Abstract
Porcine circovirus 2 (PCV2) has been prevalent in swine herds in China since 2002, causing severe economic loss to the pig industry. The number of live pigs in southeast China is > 20 million. Since information on the genetic variation of PCV2 in the Fujian province is limited, the objective of the present work was to investigate the epidemiological and evolutionary characteristics of PCV2 in southeast China from 2013 to 2017. Of the 685 samples collected from 90 different swine herds from 2013 to 2017, 356 samples from 84 different swine herds were positive for PCV2. PCV2a, PCV2b, PCV2d, and PCV2e co-existed in the Fujian province, with PCV2d being the predominant circulating strain in swineherds and PCV2e being reported for the first time in China. Strikingly, PCV2-FJ-water DNA comes from contaminated river water and not infected animals. Sequence comparison among all isolates indicated that 95 isolates shared approximately 78.7%–100% nucleotide identity and 74.5%–100% amino acid identity for open reading frame 2 (ORF2). Amino acid alignment showed that the Cap protein of PCV2e differed markedly from those of PCV2a, PCV2b, PCV2c, and PCV2d. These results indicated that various PCV2 genotypes exist in China, and that PCV2 is continuously evolving, leading to rapid emergence of new variant stains.
Keywords: Phylogenetic analysis, Recombination, PCV2, Genotype
Introduction
The porcine circovirus (PCV) is a small non-enveloped single-stranded circular DNA virus of swine. Currently, three major PCV2 genotypes have been recognized: PCV type 1 (PCV1), PCV2 type 2 (PCV2), and PCV type 3 (PCV3) (Opriessnig, Meng & Halbur, 2007; Palinski et al., 2016). PCV1 is non-pathogenic to pigs, whereas PCV2 is the primary causative agent of porcine circovirus-associated disease (PCVAD) in growing pigs, which includes systemic, respiratory and enteric manifestations, and PCV3 is associated with porcine dermatitis and nephropathy syndrome (PDNS) (Opriessnig, Meng & Halbur, 2007).
Currently, PCV2 is the primary causative agent of swine viral disease worldwide, which has resulted in considerable economic loss since it was first identified in Canada in 1991 (Harding & Clark, 1997; Krakowka et al., 2000; Bolin et al., 2001). The viral genomes range from 1,766 to 1,768 nucleotides (nt) in length and contain four major identified open reading frames (ORFs) (ORF1–ORF4). ORF2 encodes the capsid protein (Cap), which is the only structural protein of PCV2 and is related to viral antigenicity; hence, the gene encoding the Cap protein is suitable as a phylogenetic and epidemiological marker (Gomes et al., 2007; Choi & Chae, 2008; Jiang et al., 2017). Currently, at least five distinct genotypes have been described in swine based on the ORF2 sequence, which have been classified as PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e (Segalés et al., 2008; Harmon et al., 2015; Xiao, Halbur & Opriessnig, 2015; Davies et al., 2016; Karuppannan & Opriessnig, 2017). PCV2e is a new genetic group which was recently proposed by Davies et al. (2016). Currently, PCV2a, PCV2b, and PCV2d are found globally, whereas PCV2c has only been identified in Denmark and Brazil (Dupont et al., 2008; Franzo et al., 2015). PCV2e is a new genotype that has been circulating in the USA since 2015, the ORF2 sequences of which have 12 or 15 additional nts compared to those of PCV2a–PCV2d (Harmon et al., 2015; Davies et al., 2016).
PCV2 was first reported in China in 1999, and has become the causative agent of one of the country’s most serious swine diseases (Lang et al., 2000; Shuai et al., 2007; Wang et al., 2009; Guo et al., 2012; Jiang et al., 2017). Previous studies showed that various genotypes, including PCV2a, PCV2b, and PCV2d are present in China. Among these genotypes, PCV2b and PCV2d have been predominant in China since 2004 (Lang et al., 2000; Shuai et al., 2007; Wang et al., 2009; Guo et al., 2012; Jiang et al., 2017). Recently, several groups showed that PCV2 isolates in pig farms had recombined owing to the co-existence of different PCV2 genotypes (Hesse, Kerrigan & Rowland, 2008; Cai et al., 2011; Huang et al., 2013; Ramos et al., 2013; Anoopraj et al., 2015). Despite continuous reports of newly emerging strains worldwide, information on the genetic variation of PCV2 in the Fujian province is limited. Therefore, to understand the current molecular epidemiology of PCV2 in Southeast China, the objective of the present work was to investigate the epidemiological and evolutionary characteristics of PCV2 in the Fujian province from 2013 to 2017.
Materials and Methods
Animal ethics
Sampling procedures were approved by the Animal Ethics Committee of the South China Agricultural University, with a reference number of SCAU-AEC-2014-10.
