY, Song G. Avian SERPINB11 Gene: A Marker for Ovarian Endometrioid Cancer in Chickens. Exp Biol Med. 2012;237(2):150–159. doi: 10.1258/ebm.2011.011250. [DOI] [PubMed] [Google Scholar]
- 60.Lim W, Jeong W, Kim J, Ka H, Bazer FW, Han JY, Song G. Differential Expression of Secreted Phosphoprotein 1 in Response to Estradiol-17β and in Ovarian Tumors in Chickens. Biochem Biophys Res Commun. 2012;422(3):494–500. doi: 10.1016/j.bbrc.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 61.Bradaric MJ, Penumatsa K, Barua A, Edassery SL, Yu Y, Abramowicz JS, Bahr JM, Luborsky JL. Immune Cells in the Normal Ovary and Spontaneous Ovarian Tumors in the Laying Hen ( Gallus Domesticus ) Model of Human Ovarian Cancer. PLOS ONE. 2013;8(9):e74147. doi: 10.1371/journal.pone.0074147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stammer K, Edassery SL, Barua A, Bitterman P, Bahr JM, Hales DB, Luborsky J. Selenium-Binding Protein 1 Expression in Ovaries and Ovarian Tumors of in the Laying Hen, a Spontaneous Model of Human Ovarian Cancer. Gynecol Oncol. 2008;109(1):115–121. doi: 10.1016/j.ygyno.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urick ME, Johnson PA. Cyclooxygenase 1 and 2 mRNA and Protein Expression in the Gallus Domesticus Model of Ovarian Cancer. Gynecol Oncol. 2006;103(2):673–678. doi: 10.1016/j.ygyno.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 64.Giles JR, Olson LM, Johnson PA. Characterization of Ovarian Surface Epithelial Cells from the Hen: A Unique Model for Ovarian Cancer. Exp Biol Med. 2006;231(11):1718–1725. doi: 10.1177/153537020623101108. [DOI] [PubMed] [Google Scholar]
- 65.Tiwari A, Hadley JA, GLH, Elkin RG, Cooper T, Ramachandran R. Characterization of Ascites-Derived Ovarian Tumor Cells from Spontaneously Occurring Ovarian Tumors of the Chicken: Evidence for E-Cadherin Upregulation. PLOS ONE. 2013;8(2):e57582. doi: 10.1371/journal.pone.0057582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez-Burford C, Barnes MN, Berry W, Partridge EE, Grizzle WE. Immunohistochemical Expression of Molecular Markers in an Avian Model: A Potential Model for Preclinical Evaluation of Agents for Ovarian Cancer Chemoprevention. Gynecol Oncol. 2001;81(3):373–379. doi: 10.1006/gyno.2001.6191. [DOI] [PubMed] [Google Scholar]
- 67.Jackson E, Anderson K, Ashwell C, Petitte J, Mozdziak PE. CA125 Expression in Spontaneous Ovarian Adenocarcinomas from Laying Hens. Gynecol Oncol. 2007;104(1):192–198. doi: 10.1016/j.ygyno.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 68.Nowak M, Janas Ł, Stachowiak G, Stetkiewicz T, Wilczynski JR. Current Clinical Application of Serum Biomarkers to Detect Ovarian Cancer. Menopause Rev. 2015;14(4):254–259. doi: 10.5114/pm.2015.55887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kusebauch U, Campbell DS, Deutsch EW, Chu CS, Spicer DA, Brusniak M-Y, Slagel J, Sun Z, Stevens J, Grimes B, et al. Human SRMAtlas: A Resource of Targeted Assays to Quantify the Complete Human Proteome. Cell. 2016;166(3):766–778. doi: 10.1016/j.cell.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, et al. A High-Confidence Human Plasma Proteome Reference Set with Estimated Concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10(9):M110.006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA Sequencing into Clinical Diagnostics: Opportunities and Challenges. Nat Rev Genet. 2016;17(5):257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier T, Güell M, Serrano L. Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett. 2009;583(24):3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 73.Whisstock JC, Bottomley SP. Molecular Gymnastics: Serpin Structure, Folding and Misfolding. Curr Opin Struct Biol. 2006;16(6):761–768. doi: 10.1016/j.sbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 74.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PGW, Irving JA, Lomas DA, Luke CJ, Moyer RW, et al. The Serpins Are an Expanding Superfamily of Structurally Similar but Functionally Diverse Proteins. Evolution, Machanisms of Inhibition, Novel Function, and a Revised Nomenclature. J Biol Chem. 2001;276(36):33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 75.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, et al. An Overview of the Serpin Superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geiger M, Wahlmuller F, Furtmuller M. The Serpin Family: Proteins with Multiple Functions in Health and Disease. 2015 [Google Scholar]
- 77.Lim W, Kim HS, Jeong W, Ahn SE, Kim J, Kim YB, Kim MA, Kim M-K, Chung HH, Song YS, et al. SERPINB3 in the Chicken Model of Ovarian Cancer: A Prognostic Factor for Platinum Resistance and Survival in Patients with Epithelial Ovarian Cancer. PLOS ONE. 2012;7(11):e49869. doi: 10.1371/journal.pone.0049869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY, Jo H-J, Kim DH, Kim GH, Park DY. Serpin Peptidase Inhibitor Clade A Member 1 Is a Biomarker of Poor Prognosis in Gastric Cancer. Br J Cancer. 2014;111(10):1993–2002. doi: 10.1038/bjc.2014.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim W, Kim J-H, Ahn SE, Jeong W, Kim J, Bazer FW, Han JY, Song G. Avian SERPINB11 Gene: Characteristics, Tissue-Specific Expression, and Regulation of Expression by Estrogen. Biol Reprod. 2011;85(6):1260–1268. doi: 10.1095/biolreprod.111.093526. [DOI] [PubMed] [Google Scholar]
- 80.Li HWR, Leung SW, Chan CSS, Yu MM-Y, Wong YF. Expression of Maspin in Endometrioid Adenocarcinoma of Endometrium. Oncol Rep. 2007;17(2):393–398. [PubMed] [Google Scholar]
- 81.Duffy MJ, McGowan PM, Harbeck N, Thomssen C, Schmitt M. uPA and PAI-1 as Biomarkers in Breast Cancer: Validated for Clinical Use in Level-of-Evidence-1 Studies. Breast Cancer Res. 2014;16(4):428. doi: 10.1186/s13058-014-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.National Taiwan University Hospital. SERPIN D1, Its Role in Lung Cancer Invasion, Metastasis; ClinicalTrialsgov Identifier: NCT00155116. ClinicalTrials.gov [Internet] 2005 [Google Scholar]
- 83.Abdul S, Leebeek FWG, Rijken DC, de Willige SU. Natural Heterogeneity of α2-Antiplasmin: Functional and Clinical Consequences. Blood. 2016;127(5):538–545. doi: 10.1182/blood-2015-09-670117. [DOI] [PubMed] [Google Scholar]
- 84.Abdul-Rahman PS, Lim B-K, Hashim OH. Expression of High-Abundance Proteins in Sera of Patients with Endometrial and Cervical Cancers: Analysis Using 2-DE with Silver Staining and Lectin Detection Methods. Electrophoresis. 2007;28(12):1989–1996. doi: 10.1002/elps.200600629. [DOI] [PubMed] [Google Scholar]
- 85.Yang G-D, Yang X-M, Lu H, Ren Y, Ma M-Z, Zhu L-Y, Wang J-H, Song W-W, Zhang W-M, Zhang R, et al. SERPINA3 Promotes Endometrial Cancer Cells Growth by Regulating G2/M Cell Cycle Checkpoint and Apoptosis. Int J Clin Exp Pathol. 2014;7(4):1348–1358. [PMC free article] [PubMed] [Google Scholar]
- 86.Auersperg N. The Origin of Ovarian Cancers--Hypotheses and Controversies. Front Biosci. 2013;5:709–719. doi: 10.2741/s401. [DOI] [PubMed] [Google Scholar]
