Abstract
Genome rearrangements have played an important role in the evolution of Yersinia pestis from its progenitor Yersinia pseudotuberculosis. Traditional phylogenetic trees for Y. pestis based on sequence comparison have short internal branches and low bootstrap supports as only a small number of nucleotide substitutions have occurred. On the other hand, even a small number of genome rearrangements may resolve topological ambiguities in a phylogenetic tree. We reconstructed phylogenetic trees based on genome rearrangements using several popular approaches such as Maximum likelihood for Gene Order and the Bayesian model of genome rearrangements by inversions. We also reconciled phylogenetic trees for each of the three CRISPR loci to obtain an integrated scenario of the CRISPR cassette evolution. Analysis of contradictions between the obtained evolutionary trees yielded numerous parallel inversions and gain/loss events. Our data indicate that an integrated analysis of sequence-based and inversion-based trees enhances the resolution of phylogenetic reconstruction. In contrast, reconstructions of strain relationships based on solely CRISPR loci may not be reliable, as the history is obscured by large deletions, obliterating the order of spacer gains. Similarly, numerous parallel gene losses preclude reconstruction of phylogeny based on gene content.
Keywords: Phylogeny reconstruction, Bacteria evolution, Genome rearrangements
Introduction
Yersinia pestis, causing fulminant plague, has evolved clonally from an enteric pathogen, Yersinia pseudotuberculosis, that, in contrast, causes a relatively benign enteric illness. Horizontal gene acquisition, massive gene loss, and genome rearrangement events all have played important roles in the evolution of Y. pestis from its progenitor (Achtman et al., 1999). Y. pseudotuberculosis and Y. pestis differ radically in their pathogenesis despite sharing >97% identity in 75% of their chromosomal genes (Martínez-Chavarría & Vadyvaloo, 2015). As only a small number of nucleotide substitutions have occurred, traditional phylogenetic trees of Y. pestis strains based on sequence comparison have short internal branches and low bootstraps.
Gene order in prokaryotes is relatively poorly conserved making it a convenient tool for the analysis of the species and strain evolution, when changes in protein, and even gene sequences do not provide sufficient resolution (Wolf et al., 2001). In addition, genome rearrangements are less sensitive to homologous recombination and hence allow for an alternative approach to construction of phylogenetic trees, as even a small number of genome rearrangements may resolve topological ambiguities in a phylogenetic tree (Darling, Miklós & Ragan, 2008).
Among factors affecting genome rearrangement are abundance of mobile elements and the state of repair/recombination systems in the respective genomes (Novichkov et al., 2009). Newly formed pathogens such as Y. pestis are known to have a particularly high rate of rearrangements that may be caused by the prevalence of a large variety and number of insert sequences (ISs) (Liang et al., 2014).
The comparison of the Y. pestis KIM genome sequence with Y. pestis strain CO92 has divided both genomes into 27 conserved segments, and the most parsimonious series of inversions for three multiple-inversion regions has been described (Deng et al., 2002). Further, large-scale genome rearrangements have been described in strains Antiqua, Nepal and Angola (Chain et al., 2006; Eppinger et al., 2010). Comparison of pairs of bacterial genomes has revealed a characteristic “cross-like” pattern of localization of orthologous genes, indicating that inversions around the origin of replication comprise one of the dominant types of genome rearrangements (Eisen et al., 2000).
Multiple genome alignment of nine Y. pestis and Y. pseudotuberculosis genomes has featured universal Locally Collinear Blocks (LCBs) yielding seven parsimonious scenarios of the inversion history. The reconstructed pattern of genome rearrangements confirms strong preference for the replichore balance and over-representation of “symmetric inversions”—inversions with endpoints that are equally distant from the origin of chromosomal replication (Darling, Miklós & Ragan, 2008).
Several algorithms based on a variety of optimization approaches have been developed for the reconstruction of the rearrangement history (Avdeyev et al., 2016; Hu, Lin & Tang, 2014). However, reconstruction for large datasets remains a challenge, since the minimum length series of inversions (the optimal sorting path) is often not unique and equally many optimal sorting paths exist (Miklós & Darling, 2009).