Clinical samples
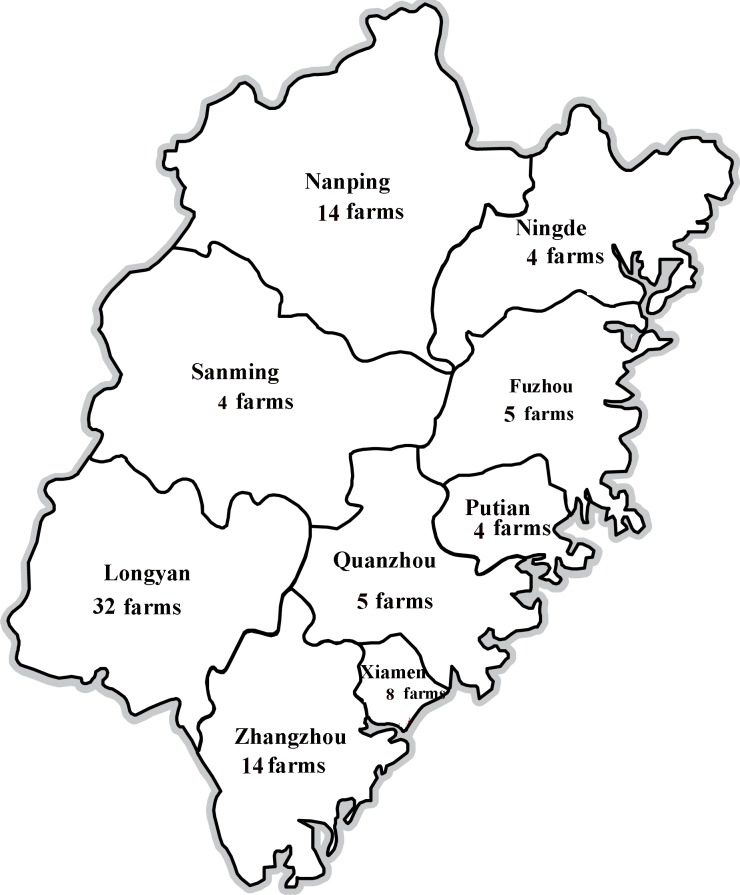
Six hundred and eighty-five clinical samples (blood, lungs, kidneys, livers, and lymph nodes) were collected from unthrifty pigs (4–10 weeks of age) in different farms of Fujian province, China, between 2013 and 2017. In particular, samples were collected from 32 farms in Longyan (196 samples), 14 farms in Zhangzhou (105 samples), eight farms in Xiamen (75 samples), five farms in Fuzhou (54 samples), four farms in Ningde (40 samples), 14 farms in Nanping (92 samples), four farms in Sanming (40 samples), five farms in Quanzhou (45 samples), and four farms in Putian (38 samples) from 2013 to 2017 (Fig. 1). These pigs were suspected to have clinical signs of Postweaning Multisystemic Wasting Syndrome (PMWS) and/or PDNS. In addition, 200 water samples were collected from the river or pool covering a geographic area of about 5 km2 near every pig farm.
Figure 1. Locations of PCV2-positive swine farms where samples were collected in Fujian Province.
Polymerase chain reaction (PCR) amplification and nucleotide sequencing
Total viral DNA was extracted from tissue samples using DNA extraction kit ver.3.0 (TaKaRa, Dalian, China) following the manufacturer’s protocol. ORF2 was amplified from the extracted DNA using specific primers according to previous studies (Fort et al., 2007). PCR products were purified using the gel extraction kit (Tiangen Biotech Co. Ltd., Beijing, China) and cloned into the pGEM-T Easy vector (Promega). Recombinant clones were sequenced by Sangon Biotech (Shanghai) Co. Ltd. Each fragment was independently sequenced at least thrice.
Phylogenetic analysis
Ninety-five ORF2 genes of PCV2 isolates were identified in the Fujian province during 2013–2017. Representative PCV2 sequences, including PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e isolates in the GenBank, were used for sequence alignments and phylogenetic analyses.
Multiplex sequencing alignments were performed using CLUSTAL X (version 1.83). The phylogenetic tree was constructed using the neighbor joining (NJ) method with MEGA 6.0, the maximum composite likelihood model, and a bootstrap confidence value of 1,000 replicates.
Recombination analysis
To detect putative recombination events in PCV2, recombinant strains were investigated on the complete genome database using a recombination detection program (RDP v.4.24) (Martin et al., 2010) as described by Ramos et al. (2013). Briefly, seven methods (RDP, GeneConv, BootScan, MaxChi, Chimera, SiScan, and 3Seq) were used for preliminary scan and only events detected by more than two methods with a highest acceptable p-value of 0.01 were considered. The recombination event was further confirmed using SimPlot program (v.3.5.1) and boot scanning analysis was performed with a 200-bp window sliding along a step size of 20 bp (Lole et al., 1999).
Results
PCV2 detection
Of the 685 samples collected from 90 different swine herds located in Fujian province from 2013 to 2017, 356 samples from 84 different swine herds were positive for PCV2. Ninety-five positive samples were selected for further analysis of PCV2 characteristics (Table 1).