- 87.Koh SCL, Khalil R, Lim F-K, Ilancheran A, Choolani M. The Association between Fibrinogen, von Willebrand Factor, Antithrombin III, and D-Dimer Levels and Survival Outcome by 36 Months from Ovarian Cancer. Clin Appl Thromb. 2006;12(1):3–8. doi: 10.1177/107602960601200102. [DOI] [PubMed] [Google Scholar]
- 88.Allingstrup M, Wetterslev J, Ravn FB, Moller AM, Afshari A. Antithrombin III for Critically Ill Patients: A Systematic Review with Meta-Analysis and Trial Sequential Analysis. Intensive Care Med. 2016;42(4):505–520. doi: 10.1007/s00134-016-4225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Al-Zghoul M-B, Dalab AES, Yahya IE, Althnaian TA, Al-ramadan SY, Ali AM, Albokhadaim IF, El-Bahr SM, Al Busadah KA, Hannon KM. Thermal Manipulation during Broiler Chicken Embryogenesis: Effect on mRNA Expressions of Hsp108, Hsp70, Hsp47 and Hsf-3 during Subsequent Post-Hatch Thermal Challenge. Res Vet Sci. 2015;103:211–217. doi: 10.1016/j.rvsc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 90.Gruys E, Toussaint MJM, Niewold TA, Koopmans SJ. Acute Phase Reaction and Acute Phase Proteins. J Zhejiang Univ Sci B. 2005;6(11):1045–1056. doi: 10.1631/jzus.2005.B1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jain S, Gautam V, Naseem S. Acute-Phase Proteins: As Diagnostic Tool. J Pharm Bioallied Sci. 2011;3(1):118–127. doi: 10.4103/0975-7406.76489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tillett WS, Francis T. Serological Reactions in Pneumonia with a Non-Protein Somatic Fraction of Pneumococcus. J Exp Med. 1930;52(4):561–571. doi: 10.1084/jem.52.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cray C, Zaias J, Altman NH. Acute Phase Response in Animals: A Review. Comp Med. 2009;59(6):517–526. [PMC free article] [PubMed] [Google Scholar]
- 94.Markanday A. Acute Phase Reactants in Infections: Evidence-Based Review and a Guide for Clinicians. Open Forum Infect Dis. 2015;2(3):ofv098. doi: 10.1093/ofid/ofv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eckersall PD, Bell R. Acute Phase Proteins: Biomarkers of Infection and Inflammation in Veterinary Medicine. Vet J. 2010;185(1):23–27. doi: 10.1016/j.tvjl.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 96.O’reilly El, Eckersall Pd. Acute Phase Proteins: A Review of Their Function, Behaviour and Measurement in Chickens. Worlds Poult Sci J. 2014;70(01):27–44. [Google Scholar]
- 97.Inoue M, Satoh W, Murakami H. Plasma Alpha 1-Acid Glycoprotein in Chickens Infected with Infectious Bursal Disease Virus. Avian Dis. 1997;41(1):164–170. [PubMed] [Google Scholar]
- 98.Sevimli A, Misirlioglu D, Polat U, Yalcin M, Akkoc A, Uguz C. The Effects of Vitamin A, Pentoxyfylline and Methylprednisolone on Experimentally Induced Amyloid Arthropathy in Brown Layer Chicks. Avian Pathol. 2005;34(2):143–149. doi: 10.1080/03079450500059149. [DOI] [PubMed] [Google Scholar]
- 99.Nazifi S, Dadras H, Hoseinian SA, Ansari-Lari M, Masoudian M. Measuring Acute Phase Proteins (Haptoglobin, Ceruloplasmin, Serum Amyloid A, and Fibrinogen) in Healthy and Infectious Bursal Disease Virus-Infected Chicks. Comp Clin Pathol. 2009;19(3):283–286. [Google Scholar]