Later, the LCB model has been used to infer the phylogenetic relationships among eight complete Y. pestis genomes from the breakpoint distance matrix, yielding the conclusion that the pattern of Y. pestis chromosome rearrangements reflects the genetic features of specific geographical areas and might be applied to distinguish Y. pestis isolates (Liang et al., 2010). A set of gene families from thirteen Yersinia species has been used to reconstruct a complete genome sequence for the ancestor, integrating information from the sequences, the species tree, and the gene order (Duchemin, Daubin & Tannier, 2015).
Being a traditional object for the spoligotyping, a special type of genotyping based on the spacer nucleotide analysis, CRISPR systems of Y. pestis strains often serve as a model for CRISPR-based evolutionary studies. All three separate genomic CRISPR loci have been described in detail (Pourcel, Salvignol & Vergnaud, 2005), including numerous strains without complete genomes (Vergnaud et al., 2007; Cui et al., 2008; Riehm et al., 2012; Barros et al., 2014; Riehm et al., 2015). Relationships between strains have been studied using the distance based on shared and differential spacers content only (Barros et al., 2014) or taking into account the principles of evolutionary cassette dynamics. In particular, the evolutionary history of Y. pestis based on CRISPR polymorphism has been reconstructed in the form of an acyclic oriented graph (Cui et al., 2008). Later, a general mathematical model of CRISPR evolution has been applied to reconstruct the relationships of strains for each of the three CRISPR loci (Kupczok & Bollback, 2013).
Here, we integrate the history at different levels of genome evolution, including gene flux, sequence divergence, chromosome segmental inversions, and spacer acquisitions and deletions in CRISPR cassettes, for genomes of twelve completely sequenced Y. pestis strains and four Y. pseudotuberculosis strains.
Materials and Methods
Genomes
Complete genome sequences of four Yersinia pseudotuberculosis and twelve Yersinia pestis, all available as of August 1st, 2013, were taken from the NCBI Genome database (Benson et al., 2015) and are listed in Table S1.
Construction of orthologs
Bidirectional best hits (BBHs) were constructed for each pair of strains using BLASTP (Zhang & Madden, 1997). BLASTP hits with identity <50% or coverage of the shorter sequence <67% were ignored. At the next step, if paralogs were more similar to each other than to either BBH partner, both paralogs were added to the orthologous group. Then, maximal connected components were constructed. This was done using ad hoc software based on the Relational Database Management System (RDBMS) Oracle Database Express Edition.
Trees based on nucleotide alignments
First we performed codon alignment and filtering for each of the 2117 orthologous groups using Mafft version v7.123b (Katoh & Standley, 2013) and Guidance 2.01 (Penn et al., 2010). Orthologous groups containing sequences with score below 0.8 were excluded from further analysis. Poorly aligned residues (guidance score below 0.8) were masked. The resulting sequences were concatenated and the tree was constructed with RAxML v8.2.9 (Stamatakis, 2014) using the GTR+Gamma model with 100 bootstrap runs.
Synteny blocks reconstruction
Synteny blocks were constructed using the Sibelia algorithm (Minkin et al., 2013) with the block length threshold 5,000 bp. To ensure robustness of the tree topology relative to this parameter we also performed calculations with the block length thresholds of 500 bp and 2,000 bp (see Figs. S2– S4). We used two approaches for the blocks construction. Splitting the chromosomes on non-repetitive common blocks was used for inversions analysis as the construction of all types of blocks allowed us to consider all type of rearrangements such as losses and gains.