Table 1. List of Fujian PCV2 samples in this study.
| Name | Geographic origin | Source | Time | Genetype | Accession No. |
|---|---|---|---|---|---|
| CN-FJ-C1-2017 | Putian | lymph node | 2017 | PCV2d | MF679546 |
| CN-FJ-C008 | Putian | serum | 2017 | PCV2a | MF679547 |
| CN-FJ-N001-2011 | Longayan | lymph node | 2011 | PCV2d | MF679548 |
| CN-FJ-C8-2017 | Longayan | Spleen/Lung | 2017 | PCV2a | MF679549 |
| CN-FJ-C3-2017 | Longayan | lymph node | 2017 | PCV2d | MF679550 |
| CN-FJ-6S-2017 | Zhangzhou | lymph node | 2017 | PCV2d | MF679551 |
| CN-FJ-7SC-2017 | Putian | serum | 2017 | PCV2d | MF679552 |
| CN-FJ-F8C-2017 | Xiamen | lymph node | 2017 | PCV2d | MF679553 |
| CN-FJ-LY01-2016 | Quanzhou | Lung | 2016 | PCV2d | MF679554 |
| CN-FJ-LY03-2016 | Ningde | lymph node | 2016 | PCV2d | MF679555 |
| PCV2-FJ-3 | Ningde | serum | 2013 | PCV2b | MG229682 |
| FJ01 | Longyan | lymph node | 2013 | PCV2a | MG252966 |
| FJ02 | Zhangzhou | Spleen/Lung | 2014 | PCV2d | MG229674 |
| FJ03 | Xiamen | lymph node | 2014 | PCV2d | MG229681 |
| FJ04 | Nanping | lymph node | 2013 | PCV2d | MG229675 |
| FJ05 | Fuzhou | lymph node | 2014 | PCV2d | MG229676 |
| FJ06 | Longyan | serum | 2013 | PCV2d | MG229677 |
| FJ07 | Quanzhou | lymph node | 2013 | PCV2d | MG229678 |
| FJ08 | Zhangzhou | Lung | 2013 | PCV2d | MG229679 |
| FJ09 | Longyan | lymph node | 2013 | PCV2b | MG252967 |
| FJ10 | Nanping | lymph node | 2014 | PCV2b | MG229680 |
| CN-FJ-LY04-2016 | Quanzhou | serum | 2016 | PCV2b | MF679556 |
| CN-FJ-NP01-2016 | Nanping | lymph node | 2016 | PCV2d | MF679557 |
| CN-FJ-NP02-2016 | Nanping | Spleen/Lung | 2016 | PCV2d | MF679558 |
| CN-FJ-NP03-2016 | Nanping | lymph node | 2016 | PCV2d | MF679605 |
| FJ-NP04-2017 | Nanping | serum | 2016 | PCV2d | MF679559 |
| CN-FJ-ZZ01-2016 | Zhangzhou | lymph node | 2016 | PCV2d | MF679560 |
| CN-FJ-ZZ02-2016 | Zhangzhou | Spleen/Lung | 2016 | PCV2d | MF679561 |
| CN-FJ-ZZ03-2016 | Zhangzhou | lymph node | 2016 | PCV2d | MF679562 |
| CN-FJ-ZZ04-2017 | Zhangzhou | Lung | 2017 | PCV2d | MF679563 |
| CN-FJ-N002-2012 | Quanzhou | lymph node | 2012 | PCV2d | MF679564 |
| CN-FJ-ZZ05-2017 | Zhangzhou | spleen | 2017 | PCV2d | MF679565 |
| CN-FJ-ZZ06-2017 | Zhangzhou | lymph node | 2017 | PCV2d | MF679566 |
| CN-FJ-C011 | Xiamen | serum | 2015 | PCV2d | MF679575 |
| CN-FJ-Z11 | Xiamen | lymph node | 2017 | PCV2d | MF679567 |
| CN-FJ-ZZ12 | Xiamen | serum | 2015 | PCV2d | MF679568 |
| CN-FJ-C12 | Xiamen | lymph node | 2016 | PCV2d | MF679569 |
| CN-FJ-C13 | Quanzhou | Lung | 2015 | PCV2d | MF679570 |
| CN-FJ-C14 | Longyan | Spleen/Lung | 2015 | PCV2d | MF679571 |
| CN-FJ-CFI | Longyan | lymph node | 2013 | PCV2d | MF679572 |
| CN-FJ-TETN | Longyan | serum | 2013 | PCV2b | MF679573 |
| CN-FJ-TEI | Quanzhou | serum | 2013 | PCV2d | MF679574 |
| CN-FJ-CP11 | Quanzhou | lymph node | 2015 | PCV2d | MF679576 |