- 100.Wicher KB, Fries E. Haptoglobin, a Hemoglobin-Binding Plasma Protein, Is Present in Bony Fish and Mammals but Not in Frog and Chicken. Proc Natl Acad Sci U S A. 2006;103(11):4168–4173. doi: 10.1073/pnas.0508723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iwasaki K, Morimatsu M, Inanami O, Uchida E, Syuto B, Kuwabara M, Niiyama M. Isolation, Characterization, and cDNA Cloning of Chicken Turpentine-Induced Protein, a New Member of the Scavenger Receptor Cysteine-Rich (SRCR) Family of Proteins. J Biol Chem. 2001;276(12):9400–9405. doi: 10.1074/jbc.M011713200. [DOI] [PubMed] [Google Scholar]
- 102.Muller-Eberhard U. Hemopexin; Enzymology. In: B-M, editor. Immunochemical Techniques Part M: Chemotaxis and Inflammation. Vol. 163. Academic Press; 1988. pp. 536–565. [Google Scholar]
- 103.Goldfarb V, Trimble RB, De Falco M, Liem H, Metcalfe SA, Wellner D, Muller-Eberhard U. An Avian Serum .alpha.1-Glycoprotein, Hemopexin, Differing Significantly in Both Amino Acid and Carbohydrate Composition from Mammalian (.beta.-Glycoprotein) Counterparts. Biochemistry (Mosc) 1986;25(21):6555–6562. doi: 10.1021/bi00369a033. [DOI] [PubMed] [Google Scholar]
- 104.Tolosano E, Fagoonee S, Morello N, Vinchi F, Fiorito V. Heme Scavenging and the Other Facets of Hemopexin. Antioxid Redox Signal. 2010;12(2):305–320. doi: 10.1089/ars.2009.2787. [DOI] [PubMed] [Google Scholar]
- 105.Grieninger G, Liang TJ, Beuving G, Goldfarb V, Metcalfe SA, Muller-Eberhard U. Hemopexin Is a Developmentally Regulated, Acute-Phase Plasma Protein in the Chicken. J Biol Chem. 1986;261(33):15719–15724. [PubMed] [Google Scholar]
- 106.Adler KL, Peng PH, Peng RK, Klasing KC. The Kinetics of Hemopexin and alpha1-Acid Glycoprotein Levels Induced by Injection of Inflammatory Agents in Chickens. Avian Dis. 2001;45(2):289–296. [PubMed] [Google Scholar]
- 107.Barnes DM, Song Z, Klasing KC, Bottje W. Protein Metabolism during an Acute Phase Response in Chickens. Amino Acids. 2002;22(1):15–26. doi: 10.1007/s726-002-8198-6. [DOI] [PubMed] [Google Scholar]
- 108.Garcia KO, Berchieri-Junior A, Santana AM, Freitas-Neto OC, Fagliari JJ. Experimental Infection of Commercial Layers Using a Salmonella Enterica Serovar Gallinarum Strain: Leukogram and Serum Acute-Phase Protein Concentrations. Rev Bras Cienc Avicola. 2009;11(4):263–270. [Google Scholar]
- 109.Wellner D, Cheng K-C, Muller-Eberhard U. N-Terminal Amino Acid Sequences of the Hemopexins from Chicken, Rat and Rabbit. Biochem Biophys Res Commun. 1988;155(2):622–625. doi: 10.1016/s0006-291x(88)80540-0. [DOI] [PubMed] [Google Scholar]
- 110.Apweiler R, Hermjakob H, Sharon N. On the Frequency of Protein Glycosylation, as Deduced from Analysis of the SWISS-PROT Database. Biochim Biophys Acta. 1999;1473(1):4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 111.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical Analysis of the Protein Environment of N-Glycosylation Sites: Implications for Occupancy, Structure, and Folding. Glycobiology. 2004;14(2):103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 112.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew Chem Int Ed Engl. 2005;44(45):7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 113.Kuster B, Mann M. 18O-Labeling of N-Glycosylation Sites To Improve the Identification of Gel-Separated Glycoproteins Using Peptide Mass Mapping and Database Searching. Anal Chem. 1999;71(7):1431–1440. doi: 10.1021/ac981012u. [DOI] [PubMed] [Google Scholar]
- 114.Nepomuceno AI, Gibson RJ, Randall SM, Muddiman DC. Accurate Identification of Deamidated Peptides in Global Proteomics Using a Quadrupole Orbitrap Mass Spectrometer. J Proteome Res. 2014;13(2):777–785. doi: 10.1021/pr400848n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Degen SJ. The Prothrombin Gene and Its Liver-Specific Expression. Semin Thromb Hemost. 1992;18(2):230–242. doi: 10.1055/s-2007-1002429. [DOI] [PubMed] [Google Scholar]