Trees based on gene order
Trees based on gene order were build using two approaches. The trees using the Maximum Likelihood approach for the gene order were constructed using the MLGO software (Hu, Lin & Tang, 2014) for two data sets, synteny blocks having only one copy in every genome, and all synteny blocks found in these genomes. For both datasets we performed 1,000 replicates for the bootstrap analysis. The phylogenetic network was obtained using the Bayesian model of genome rearrangements by inversions implemented in the BADGER software (Larget, Simon & Kadane, 2002). We calculated 1,510,000 modification proposal steps, discarded the first 10,000 steps of each chain as burn-in and then subsampled every 50 steps as described in (Darling, Miklós & Ragan, 2008). The convergence of the Markov chain was assessed across multiple independent runs of BADGER as recommended in the BADGER manual. We used SplitsTree v4 (Huson & Bryant, 2006) for the network visualization.
Trees based on CRISPR cassettes composition
CRISPR cassettes were downloaded from CRISPRdb (Grissa, Vergnaud & Pourcel, 2007). Phylogenetic trees were reconstructed manually based on the CRISPR cassettes evolution rules. At that, two types of events were allowed, addition of a new spacer at the leader end, and deletion of one or several adjacent spacers from any part of a cassette. We further assumed (1) no independent additions of the same spacer to two different cassettes; (2) rare, but possible independent deletions of the same cassette segments; and (3) more probable single deletion of a segment including several adjacent spacers compared to several subsequent deletions of the segment parts.
Results and Discussion
Phylogenetic trees based on sequences alignments
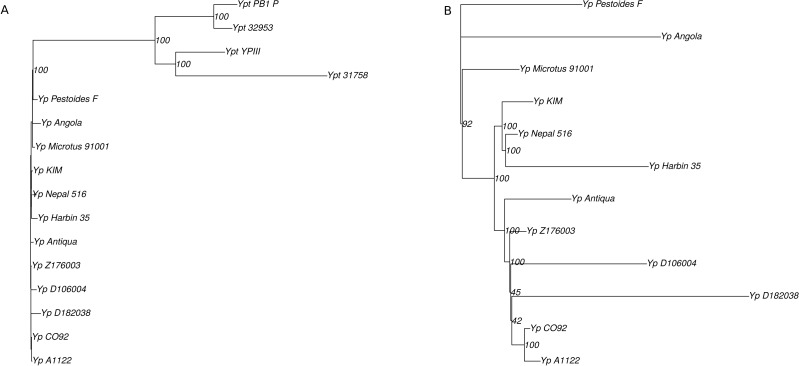
The phylogenetic tree for the analyzed Y. pseudotuberculosis and Y. pestis was constructed based on 2408 single-copy universal genes using a concatenation of individual nucleotide alignments (Fig. 1). We used the Y. enterocolitica genome to root the tree. We observed that Y. pestis strains formed a clade within the Y. pseudotuberculosis subtree, in agreement with previous genome analyses (Chain et al., 2006; Rasmussen et al., 2015).
Figure 1. (A) Phylogenetic tree of the Yersinia spp., based on nucleotide alignments of 2408 single-copy universal genes; (B) phylogenetic tree of the Y. pestis branch only.
There seemed to be several key noise factors. A small number of nucleotide substitutions resulted in low bootstrap values in several vertices, e.g., for Z176003, CO92, and A1122. Also, homologous recombination events might dramatically influence the tree topology reconstruction and lead to low level of bootstrap supports.
Phylogenetic trees based on gene order
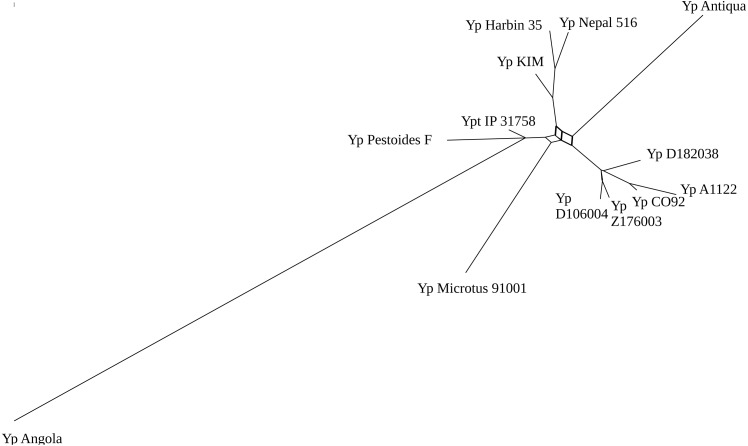
As genome rearrangements had played an important role in the evolution of Yersinia pestis, we constructed phylogenetic trees based on the gene order to check the topology and resolve nodes with low-level bootstrap support. Based on whole-genome alignments, 123 synteny blocks that were common for all strains under consideration with length more than 5,000 bp were obtained. We applied the Bayesian model of genome rearrangements by inversions and visualized phylogenetic tree signal as a consensus network (Fig. 2). This network has a complicated structure with ambiguous positions of the long branches but it has well-resolved clades with closely related strains that are not resolved in the alignment-based tree (Fig. 1B).