| CN-FJ-CA01 | Quanzhou | lymph node | 2016 | PCV2d | MF679577 |
| CN-FJ-CAE6 | Quanzhou | spleen | 2017 | PCV2d | MF679578 |
| CN-FJ-C0113 | Zhangzhou | lymph node | 2015 | PCV2d | MF679579 |
| CN-FJ-C1134 | Xiamen | serum | 2016 | PCV2d | MF679580 |
| CN-FJ-CTS02 | Longyan | lymph node | 2014 | PCV2d | MF679581 |
| CN-FJ-CTS03 | Longyan | lymph node | 2016 | PCV2d | MF679582 |
| CN-FJ-CTS04 | Longyan | Spleen/Lung | 2015 | PCV2d | MF679583 |
| CN-FJ-FZT01 | Xiamen | lymph node | 2014 | PCV2b | MF679584 |
| CN-FJ-FZC41 | Quanzhou | serum | 2015 | PCV2b | MF679585 |
| CN-FJ-FZOU | Ningde | lymph node | 2013 | PCV2d | MF679586 |
| CN-FJ-FZCP4 | Ningde | lymph node | 2014 | PCV2d | MF679587 |
| CN-FJ-FZ01134 | Ningde | Spleen/Lung | 2014 | PCV2d | MF679588 |
| CN-FJ-FCP0712 | Ningde | lymph node | 2015 | PCV2d | MF679589 |
| PCV2-FJ-FJ5 | Quanzhou | spleen | 2016 | PCV2d | MF679590 |
| PCV2-FJ-TW01 | Quanzhou | lymph node | 2017 | PCV2d | MF679591 |
| PCV2-FJ-FT02 | Putian | lymph node | 2015 | PCV2b | MF679592 |
| PCV2-FJ-4129 | Putian | Spleen/Lung | 2016 | PCV2d | MF679593 |
| PCV2-FJ-17117 | Putian | lymph node | 2016 | PCV2d | MF679594 |
| PCV2-FJ-20131 | Putian | lymph node | 2016 | PCV2d | MF679595 |
| PCV2-FJ-water | Longyan | Water | 2017 | PCV2d | MF679596 |
| PCV2-FJ-7117P | Putian | lymph node | 2016 | PCV2d | MF679597 |
| CN-FJ-N005-2013 | Zhangzhou | serum | 2013 | PCV2d | MF679598 |
| CN-FJ-N004-2012 | Fuzhou | lymph node | 2012 | PCV2d | MF679599 |
| CN-FJ-N003-2012 | Nanping | Lung | 2012 | PCV2d | MF679600 |
| CN-FJ-3M-2017 | Nanping | lymph node | 2017 | PCV2a | MF679601 |
| CN-FJ-DD6-2017 | Fuzhou | lymph node | 2017 | PCV2b | MF679602 |
| CN-FJ-LG-2017 | Longyan | Lung | 2017 | PCV2d | MF679603 |
| PCV2-CN/FuJian-612-2017 | Longyan | lymph node/spleen | 2017 | PCV2e | MF589523 |
| PCV2-CN/FuJian-625-2017 | Longyan | lymph node/spleen | 2017 | PCV2e | MF589524 |
| PCV2-FJ-SS | Fuzhou | lymph node | 2013 | PCV2d | MF589525 |
| PCV2-FJ-S2 | Nanping | Spleen/Lung | 2014 | PCV2a | MF589526 |
| PCV2-FJ-YE | Longyan | lymph node | 2012 | PCV2d | MF589527 |
| PCV2-FJ-65 | Longyan | Lung | 2017 | PCV2b | MF589528 |
| PCV2-FJ-SO | Longyan | lymph node | 2016 | PCV2d | MF589529 |
| PCV2-FJ-02 | Longyan | Spleen/Lung | 2013 | PCV2d | MF589530 |
| PCV2-FJ-03 | Longyan | lymph node | 2014 | PCV2b | MF589531 |
| PCV2-FJ-S3 | Longyan | serum | 2015 | PCV2b | MF589532 |
| PCV2-FJ-S4 | Longyan | lymph node | 2014 | PCV2d | MF589533 |
| PCV2-FJ-S1 | Fuzhou | lymph node | 2014 | PCV2a | MF589534 |
| PCV2-FJ-MAO | Fuzhou | Lung | 2013 | PCV2b | MF589535 |
| PCV2-FJ-T10 | Fuzhou | lymph node | 2014 | PCV2d | MF589536 |
| PCV2-FJ-S22 | Fuzhou | Spleen/Lung | 2016 | PCV2d | MF589537 |
| PCV2-FJ-06 | Fuzhou | Lung | 2017 | PCV2a | MF589538 |
| PCV2-FJ-LY6 | Zhangzhou | lymph node | 2014 | PCV2d | MF589539 |
| PCV2-FJ-TI | Zhangzhou | Spleen | 2014 | PCV2d | MF589540 |