- 116.Webber D, Rodgers AL, Sturrock ED. Glycosylation of Prothrombin Fragment 1 Governs Calcium Oxalate Crystal Nucleation and Aggregation, but Not Crystal Growth. Urol Res. 2007;35(6):277–285. doi: 10.1007/s00240-007-0119-z. [DOI] [PubMed] [Google Scholar]
- 117.Wu W, Suttie JW. N-Glycosylation Contributes to the Intracellular Stability of Prothrombin Precursors in the Endoplasmic Reticulum. Thromb Res. 1999;96(2):91–98. doi: 10.1016/s0049-3848(99)00070-5. [DOI] [PubMed] [Google Scholar]
- 118.Lim W, Jeong W, Kim J-H, Lee J-Y, Kim J, Bazer FW, Han JY, Song G. Differential Expression of Alpha 2 Macroglobulin in Response to Dietylstilbestrol and in Ovarian Carcinomas in Chickens. Reprod Biol Endocrinol. 2011;9:137. doi: 10.1186/1477-7827-9-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beyazit F, Ayhan S, Celik HT, Gungor T. Assessment of Serum Angiotensin-Converting Enzyme in Patients with Epithelial Ovarian Cancer. Arch Gynecol Obstet. 2015;292(2):415–420. doi: 10.1007/s00404-015-3661-x. [DOI] [PubMed] [Google Scholar]
- 120.Kozak KR, Amneus MW, Pusey SM, Su F, Luong MN, Luong SA, Reddy ST, Farias-Eisner R. Identification of Biomarkers for Ovarian Cancer Using Strong Anion-Exchange ProteinChips: Potential Use in Diagnosis and Prognosis. Proc Natl Acad Sci U S A. 2003;100(21):12343–12348. doi: 10.1073/pnas.2033602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Avall-Lundqvist EH, Peterson CO. Serum Cholesterol and Apolipoprotein B Levels May Reflect Disease Activity in Ovarian Cancer Patients. Acta Oncol. 1996;35(8):1007–1010. doi: 10.3109/02841869609100719. [DOI] [PubMed] [Google Scholar]
- 122.Yang HS, Li Y, Deng HX, Peng F. Identification of beta2-Microglobulin as a Potential Target for Ovarian Cancer. Cancer Biol Ther. 2009;8(24):2323–2328. doi: 10.4161/cbt.8.24.9982. [DOI] [PubMed] [Google Scholar]
- 123.Sundfeldt K, Piontkewitz Y, Ivarsson K, Nilsson O, Hellberg P, Brannstrom M, Janson PO, Enerback S, Hedin L. E-Cadherin Expression in Human Epithelial Ovarian Cancer and Normal Ovary. Int J Cancer. 1997;74(3):275–280. doi: 10.1002/(sici)1097-0215(19970620)74:3<275::aid-ijc7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 124.Hynes NE. ErbB2 Activation and Signal Transduction in Normal and Malignant Mammary Cells. J Mammary Gland Biol Neoplasia. 1996;1(2):199–206. doi: 10.1007/BF02013643. [DOI] [PubMed] [Google Scholar]
- 125.Bignotti E, Tassi RA, Calza S, Ravaggi A, Bandiera E, Rossi E, Donzelli C, Pasinetti B, Pecorelli S, Santin AD. Gene Expression Profile of Ovarian Serous Papillary Carcinomas: Identification of Metastasis-Associated Genes. Am J Obstet Gynecol. 2007;196(3):245.e1–11. doi: 10.1016/j.ajog.2006.10.874. [DOI] [PubMed] [Google Scholar]
- 126.Moll F, Katsaros D, Lazennec G, Hellio N, Roger P, Giacalone P-L, Chalbos D, Maudelonde T, Rochefort H, Pujol P. Estrogen Induction and Overexpression of Fibulin-1C mRNA in Ovarian Cancer Cells. Oncogene. 2002;21(7):1097–1107. doi: 10.1038/sj.onc.1205171. [DOI] [PubMed] [Google Scholar]
- 127.Garzetti GG, Ciavattini A, Lucarini G, Goteri G, de e Nictolis M, Garbisa S, Masiero L, Romanini C, Graziella B. Tissue and Serum Metalloproteinase (MMP-2) Expression in Advanced Ovarian Serous Cystoadenocarcinomas: Clinical and Prognostic Implications. Anticancer Res. 1995;15(6B):2799–2804. [PubMed] [Google Scholar]