Figure 2. Phylogenetic trees network of the Yersinia pestis with Bayesian posterior probability threshold = 0.1.
Application of the Maximum Likelihood approach (Hu, Lin & Tang, 2014) to synteny blocks common for all Y. pestis strains revealed the optimal tree topology (Fig. 3A). Strains D106004 and Z176003 formed a separate branch in the inversions-based tree due to the same inversion with length about 150 kB that had occurred in these strains and D182038 that was an outgroup; at that, in the sequence-based tree Z176003 was an outgroup with a low bootstrap support of this node (Fig. 1B). One more parallel inversion with length about 350 kB was found in A1122 and D182038 that had lead to a low support in this node in the inversions-based tree. The boundaries of the inversions are formed by repeated sequences (transposases).
Figure 3. Phylogenetic trees of the Yersinia spp. based on gene order.
(A) Optimal topology based on inversions. (B) Optimal topology based on all types of rearrangements. Nodes that produce differences in the trees are labeled in red.
The observed parallel inversions could be explained by homologous recombination (horizontal transfer between strains) involving a segment containing the inverted fragments. If this were the case, sequence trees constructed using the genes from the inverted fragments would cluster together strains with the parallel inversions. However, the trees for both inversions are poorly resolved and hence provide no information about possible horizontal transfer ( Fig. S1).
One more short parallel inversion was found in Y. pestis KIM and Y. pestis Nepal at decreased synteny length threshold ( Fig. S4). The inverted block of length 3,500 bp contained integrase, antibiotic biosynthesis monooxygenase, dihydroorotase, DNA damage-inducible protein I, biofilm formation regulatory protein BssS, and IS256 family transposase. A possible explanation could be an incorporation of a mobile element in different orientations.
Adding the information about non-common blocks leads to decrease of the bootstrap supports (Fig. 3B). This may be explained by numerous parallel gains and losses natural to fast-evolving bacterial genomes such as recently formed pathogens. In particular, the Antiqua strain moves to the Microtus node, in agreement with the fact that, according to the ability to ferment glycerol and to reduce nitrate, strains Antiqua, Pestoides, Microtus, and Angola belong to the Antiqua biovar (Chain et al., 2006).
Based on the phylogenetic patterns, most events are losses, with only three blocks likely to have been inserted and three blocks having mosaic patterns that cannot be interpreted. However, as the latter have the same position in the genomes, they probably represent parallel losses. The inserted blocks are a prophage insertion, a fragment with a gene encoding a penicillin-binding protein and a transposase, and a gene encoding domains of an invasin-like inverse autotransporter protein.