| PCV2-FJ-EX | Zhangzhou | lymph node | 2015 | PCV2d | MF589541 |
| PCV2-FJ-TN | Zhangzhou | lymph node | 2015 | PCV2d | MF589542 |
| PCV2-FJ-SY | Longyan | lymph node | 2015 | mPCV2d | MF589543 |
| PCV2-FJ-01 | Quanzhou | serum | 2016 | PCV2d | MF589544 |
| PCV2-FJBZ05 | Longyan | lymph node | 2015 | PCV2b | MG011720 |
| CN-PCV2-FJ1145 | Longyan | Spleen/Lung | 2016 | mPCV2b | MG011721 |
| PCV2-FJLY10 | Longyan | lymph node | 2016 | PCV2b | MG011722 |
Phylogenetic analysis of ORF2
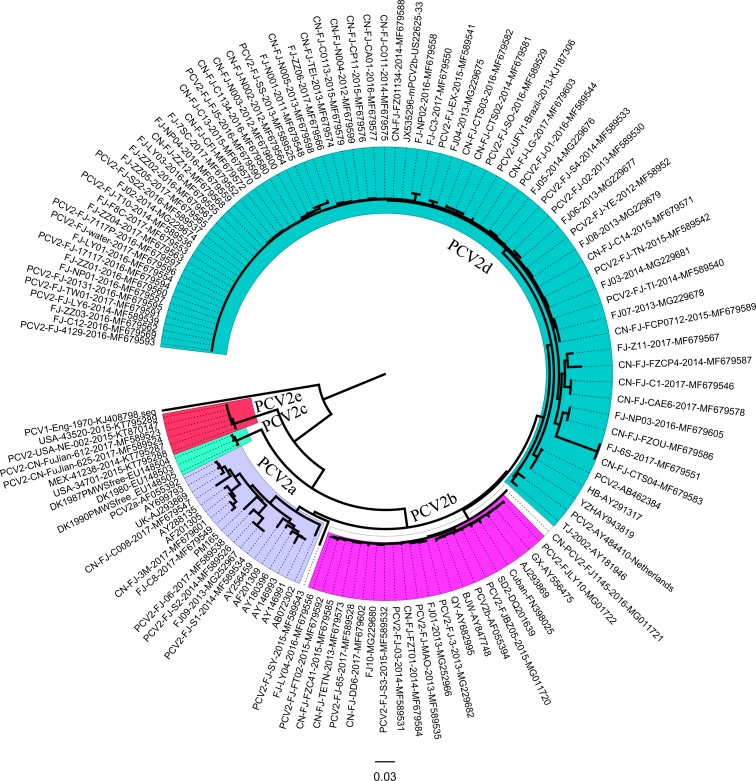
Phylogenetic analysis based on ORF2 showed that Fujian PCV2 strains were classified into five subtypes (PCV2a, PCV2b, PCV2d, and PCV2e) (Fig. 2). Among Fujian PCV2 strains, 7.37% (7/95) of the sequences corresponded to PCV2a, 15.8% (15/95) to PCV2b, 72.6% (69/95) to PCV2d, and 2.1% (2/95) to PCV2e. PCV2-CN-FuJian-612-2017 and PCV2-CN-FuJian-625-2017 were classified as PCV2e, which has been reported for the first time in China. Interestingly, PCV2-FJ-water DNA comes from contaminated water instead of from infected animals.
Figure 2. Neighbor-joining (NJ) phylogenetic analysis of Fujian PCV2 ORF2 genes.
Reliability of the tree was assessed by bootstrap analysis of 1,000 replications. PCV2b (GenBank accession no. AF055394).
Amino acid analysis of ORF2
The lengths of the Fujian PCV2 ORF2 sequences were 702 nt (PCV2a, PCV2b, or PCV2d), 705 nt (PCV2c and PCV2d), and 717 nt (PCV2e), which encoded Cap proteins of 233, 234, and 238 amino acids, respectively. These 95 isolates shared approximately 78.7%–100% nucleotide identity and 74.5%–100% amino acid identity for ORF2. Amino acid alignments showed that the Cap protein of PCV2e and two strains (CN-FJ-CTS04 and FJ-6S-2017) differed markedly from those of PCV2a, PCV2b, PCV2c, and PCV2d. Unlike the sequence of other known PCV2 ORF2, the PCV2e ORF2 possesses 12 or 15 additional nts, which generate a terminal seven amino acid sequence (PPPLSYM) that is different from the PK and NPK termini of the 233 and 234 amino acid capsids, respectively (Davies et al., 2016). The PCV2 strains in genotype PCV2d harbored four unique amino acid mutations (Y8 → F/T8, A68 → N68, F53 → I53, and V215 → I215), whereas PCV2 in genotype PCV2e harbored ten unique amino acid mutations (K58 → V8, D115 → E115, A133 → T133, A135 → N135, T137 → S137, T170 → I170, V196 → T196, N204 → H204, Y207 → T207 and L226 → M226).
Recombination analysis
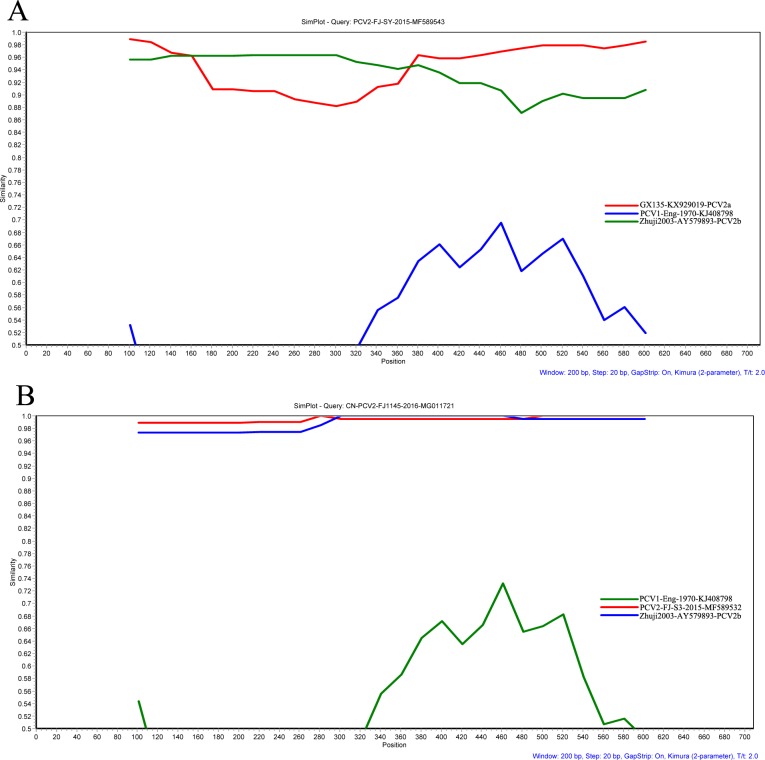
Seven methods (RDP, GeneConv, BootScan, MaxChi, Chimera, SiScan, and 3Seq) were implemented in RDP 4.24 to detect potential recombinants. The results showed that inter-genotypic (PCV2-FJ-SY) and intra-genotypic (CN-PCV2-FJ1145) recombination events of PCV2 have been detected (Fig. 3). Additionally, the results of SIMPLOT software (v3.5.1) confirmed the putative recombinant events obtained with the RDP 4.24 software.