- 128.Kufe DW. Mucins in Cancer: Function, Prognosis and Therapy. Nat Rev Cancer. 2009;9(12):874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giles JR, Shivaprasad HL, Johnson PA. Ovarian Tumor Expression of an Oviductal Protein in the Hen: A Model for Human Serous Ovarian Adenocarcinoma. Gynecol Oncol. 2004;95(3):530–533. doi: 10.1016/j.ygyno.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 130.Nagase H, Harris ED. Ovostatin: A Novel Proteinase Inhibitor from Chicken Egg WhiteII. Mechanism of Inhibition Studied with Collagenase and Thermolysin. J Biol Chem. 1983;258(12):7490–7498. [PubMed] [Google Scholar]
- 131.Duggan C, Kennedy S, Kramer MD, Barnes C, Elvin P, McDermott E, O’Higgins N, Duffy MJ. Plasminogen Activator Inhibitor Type 2 in Breast Cancer. Br J Cancer. 1997;76(5):622–627. doi: 10.1038/bjc.1997.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Z, Bast RC, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng X-Y, et al. Three Biomarkers Identified from Serum Proteomic Analysis for the Detection of Early Stage Ovarian Cancer. Cancer Res. 2004;64(16):5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 133.Satelli A, Li S. Vimentin as a Potential Molecular Target in Cancer Therapy Or Vimentin, an Overview and Its Potential as a Molecular Target for Cancer Therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Disilvestro RA, Harris ED. Purification and Partial Characterization of Ceruloplasmin from Chicken Serum. Arch Biochem Biophys. 1985;241(2):438–446. doi: 10.1016/0003-9861(85)90568-5. [DOI] [PubMed] [Google Scholar]
- 135.Xie H, Newberry L, Clark FD, Huff WE, Huff GR, Balog JM, Rath NC. Changes in Serum Ovotransferrin Levels in Chickens with Experimentally Induced Inflammation and Diseases. Avian Dis. 2002;46(1):122–131. doi: 10.1637/0005-2086(2002)046[0122:CISOLI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 136.Millet S, Bennett J, Lee KA, Hau M, Klasing KC. Quantifying and Comparing Constitutive Immunity across Avian Species. Dev Comp Immunol. 2007;31(2):188–201. doi: 10.1016/j.dci.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 137.Laursen SB, Nielsen OL. Mannan-Binding Lectin (MBL) in Chickens: Molecular and Functional Aspects. Dev Comp Immunol. 2000;24(2–3):85–101. doi: 10.1016/s0145-305x(99)00066-x. [DOI] [PubMed] [Google Scholar]
- 138.Buyse J, Swennen Q, Niewold TA, Klasing KC, Janssens GPJ, Baumgartner M, Goddeeris BM. Dietary L-Carnitine Supplementation Enhances the Lipopolysaccharide-Induced Acute Phase Protein Response in Broiler Chickens. Vet Immunol Immunopathol. 2007;118(1–2):154–159. doi: 10.1016/j.vetimm.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 139.Hrubec TC, Whichard JM, Larsen CT, Pierson FW. Plasma versus Serum: Specific Differences in Biochemical Analyte Values. J Avian Med Surg. 2002;16(2):101–105. [Google Scholar]
- 140.Amrani DL, Mauzy-Melitz D, Mosesson MW. Effect of Hepatocyte-Stimulating Factor and Glucocorticoids on Plasma Fibronectin Levels. Biochem J. 1986;238(2):365–371. doi: 10.1042/bj2380365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S-1: BLAST sequence alignment of P20057 and H9L385
Figure S-2: BLAST sequence alignment of Q90WR3 and H9L385
Figure S-3: BLAST sequence alignment of F1NXV6 and P00734 with annotated glycosylation sites (sequence motif N-X-(S/T), X≠P).
Table S-1: Complete list of experimental data incorporated into the PeptideAtlas