CRISPR analysis
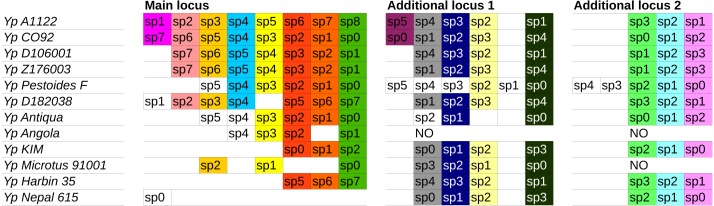
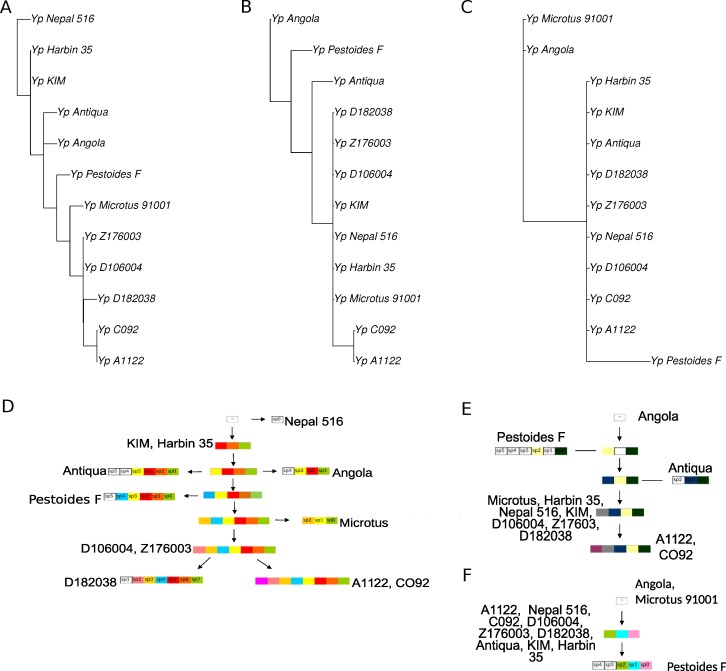
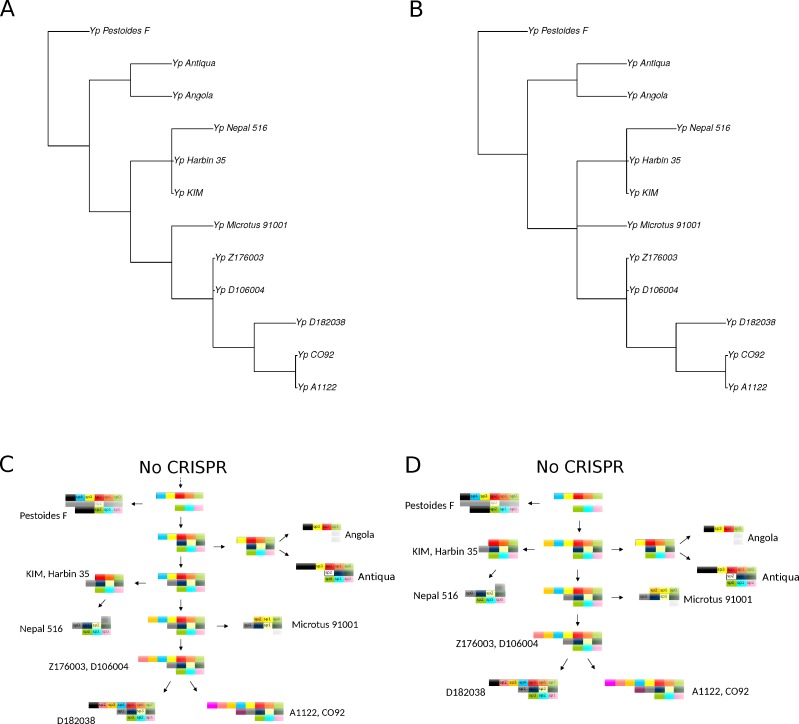
CRISPR cassettes of the considered Y. pestis strains are shown in Fig. 4. Initially, we constructed separate phylogenetic trees for each of the three CRISPR loci using the parsimony approach (Fig. 5, see ‘Methods’). As the number of events in each locus was small, the history of each locus could be reconstructed unambiguously.
Figure 4. CRISPR cassettes of completely sequenced Y. pestis strains. Cassette IDs and spacer numbers are given according to CRISPRdb (Grissa, Vergnaud & Pourcel, 2007).