Figure 3. Recombination analysis of the PCV2 ORF2 gene of PCV2-FJ-SY (A) and CN-PCV2-FJ1145 (B) using a sliding window of 200 nt moving in 20 nt steps.
Recombination events were analyzed by Simplot software (v. 3.5.1). A PCV1 isolate (KJ408798) was used as outgroup.
Discussion
PCV2 has been prevalent in Chinese swine herds since it was first identified in China in 1999. Previous studies demonstrated that various PCV2 genotypes exist in China, including PCV2a, PCV2b, PCV2c, PCV2d, and a minor recombinant group. Surprisingly, different PCV2 genotypes or strains have co-existed in Chinese swine herds. Currently, the amount of live pigs in the Fujian province (Southeast China) is >20 million heads, although the epidemic status and genetic diversity of PCV2 is still unknown. Thus, we sought to investigate the genetic diversity and epidemiology of PCV2 in southeast China in 2013–2017.
On the basis of ORF2 nucleotide sequences, the genotypes of Fujian PCV2 have been classified as PCV2a, PCV2b, PCV2d, and PCV2e (Opriessnig, Meng & Halbur, 2007; Guo et al., 2010; Guo et al., 2012; Harmon et al., 2015; Davies et al., 2016), of which, PCV2d was predominant in Fujian from 2014 to 2017. The present data indicate that the prevalence of PCV2d increased from 57.1% in 2013 to 87.5% in 2016, suggesting that PCV2d has replaced PCV2b and has now become the predominant PCV2 genotype in Fujian since 2013. Additionally, CN-FJ-CTS04 and FJ-6S-2017 strains emerged in cases of in cases of apparent vaccine failure. Overuse of vaccine has probably contributed to the variation of PCV2. PCV2e, a new PCV2 genotype, was identified in the USA in 2015, the ORF2 sequence of which contains 12 or 15 additional nt compared to other known PCV2 ORF2 sequences and shows approximately 85% identity with other known PCV2 ORF2 sequences (Harmon et al., 2015; Davies et al., 2016). In the present study, only two PCV2e sequences were detected in Fujian, suggesting that PCV2e replication is low in the swine herds. The group members encoded a capsid protein of 238 amino acids, which is markedly different from all known capsid proteins of 233 or 234 amino acids (Xiao et al., 2016). The pathogenicity of PCV2e warrants further investigation.
Recently, several groups detected PCV2 DNA in water samples, human stool, beef, calf bone marrow, farm air, house flies, and mosquito (Verreault et al., 2010; Blunt et al., 2011; Garcia et al., 2012; Yang et al., 2012). In the present study, PCV2 DNA was also detected in river water, which is indicative of a potential human health hazard.
PCV2 nucleotide substitution rate has been calculated to vary between 1.2 × 10−3 and 6.6 × 10−3 substitutions/site/year, which is close to that of RNA viruses (Firth et al., 2009; Pérez et al., 2011). Recombination is critical for viral evolution, and recombination events between PCV2 strains could generate PCV2 of varying pathogenicity (Olvera, Cortey & Segales, 2007; Hesse, Kerrigan & Rowland, 2008; Wang et al., 2009). Both inter-genotypic and intra-genotypic recombination events of PCV2 have been recorded; inter-genotypic recombination occurs within ORF1 and ORF2, whereas intra-genotypic recombination mainly occurs within ORF2. Recently, a PCV2c genotype strain (GenBank accession no. KC823058) was reported in China, and three strains were generated from PCV2b and PCV2c recombination (Liu et al., 2016). In the present study, we detected that one natural strain (PCV2-FJ-SY) was generated by recombination between different lineages of PCV2, while CN-PCV2-FJ1145 was generated by intra-genotypic recombination. These results indicated that PCV2 is continuously evolving through genome recombination, which leads to rapid emergence of new variant stains.
In summary, PCV2a, PCV2b, PCV2d, and PCV2e co-exist in Fujian, and PCV2d-2 is the predominant strain that has been circulating in Fujian since 2013. Our results enhance the understanding of the PCV2 epidemic in Fujian, and may assist veterinary workers in establishing suitable prevention and control policies for the novel strains emerging from viral evolution.
Conclusions
The findings of this study demonstrated that PCV2 is ubiquitous in China, with PCV2d being the predominant strain circulating from 2013 to 2017. PCV2 is continuously evolving through genome recombination or new viral introductions from distant geographic regions, which leads to rapid emergence of new variant stains.