Identical spacers are shown by the same color; unique spacers are set in frames.
Figure 5. Cladograms (A, B, C) and schemas of evolution (D, E, F) of three CRISPR loci of Y. pestis. (A, D) The main, most variable, locus; (B, E) additional locus 1; (C, D) additional locus 2.
However, the genome of Y. pestis evolves as a whole and the individual histories of the loci should be reconciled. In this case the reconstruction is ambiguous, as there are two equivalent reconstructions of the common ancestors and five equal positions of the Nepal strain on the maximum parsimony tree. Two maximum parsimony trees most compatible with the sequence tree are shown in Fig. 6. The trees constructed based on nucleotide sequences or rearrangements satisfy the rules of CRISPR cassette evolution (see Methods), but each of them implies two additional losses of cassette segments in comparison with the maximum parsimony tree. In particular, the sequence-based tree implies two independent parallel losses of the same segments of the main locus in the Angola and Antiqua strains branches.
Figure 6. Cladograms (A, B) and schemas of evolution (C, D) of two integrated CRISPR-based maximum parsimony phylogenetic trees most compatible with the sequence tree.
No direct evidence for homologous recombination or horizontal transfer of complete CRISPR loci or smaller groups of spacers was observed. While the parallel losses could be interpreted as a sign of homologous recombination/horizontal transfer, parallel events seem more likely, given the overall high rate of spacer loss. Generally, the problem of horizontal transfer vs. parallel events, duplications, and losses is a difficult one in the comparative genomics of prokaryotes (Koonin, 2016).
Conclusions
Detailed reconstruction of evolution of bacterial strains provides a framework for epidemiological studies and analysis of acquired pathogenesis loci and drug resistance determinants.
Using Y. pestis as an example, we demonstrate that integrated analysis of sequence-based and inversion-based trees enhances the resolution of the phylogenetic reconstruction. At that, inversions may resolve branches with low bootstrap support.
In contrast, reconstructions of strain relationships based on solely CRISPR loci may not be reliable, as the history is greatly obscured by large deletions, obliterating the order of spacer gains. Even less reliable seem to be reconstructions based on shared spacer content. Similarly, numerous parallel gene losses preclude reconstruction of phylogeny based on gene content.
Supplemental Information
(A) in A1122 and D182038 and (B) in Z176003 and D106004.
Calculations with the block length thresholds of 500 bp.
Calculations with the block length thresholds of 500 bp. (A) Optimal topology based on inversions; (B) Optimal topology based on all types of rearrangements.
Calculations with the block length thresholds of 2,000 bp. (A) Optimal topology based on inversions; (B) Optimal topology based on all types of rearrangements.
Acknowledgments
It was initiated at the Summer School of Molecular and Theoretical Biology. We thank Iakov I. Davydov for helpful discussions.
Funding Statement
This study was supported by the Russian Science Foundation under grant 14-50-00150. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Mikhail S. Gelfand is an Academic Editor for PeerJ.
Author Contributions
Olga O. Bochkareva and Irena I. Artamonova conceived and designed the experiments, analyzed the data, prepared figures and/or tables, approved the final draft.
Natalia O. Dranenko analyzed the data, prepared figures and/or tables.
Elena S. Ocheredko, German M. Kanevsky, Yaroslav N. Lozinsky and Vera A. Khalaycheva analyzed the data.
Mikhail S. Gelfand conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Github: https://github.com/OlgaBochkaryova/yersinia-phylogeny-analysis.git.
References
- Achtman et al. (1999).Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carnie E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(24):14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdeyev et al. (2016).Avdeyev P, Jiang S, Aganezov S, Hu F, Alekseyev MA. Reconstruction of ancestral genomes in presence of gene gain and loss. Journal of Computational Biology. 2016;23(3):150–164. doi: 10.1089/cmb.2015.0160. [DOI] [PubMed] [Google Scholar]
- Barros et al. (2014).Barros M, Francą C, Lins R, Santos M, Silva E, Oliveira M, Silveira-Filho V, Rezende A, Balbino V, Leal-Balbino T. Dynamics of CRISPR loci in microevolutionary process of Yersinia pestis strains. PLOS ONE. 2014;9(9):e108353. doi: 10.1371/journal.pone.0108353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson et al. (2015).Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2015;43(Database issue):D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain et al. (2006).Chain PSG, Hu P, Malfatti SA, Radnedge L, Larimer F, Vergez LM, Worsham P, Chu MC, Andersen GL. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. Journal of Bacteriology. 2006;188(12):4453–4463. doi: 10.1128/JB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui et al. (2008).Cui Y, Li Y, Gorgé O, Platonov M, Yan Y, Guo Z, Pourcel C, Dentovskaya S, Balakhonov S, Wang X, Song Y, Anisimov A, Vergnaud G, Yang R. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLOS ONE. 2008;3(7):e2652. doi: 10.1371/journal.pone.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling, Miklós & Ragan (2008).Darling AE, Miklós I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLOS Genetics. 2008;4(7):e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng et al. (2002).Deng W, Burland V, Plunkett GI, Boutin A, Mayhew G, Liss P, Perna N, Rose D, Mau B, Zhou S, Schwartz D, Fetherston J, Lindler L, Brubaker R, Plano G, Straley S, McDonough K, Nilles M, Matson J, Blattner F, Perry R. Genome sequence of Yersinia pestis KIM. Journal of Bacteriology. 2002;184(16):4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchemin, Daubin & Tannier (2015).Duchemin W, Daubin V, Tannier E. Reconstruction of an ancestral Yersinia pestis genome and comparison with an ancient sequence. BMC Genomics. 2015;16(Suppl 10) doi: 10.1186/1471-2164-16-S10-S9. Article S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen et al. (2000).Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biology. 2000;1(6) doi: 10.1186/gb-2000-1-6-research0011. Article research0011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger et al. (2010).Eppinger M, Worsham PL, Nikolich MP, Riley DR, Sebastian Y, Mou S, Achtman M, Lindler LE, Ravel J. Genome sequence of the deep-rooted Yersinia pestis strain angola reveals new insights into the evolution and pangenome of the plague bacterium. Journal of Bacteriology. 2010;192(6):1685–1699. doi: 10.1128/JB.01518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa, Vergnaud & Pourcel (2007).Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Lin & Tang (2014).Hu F, Lin Y, Tang J. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics. 2014;15:354. doi: 10.1186/s12859-014-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson & Bryant (2006).Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Katoh & Standley (2013).Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin (2016).Koonin E. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Research. 2016;5 doi: 10.12688/f1000research.8737.1. Article 1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupczok & Bollback (2013).Kupczok A, Bollback J. Probabilistic models for CRISPR spacer content evolution. BMC Evolutionary Biology. 2013;13 doi: 10.1186/1471-2148-13-54. Article 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget, Simon & Kadane (2002).Larget B, Simon DL, Kadane J. On a Bayesian approach to phylogenetic inference from animal mitochondrial genome arrangements (with discussion) Journal of the Royal Statistical Society, B. 2002;64:681–693. doi: 10.1111/1467-9868.00356. [DOI] [Google Scholar]