Supplemental Information
Funding Statement
This work was supported by Major Project of the Science and Technology Program of Fujian Province, China (2014NZ0002-3), the Fujian Natural Science Foundation, Fujian Province, China (2016J01168), and the Collaborative Innovation Center of Animal Health and Food Safety Application Technology in Fujian, Fujian Vocational College of Agriculture (Dongxie 201701). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Jiankui Liu, Email: liujiankui@lyun.edu.cn, liujiankui99@126.com.
Xiaoyan Yang, Email: lyyxy1988@126.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Jiankui Liu conceived and designed the experiments, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Chunhua Wei conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Ailing Dai and Xiaoyan Yang analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Zhifeng Lin and Manlin Luo Jiayue Liu performed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Kewei Fan contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jianlin Fan performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Manlin Luo performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
Sampling procedures were approved by the Animal Ethics committee of the South China Agricultural University, reference number SCAU-AEC-2014-10.
DNA Deposition
Data Availability
The following information was supplied regarding data availability:
Sequence data can be accessed through the GenBank accession numbers in Table 1 and in the Supplemental Information.
References
- Anoopraj et al. (2015).Anoopraj R, Rajkhowa TK, Cherian S, Arya RS, Tomar N, Gupta A, Ray PK, Somvanshi R, Saikumar G. Genetic characterisation and phylogenetic analysis of PCV2 isolates from India: indications for emergence of natural inter-genotypic recombinants. Infection Genetics and Evolution. 2015;31:25–32. doi: 10.1016/j.meegid.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Blunt et al. (2011).Blunt R, McOrist S, McKillen J, McNair I, Jiang T, Mellits K. House fly vector for porcine circovirus 2b on commercial pig farms. Veterinary Microbiology. 2011;149:452–455. doi: 10.1016/j.vetmic.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Bolin et al. (2001).Bolin SR, Stoffregen WC, Nayar GP, Hamel AL. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. Journal of Veterinary Diagnostic Investigation. 2001;13(3):185–194. doi: 10.1177/104063870101300301. [DOI] [PubMed] [Google Scholar]
- Cai et al. (2011).Cai L, Han X, Ni J, Yu X, Zhou Z, Zhai X, Chen X, Tian K. Natural recombinants derived from different patterns of recombination between two PCV2b parental strains. Virus Research. 2011;158(1–2):281–288. doi: 10.1016/j.virusres.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Choi & Chae (2008).Choi KS, Chae JS. Genetic characterization of porcine circovirus type 2 in Republic of Korea. Research in Veterinary Science. 2008;84:497–501. doi: 10.1016/j.rvsc.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Davies et al. (2016).Davies B, Wang X, Dvorak CM, Marthaler D, Murtaugh MP. Diagnostic phylogenetics reveals a new porcine circovirus 2 cluster. Virus Resarch. 2016;217:32–37. doi: 10.1016/j.virusres.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Dupont et al. (2008).Dupont K, Nielsen EO, Baekbo P, Larsen LE. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Veterinary Microbiology. 2008;128(1–2):56–64. doi: 10.1016/j.vetmic.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Firth et al. (2009).Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. Journal of Virology. 2009;83:12813–12821. doi: 10.1128/JVI.01719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort et al. (2007).Fort M, Olvera A, Sibila M, Segalés J, Mateu E. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS affected pigs. Veterinary Microbiology. 2007;125:244–255. doi: 10.1016/j.vetmic.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Franzo et al. (2015).Franzo G, Cortey M, De Castro AM, Piovezan U, Szabo MP, Drigo M, Segalés J, Richtzenhain LJ. Genetic characterisation of Porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: an opportunity to reconstruct the history of PCV2 evolution. Veterinary Microbiology. 2015;178(1–2):158–162. doi: 10.1016/j.vetmic.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Garcia et al. (2012).Garcia LA, Viancelli A, Rigotto C, Pilotto MR, Esteves PA, Kunz A, Barardi CR. Surveillance of human and swine adenovirus, human norovirus and swine circovirus in water samples in Santa Catarina, Brazil. Journal of Water and Health. 2012;10:445–452. doi: 10.2166/wh.2012.190. [DOI] [PubMed] [Google Scholar]
- Gomes et al. (2007).Gomes M, De Castro AM, Cortez A, Heinemann MB, Brandao PE, Richtzenhain LJ. Genetic diversity of Brazilian strains of porcine circovirus type 2 (PCV-2) revealed by analysis of the cap gene (ORF-2) Archives of Virology. 2007;152:1435–1445. doi: 10.1007/s00705-007-0976-3. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2012).Guo L, Fu Y, Wang Y, Lu Y, Wei Y, Tang Q, Fan P, Liu J, Zhang L, Zhang F, Huang L, Liu D, Li S, Wu H, Liu C. A porcine circovirus type 2 (PCV2) mutant with 234 amino acids in capsid protein showed more virulence in vivo, compared with classical PCV2a/b strain. PLOS ONE. 2012;7(7):e41463. doi: 10.1371/journal.pone.0041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2010).Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virology Journal. 2010;7 doi: 10.1186/1743-422X-7-273. Article 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding & Clark (1997).Harding JC, Clark EG. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) Journal of Swine Health and Production. 1997;5:201–203. [Google Scholar]