- Liang et al. (2010).Liang Y, Hou X, Wang Y, Cui Z, Zhang Z, Zhu X, Xia L, Shen X, Cai H, Wang J, Xu D, Zhang E, Zhang H, Wei J, He J, Song Z, Yu X, Yu D, Hai R. Genome rearrangements of completely sequenced strains of Yersinia pestis. Journal of Clinical Microbiology. 2010;48(5):1619–1623. doi: 10.1128/JCM.01473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang et al. (2014).Liang Y, Xie F, Tang X, Wang M, Zhang E, Zhang Z, Cai H, Wang Y, Shen X, Zhao H, Yu D, Xia L, Hai R. Chromosomal rearrangement features of Yersinia pestis strains from natural plague foci in China. American Journal of Tropical Medicine and Hygiene. 2014;91(4):722–728. doi: 10.4269/ajtmh.13-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Chavarría & Vadyvaloo (2015).Martínez-Chavarría LC, Vadyvaloo V. Yersinia pestis and Yersinia pseudotuberculosis infection: a regulatory RNA perspective. Frontiers in Microbiology. 2015;6 doi: 10.3389/fmicb.2015.00956. Article 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklós & Darling (2009).Miklós I, Darling AE. Efficient sampling of parsimonious inversion histories with application to genome rearrangement in Yersinia. Genome Biology and Evolution. 2009;1:153–164. doi: 10.1093/gbe/evp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkin et al. (2013).Minkin I, Patel A, Kolmogorov M, Vyahhi N, Pham S. Sibelia: a scalable and comprehensive synteny block generation tool for closely related microbial genomes. 13th workshop on algorithms in bioinformatics (WABI2013); 2013. 1307.7941 [Google Scholar]
- Novichkov et al. (2009).Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. Journal of Bacteriology. 2009;191(1):65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn et al. (2010).Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Research. 2010;38(Web Server issue):W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel, Salvignol & Vergnaud (2005).Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151(3):653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Rasmussen et al. (2015).Rasmussen S, Allentoft ME, Nielsen K, Orlando L, Sikora M, Sjögren K-G, Pedersen AG, Schubert M, Dam AV, Kapel CMO, Nielsen HB, Brunak S, Avetisyan P, Epimakhov A, Khalyapin MV, Gnuni A, Kriiska A, Lasak I, Metspalu M, Moiseyev V, Gromov A, Pokutta D, Saag L, Varul L, Yepiskoposyan L, Sicheritz-Pontén T, Foley RA, Lahr MM, Nielsen R, Kristiansen K, Willerslev E. Early divergent strains of Yersinia pestis in Eurasia 5,000 Years Ago. Cell. 2015;163(3):571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehm et al. (2015).Riehm J, Projahn M, Vogler A, Rajerison M, Andersen G, Hall C, Zimmermann T, Soanandrasana R, Andrianaivoarimanana V, Straubinger R, Nottingham R, Keim P, Wagner D, Scholz H. Diverse genotypes of Yersinia pestis caused plague in Madagascar in 2007. PLOS Neglected Tropical Diseases. 2015;9(6):e0003844. doi: 10.1371/journal.pntd.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehm et al. (2012).Riehm J, Vergnaud G, Kiefer D, Damdindorj T, Dashdavaa O, Khurelsukh T, Zöller L, Wölfel R, Fléche PL, Scholz H. Yersinia pestis lineages in Mongolia. PLOS ONE. 2012;7(2):e30624. doi: 10.1371/journal.pone.0030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis (2014).Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnaud et al. (2007).Vergnaud G, Li Y, Gorgé O, Cui Y, Song Y, Zhou D, Grissa I, Dentovskaya S, Platonov M, Rakin A, Balakhonov S, Neubauer H, Pourcel C, Anisimov A, Yang R. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Advances in Experimental Medicine and Biology. 2007;603:327–338. doi: 10.1007/978-0-387-72124-8_30. [DOI] [PubMed] [Google Scholar]
- Wolf et al. (2001).Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Research. 2001;11(3):356–372. doi: 10.1101/gr.GR-1619R. [DOI] [PubMed] [Google Scholar]
- Zhang & Madden (1997).Zhang J, Madden T. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Research. 1997;7(6):649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) in A1122 and D182038 and (B) in Z176003 and D106004.
Calculations with the block length thresholds of 500 bp.
Calculations with the block length thresholds of 500 bp. (A) Optimal topology based on inversions; (B) Optimal topology based on all types of rearrangements.
Calculations with the block length thresholds of 2,000 bp. (A) Optimal topology based on inversions; (B) Optimal topology based on all types of rearrangements.
Data Availability Statement
The following information was supplied regarding data availability:
Github: https://github.com/OlgaBochkaryova/yersinia-phylogeny-analysis.git.