- Harmon et al. (2015).Harmon KM, Gauger PC, Zhang J, Piñeyro PE, Dunn DD, Chriswell AJ. Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announcements. 2015;3(6):e01315–15. doi: 10.1128/genomeA.01315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse, Kerrigan & Rowland (2008).Hesse R, Kerrigan M, Rowland RR. Evidence for recombination between PCV2a and PCV2b in the field. Virus Research. 2008;132(1–2):201–207. doi: 10.1016/j.virusres.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2013).Huang Y, Shao M, Xu X, Zhang X, Du Q, Zhao X, Zhang W, Lyu Y, Tong D. Evidence for different patterns of natural inter-genotype recombination between two PCV2 parental strains in the field. Virus Research. 2013;175(1):78–86. doi: 10.1016/j.virusres.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2017).Jiang CG, Wang G, Tu YB, Liu YG. Genetic analysis of porcine circovirus type 2 in China. Archives of Virology. 2017;162(9):2715–2726. doi: 10.1007/s00705-017-3414-1. [DOI] [PubMed] [Google Scholar]
- Karuppannan & Opriessnig (2017).Karuppannan AK, Opriessnig T. Porcine circovirus type 2 (PCV2) vaccines in the context of current molecular epidemiology. Viruses. 2017;9(5) doi: 10.3390/v9050099. Article E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowka et al. (2000).Krakowka S, Ellis JA, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Veterinary Pathology. 2000;37(3):254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- Lang et al. (2000).Lang H, Zhang G, Wu F, Zhang C. Detection of serum antibody against postweaning multisystemic wasting syndrome in pigs. Chinese Journal of Veterinary Science and Technology. 2000;30:3–5. [Google Scholar]
- Liu et al. (2016).Liu X, Wang FX, Zhu HW, Sun N, Wu H. Phylogenetic analysis of porcine circovirus type 2 (PCV2) isolates from China with high homology to PCV2c. Archives of Virology. 2016;161(6):1591–1599. doi: 10.1007/s00705-016-2823-x. [DOI] [PubMed] [Google Scholar]
- Lole et al. (1999).Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin et al. (2010).Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera, Cortey & Segales (2007).Olvera A, Cortey M, Segales J. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. 2007;357:175–185. doi: 10.1016/j.virol.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Opriessnig, Meng & Halbur (2007).Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. Journal of Veterinary Diagnostic Investigation. 2007;19(6):591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- Palinski et al. (2016).Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Journal of Virology. 2016;91(1):e01879–16. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez et al. (2011).Pérez LJ, De Arce HD, Cortey M, Domínguez P, Percedo MI, Perera CL, Tarradas J, Frías MT, Segalés J, Ganges L, Núñez JI. Phylogenetic networks to study the origin and evolution of porcine circovirus type2 (PCV2) in Cuba. Veterinary Microbiology. 2011;151:245–254. doi: 10.1016/j.vetmic.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Ramos et al. (2013).Ramos N, Mirazo S, Castro G, Arbiza J. Molecular analysis of porcine circovirus type 2 strains from Uruguay: evidence for natural occurring recombination. Infection Genetics and Evolution. 2013;19:23–31. doi: 10.1016/j.meegid.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Segalés et al. (2008).Segalés J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, Dupont K, McCullough K, Ellis J, Krakowka S, Mankertz A, Fredholm M, Fossum C, Timmusk S, Stockhofe-Zurwieden N, Beattie V, Armstrong D, Grassland B, Baekbo P, Allan G. PCV-2 genotype definition and nomenclature. Veterinary Record. 2008;162:867–868. doi: 10.1136/vr.162.26.867. [DOI] [PubMed] [Google Scholar]
- Shuai et al. (2007).Shuai J, Wei W, Li X, Chen N, Zhang Z, Chen X, Fang W. Genetic characterization of porcine circovirus type 2 (PCV2) from pigs in high-seroprevalence areas in southeastern China. Virus Genes. 2007;35:619–627. doi: 10.1007/s11262-007-0121-0. [DOI] [PubMed] [Google Scholar]
- Verreault et al. (2010).Verreault D, Létourneau V, Gendron L, Massé D, Gagnon CA, Duchaine C. Airborne porcine circovirus in Canadian swine confinement buildings. Veterinary Microbiology. 2010;141:224–230. doi: 10.1016/j.vetmic.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2009).Wang F, Guo X, Ge X, Wang Z, Chen Y, Cha Z, Yang H. Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Research. 2009;145:151–156. doi: 10.1016/j.virusres.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Xiao, Halbur & Opriessnig (2015).Xiao CT, Halbur PG, Opriessnig T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. Journal of General Virology. 2015;96(7):1830–1841. doi: 10.1099/vir.0.000100. [DOI] [PubMed] [Google Scholar]
- Xiao et al. (2016).Xiao CT, Harmon KM, Halbur PG, Opriessnig T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the US during 2014-2016. Veterinary Microbiology. 2016;197:72–77. doi: 10.1016/j.vetmic.2016.11.009. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2012).Yang X, Hou L, Ye J, He Q, Cao S. Detection of porcine circovirus type 2 (PCV2) in mosquitoes from pig farms by PCR. Pakistan Veterinary Journal. 2012;32:134–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Sequence data can be accessed through the GenBank accession numbers in Table 1 and in the Supplemental Information.